Abstract
Natural killer (NK) cells perform immunosurveillance of virally infected and transformed cells, and their activation depends on the balance between signaling by inhibitory and activating receptors. Cytokine receptor signaling can synergize with activating receptor signaling to induce NK cell activation. We investigated the interplay between the signaling pathways stimulated by the cytokine interleukin-2 (IL-2) and the activating receptor NKG2D in immature (CD56bright) and mature (CD56dim) subsets of human primary NK cells using mass cytometry experiments and in silico modeling. Our analysis revealed that IL-2 changed the abundances of several key proteins, including NKG2D and the phosphatase CD45. Furthermore, we found differences in correlations between protein abundances, which were associated with the maturation state of the NK cells. The mass cytometry measurements also revealed that the signaling kinetics of key protein abundances induced by NKG2D stimulation depended on the maturation state and the pretreatment condition of the NK cells. Our in silico model, which described the multidimensional data with coupled first-order reactions, predicted that the increase in CD45 abundance was a major enhancer of NKG2D-mediated activation in IL-2–treated CD56bright NK cells but not in IL-2–treated CD56dim NK cells. This dependence on CD45 was verified by measurement of CD107a mobilization to the NK cell surface (a marker of activation). Our mathematical framework can be used to glean mechanisms underlying synergistic signaling pathways in other activated immune cells.
INTRODUCTION
Natural killer (NK) cells are lymphocytes of the innate immune system (1, 2). Unlike lymphocytes of the adaptive immune system, such as T and B cells, activation of NK cells is not dominated by a single primary receptor but by a diverse set of germline-encoded activating and inhibitory NK receptors (NKRs) (1, 2). Cognate ligands on target cells (such as virally infected cells or tumor cells) disrupt the balance between activating and inhibitory NKRs that initiate opposing signals and generate a bias toward activating signals. This results in NK cell activation, which then leads to the lysis of target cells through the release of the contents of cytolytic granules (a process called cytotoxicity), the secretion of cytokines such as interferon-γ (IFN-γ), or both (1, 2).
An intriguing aspect of NK cell activation is the inability of many activating NKRs to stimulate robust NK cell activation when these receptors are engaged individually (3). However, pretreating NK cells with cytokines, such as interleukin-2 (IL-2), that are often produced in the host during an infection (3), removes this constraint. For example, cross-linking of the activating receptor NK group 2, member D (NKG2D) with agonistic monoclonal antibodies (mAbs) fails to stimulate any appreciable activation of primary NK cells unless the NK cells are pretreated with IL-2 (3). An added complexity arises because of the differences in NK cell responses during different stages of NK cell development (4, 5). For example, activation of immature human NK cells that have increased amounts of the cell surface marker protein CD56 (so-called CD56bright NK cells) by treatment with IL-12 in combination with IL-18 predominantly stimulates the production of cytokines (6), whereas the activation through activating NKRs of more mature primary human NK cells, which have reduced cell surface abundances of CD56 (that is, CD56dim NK cells), generates a more robust cytotoxic response (4). This type of behavior also opens up the interesting possibility that cytokine-NKR synergies are executed differently during different stages of NK cell development. Although such synergies between cytokines and NKRs are well documented in multiple experimental systems, the underlying mechanisms are not well understood.
The roadblocks in obtaining a mechanistic understanding underlying cytokine-NKR synergy arise because of experimental challenges in investigating the signaling kinetics and the difficulty in setting up mechanistic in silico models in a signaling system with many poorly characterized signaling events. The NKRs transmit signals through diverse signaling pathways, which involve distinct sets of adaptors, kinases, and phosphatases (1, 7, 8). Even a given NKR can activate key downstream proteins through different signaling pathways. For example, upon activation, the complex formed between NKG2D and DNAX-activating protein of 10 kDa (DAP10) binds to the kinase phosphatidylinositol 3-kinase (PI3K) or to the adaptor growth factor receptor–bound protein 2 (Grb2), which initiates different sets of intermediate signaling events that lead to the activation of key proteins, such as Rac1 (9–11). As a result, it becomes difficult to experimentally characterize the signaling events for the synergy between a particular cytokine and NKR by assaying a small number of activation markers. A technical challenge in assaying many signaling proteins in NK cells by standard experimental techniques (for example, Western blotting analysis) is the low abundance of these proteins in NK cells, as well as the heterogeneity of the NK cell population, which contains subsets of cells with different developmental states. Another factor that impedes the generalization of a known NK cell signaling mechanism in one species (for example, mice) to another (for example, humans) is the interspecies differences between the type of NKRs and their associated signaling pathways (7, 8). As a result, many of the events in NK cell signaling are not well characterized (7, 12). This also makes it difficult to establish mechanistic in silico models based on standard modeling techniques (for example, differential equations or Master equation) (13–15), which require detailed knowledge regarding the interactions between the involved molecular species.
Here, we investigated the synergy between IL-2 and NKG2D signaling pathways in primary human CD56bright and CD56dim NK cells. We combined mass cytometry analysis with a data-driven in silico modeling approach that has predictive powers to derive a mechanistic understanding of synergy. The mass cytometry technique enabled us to assay 37 different signaling proteins in single cells across multiple time points, producing a detailed description of the signaling kinetics, which included cell-to-cell variations of the signaling kinetics. The multidimensional single-cell data were analyzed with a data-driven in silico framework to quantify the flux between two signaling proteins in a time interval. The flux is a measure of the rate of net flow of molecules between proteins during signaling. If A and B are two proteins influencing each other’s abundances, then the flux from A→B (fA→B) represents how fast the abundance of A is changing to generate B. The analysis provided a dynamic description of the signaling kinetics, which quantitatively elucidated roles of signaling components in regulating the temporal evolution of the measured protein abundances. This combined approach enabled us to predict the main regulators of the synergy between IL-2 and NKG2D signaling and to quantify the differences in the synergy between IL-2 and NKG2D exhibited by immature (CD56bright) and mature (CD56dim) primary human NK cells. In IL-2–treated CD56bright NK cells, our analysis predicted the phosphatase CD45 to be a main regulator of increased NK cell activation. In contrast, our analysis indicated the involvement of additional regulators other than CD45 in the increased activation of the IL-2–treated CD56dim NK cells. Together, the results suggest that the developed data-driven framework can be used to delineate synergistic signaling pathways in immune cells.
RESULTS
IL-2 promotes NKG2D-mediated activation in both CD56bright and CD56dim NK cells but with different signaling kinetics
We assessed changes in the single-cell abundances of 37 different proteins in two subsets (CD56bright and CD56dim) of primary human NK cells that were stimulated with plate-bound agonistic anti-NKG2D antibodies. The NK cells were left untreated or pretreated with IL-2, previously titrated to achieve maximal activation (“priming”), for 24 hours before the cells were stimulated through NKG2D. The cells were assayed before (at time t = 0) and after NKG2D-mediated stimulation at multiple times (t = 4, 8, 16, 32, 64, 128, and 256 min). We compared changes in protein abundances between the different pretreatment conditions (medium alone or IL-2) for a given CD56+ NK cell subset (CD56bright or CD56dim) and between the CD56bright and CD56dim NK cell subsets for a given pretreatment condition (medium or IL-2). These experiments were performed with blood samples derived from three different healthy human donors (donors #1 to #3). Throughout the text, we describe the results for donor #1. Although some donor-to-donor variances were observed, experiments with cells from the two other donors produced results that were qualitatively similar, to a large extent (see figs. S1 to S12).
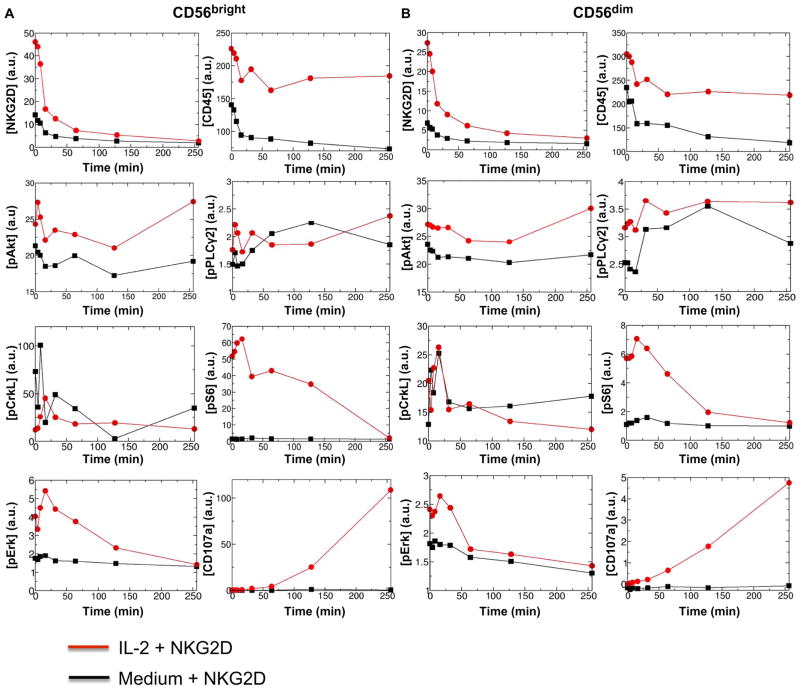
We found that IL-2 stimulated changes in the abundances of multiple proteins in both the CD56bright and CD56dim NK cell subsets. Most notably, the average abundances of NKG2D, CD45, phosphorylated S6 (pS6), and phosphorylated signal transducer and activator of transcription 5 (pSTAT5) were increased more than 3-, 1.5-, 5-, and 9-fold, respectively, in IL-2–treated CD56bright and CD56dim NK cell subsets compared to those in untreated cells (Fig. 1, figs. S1 and S12, and table S1). However, several other proteins that mark activation in lymphocytes, including phosphorylated extracellular signal–regulated kinase (pErk) (Fig. 1 and table S1) and CD69 (table S1), were also more abundant in the IL-2–treated NK cells than in the untreated cells. Furthermore, we observed differences in the average abundances of multiple proteins between the CD56bright and CD56dim NK cell populations under the same pretreatment condition (medium or IL-2). For example, and consistent with previous observations, the average abundances of CD16 and CD57 were substantially greater (>3- to 90-fold), and the average abundance of CD62L was less in untreated and IL-2–treated CD56dim NK cells compared to that in the CD56bright NK cells (table S1 and fig. S12). In addition, regardless of the pre-treatment condition, the average abundances of the activating receptor NKG2D and the inhibitory receptor NKG2A were reduced (>1.5-fold) in the CD56dim NK cells compared to those in the CD56bright NK cells (table S1 and fig. S12).
Fig. 1. Kinetics of average protein abundances in mass cytometry measurements in IL-2– or medium-treated CD56bright and CD56dim primary human NK cells stimulated by anti-NKG2D.
(A and B) Analysis of the kinetics of changes in the average abundances of the indicated proteins in CD56bright (A) and CD56dim (B) primary human NK cells at time 0 and at the indicated times after stimulation through NKG2D. a.u., arbitrary units. Before stimulation through NKG2D, the cells were pretreated with either medium (black lines) or IL-2 (red lines). Protein abundances were measured by mass cytometry in duplicate samples from each donor. The average protein abundances were calculated using Eq. 3A for the cell population in the duplicate samples. Data are from a single donor and are representative of two independent donors. The data from the second donor are shown in fig. S2. Cells from a third donor were analyzed to confirm the effect of pretreatment with IL-2 (fig. S7). For analysis of the kinetics of other proteins of interest, see fig. S1 and table S1.
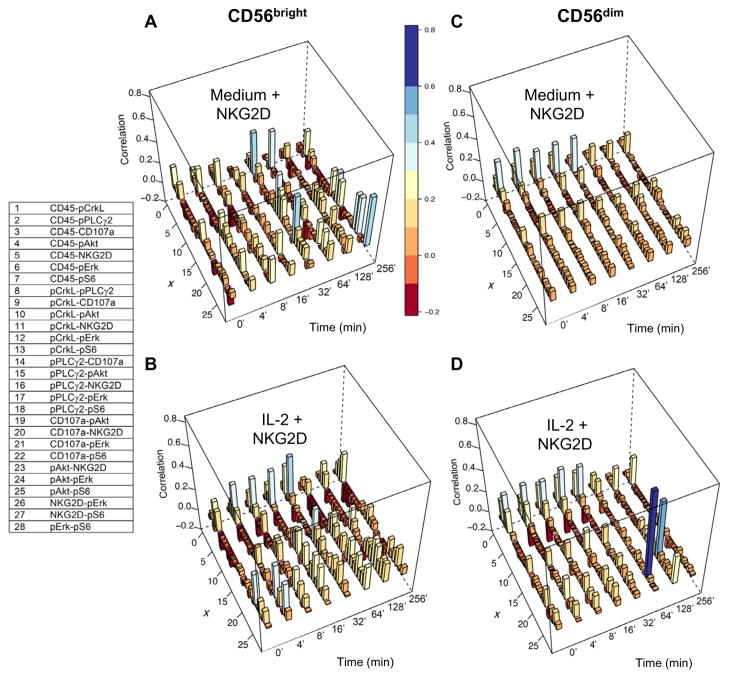
Variations in protein abundances under the NKG2D-unstimulated condition (for example, cells treated with medium or IL-2 alone) could affect the signaling kinetics that follow NKG2D-mediated stimulation, because the abundances of interacting proteins determine the propensity of the associated biochemical reactions. Thus, increased (or decreased) protein abundances in the NKG2D-unstimulated state could result in faster (or slower) changes in protein abundances involved in biochemical signaling events. We calculated the covariance between a pair of protein abundances. The covariance between two proteins indicates if an increase in abundance of a protein is associated on average with an increase (positive covariance) or decrease (negative covariance) of the abundance for the other protein. We found protein abundances that displayed both positive covariances (for example, CD45 and pAkt in IL-2–treated CD56bright NK cells) and negative covariances [for example, NKG2D and phosphorylated CrkL (pCrkL) in IL-2–treated CD56bright NK cells], with the magnitudes of the correlations varying from low (~0.01) to moderate (~0.4) values in the NK cell subsets before they underwent NKG2D stimulation (Fig. 2, A to D). The protein abundances covaried differently between the CD56bright and CD56dim NK cell subsets in an IL-2–dependent manner even before the cells underwent NKG2D stimulation (Fig. 2). The nature of covariations in protein abundances in the NKG2D-unstimulated state could also affect the kinetics of signaling. For example, a positive covariance between a kinase and a substrate would favor enhanced phosphorylation of the substrate in a cell population. We obtained similar results for donor #2 (figs. S2 and S3).
Fig. 2. Kinetics of correlations after NKG2D stimulation between protein abundances measured in cytometry experiments for the IL-2– or medium-treated CD56bright and CD56dim primary human NK cells.
(A to D) Correlations between the measured protein abundances (donor #1) were calculated by scaling the covariance matrix {Jij} in Eq. 3B by the variances of the proteins involved in the correlation: (Correlation)ij = Jij/(Jii × Jjj)1/2. The correlations for NKG2D, CD45, pAkt, pPLCγ2, pCrkL, pS6, pErk1/2, and CD107a are shown using three-dimensional bar graphs for medium-treated (A) and IL-2–treated (B) CD56bright NK cells and for medium-treated (C) and IL-2–treated (D) CD56dim NK cells. The correlations are indexed along the x axis, with integers from 1 to 28 corresponding to the indicated protein pairs (left table). The magnitudes of the correlations are shown along the z axis, and the bars are color-coded on the basis of their heights for better visualization. The correlations for donor #2 are shown in fig. S3.
The single-cell kinetics of signaling after NKG2D engagement depended on the NK cell subset (CD56bright or CD56dim) and the pretreatment condition (medium or IL-2). Stimulation of NKG2D on IL-2–treated CD56bright and CD56dim NK cells resulted in substantially increased amounts (10- to 100-fold in the cell population average) of CD107a (a marker for the release of cytotoxic and secretory granules) at late times (>128 min) compared to those in their medium-treated counterparts (Fig. 1 and fig. S2). The IL-2–treated CD56bright NK cells had increased amounts of CD107a compared to those in their IL-2–treated CD56dim counterparts (Fig. 1). Furthermore, the IL-2–treated CD56bright and CD56dim NK cells had increased amounts of pAkt and pS6 after NKG2D stimulation compared to those of the medium-treated cells (Fig. 1). The qualitative features of the averages of the protein abundances in the cell population were similar between the CD56bright and CD56dim NK cell subsets for most of the proteins analyzed (Fig. 1). However, the covariances between the protein abundances changed differently in the CD56bright and CD56dim NK cell subsets in response to NKG2D stimulation, and these changes were dependent on the pretreatment condition (Fig. 2). Therefore, the signaling kinetics in individual cells depended on both the developmental stage and the pretreatment condition. We performed additional control experiments by analyzing NK cells from a third donor that were incubated with isotype-matched control immunoglobulin G (IgG) (stimulation with IL-2 and IgG). The incubation of CD56bright and CD56dim NK cells with IgG resulted in negligible changes in CD107a abundance, even at later times (t = 264 min), and reduced abundances of pAkt and pErk compared to their NKG2D-stimulated counterparts (fig. S7). Thus, these data suggest that the increased activation in the IL-2–treated NK cells upon NKG2D stimulation arose from the synergy between IL-2 and NKG2D signaling. Next, we quantified the dependencies in the signaling kinetics with a data-driven in silico scheme.
Using the mass cytometry data, the underlying signaling kinetics is analyzed in silico
NK cell signaling kinetics are composed of different types of biochemical reactions, including binding-unbinding processes and phosphorylation and dephosphorylation events, as well as physical processes, including diffusion and cytoskeletal movements, which result in changes in the single-cell abundances of molecular species (7, 16, 17). Because these processes are affected by multiple, cell-specific properties, such as the total numbers of the involved molecular species (for example, the total numbers of NKG2D molecules) and the cell size, as well as by the intrinsic stochastic nature of biochemical processes (18, 19), each individual cell gives rise to a distinct signaling kinetic trajectory. The mass cytometry technique analyzes the single-cell abundances of a large number of signaling species (for example, proteins), thus providing a detailed description of the kinetics in terms of time-stamped snapshot data (20). The details of the underlying signaling interactions are implicitly contained in average values (Fig. 1) and the pairwise covariances (Fig. 2) for the protein abundances calculated with the mass cytometry data. Developing fully mechanistic in silico models composed of physically interacting signaling proteins to analyze such data is difficult because the detailed knowledge regarding the protein-protein interactions (for example, the precise nature of reaction propensities) required to establish such models is not available for most of the proteins pertinent to NK cell signaling. Although we were able to analyze many more markers than can be analyzed with conventional flow cytometry methods, the number of proteins analyzed by mass cytometry represented only a small percentage of the total number of physically interacting proteins and protein complexes involved in such signaling networks. Thus, in most cases, the measured proteins interact with each other through effective interactions that are modulated by the abundances of many unmeasured intermediate complexes.
We used a data-driven approach to model such effective interactions between the measured proteins. In our scheme, the measured signaling kinetics during a particular time interval were described by a system of coupled, first-order chemical reactions (Fig. 3). The reaction rates and the associated flux of molecules between pairs of proteins were estimated using the average values and covariances given by the mass cytometry data. The reaction rates describe the strengths of the effective causal interactions between the pairs of molecular species (for a detailed description regarding the framework and implementation, see Fig. 3, Materials and Methods, and fig. S4). The framework has several advantages. First, it provides a mechanistic description of the signaling kinetics during a time interval. Second, the modeled kinetics separate the contributions of basal (tonic) and IL-2 signaling before NKG2D stimulation from those that result due to NKG2D stimulation in the single-cell protein abundances measured after NKG2D stimulation. Third, the mathematical solution of the model kinetics can be obtained analytically in a closed expression, which enables precise estimation of the rate constants.
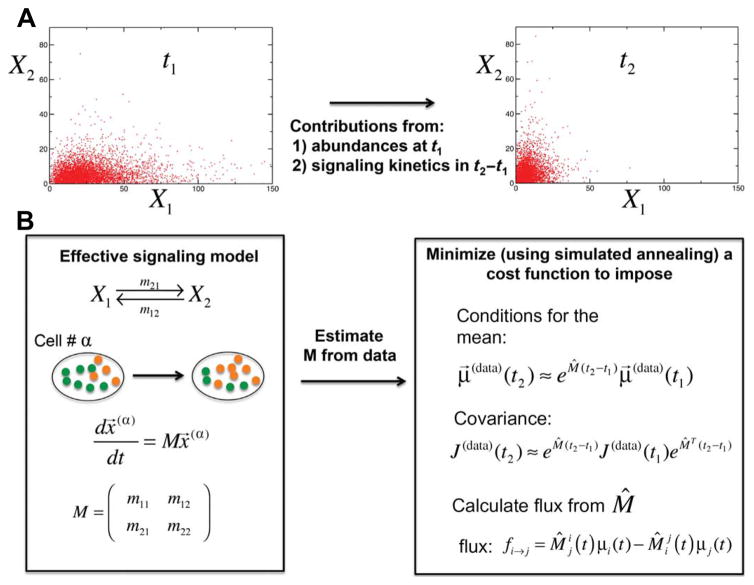
Fig. 3. Data-driveninsilicoschemeforquantifyingthesignalingkineticsunderlying the mass cytometry data.
(A) Mass cytometry measurements were used to measure the single-cell abundances of multiple proteins (for example, protein X1 and protein X2) simultaneously at different time points. X1 and X2 do not need to interact physically with each other but may be able to change each other’s abundances through intermediate signaling complexes. Individual cells display distinct trajectories of signaling kinetics because of cell-to-cell variations in total protein abundances. Because individual cells are destroyed upon each measurement, mass cytometry experiments provide time-stamped snapshot data for the underlying signaling kinetics. Thus, the data cannot be used to quantify changes in protein abundances in an individual cell that solely arise as a result of NKG2D signaling. The average values, covariances, and higher moments (for example, skewness and kurtosis) calculated from such snapshot data at a given time (for example, t2) are influenced by two factors: (i) the distribution of the abundances at an earlier time (for example, t1) and (ii) the changes in the abundances that occurred as a result of NKG2D signaling during the time interval t1 to t2. It is difficult to separate these factors from each other because of nontrivial relationships between these quantities (see Eq. 4B and the related discussion in Materials and Methods). This fact makes it difficult to infer signaling mechanisms. For example, if t1 represents a time point in the unstimulated state (cells treated with medium or IL-2) before NKG2D stimulation and t2 represents a time point after NKG2D stimulation, then a positive correlation between X1 and X2 could arise because of the anti-NKG2D–induced co-regulation of X1 and X2 in the time interval t1 to t2 or because of the increased abundance of both proteins (for example, as a result of exposure to IL-2) at the NKG2D unstimulated state (at t1). (B) We addressed these concerns by constructing a data-driven model based on first-order chemical reactions. The scheme is described with a simple setup with two measured proteins, X1 and X2, that are assumed to interact through first-order reactions with the rates (m12, m21, m11, and m22). The magnitude of these rates describe the strengths of the interactions between the proteins X1 and X2. The reaction rates m12 (propensity, m12 x2) and m21 (propensity, m21 x1) are associated with the reactions X2→X1 and X1→X2 to X2 and X1, respectively. Note that { x1, x2 } ≡ x⃗ in the propensity expressions represents the single-cell abundances of the proteins X1 and X2. The rates m11 (propensity, m11x1) and m22 (propensity, m22 x2) denote any self-decay (when m11 < −m21 or m22 < −m12) or self-production when m11 > −m21 or m22 > −m12 (for example, by auto-catalysis) or conservation (when m11 = −m21 and m22 = −m12) of X1 and X2, respectively. The kinetics of the system is described by deterministic mass action kinetics, dx⃗/dt = Mx⃗. The average abundances (μ→ ≡ {μ1,μ2}) and the covariance ({Jij}) between the abundances at times t1 and t2 are related by Eqs. 4A and 4B. We assumed that the quantities {μ→(t1), {Jij (t1)}} and {μ→ (t2),{Jij (t2)}} are determined by the mass cytometry–measured single-cell abundances μ→(data), J(data), and we then estimated the reaction rates (M̂) in the model by minimizing a cost function given by Eq. 7. The cost function ensures that the rates minimize the error between the model and the data corresponding to the average abundances and covariances. We also used a constraint that minimizes the total external flux to the system to reduce the effect of self-decay and self-production in describing the data. A simulated annealing scheme is used to perform the minimization of the cost function (for details, see Materials and Methods). Once the rates were estimated, the instantaneous average flux at time t1, the flux from i→j, or fi→j is calculated with Eq. 8.
We validated our computational scheme with snapshot data acquired from in silico networks composed of first-order (fig. S4) and nonlinear (fig. S5) reactions. This method generated results that are qualitatively similar to those published in another study (21) in quantifying relationships between signaling proteins with mass cytometry data from experiments with T cells (fig. S6). Furthermore, our scheme described protein-protein relationships in the activation of Ras by the guanine nucleotide exchange factor (GEF) SOS through an experimentally validated signaling network with nonlinear interactions (14). Using our scheme, the fluxes between the proteins at different times correctly captured the known involvement of the enzymes SOS and RasGRP1 in the activation of Ras (fig. S5). Thus, we used this method to quantify fluxes between pairs of signaling components at different times, which provided a quantitative description of the role of major players in regulating the signaling kinetics during a time interval. This approach could then be used to generate predictions to further characterize signaling mechanisms.
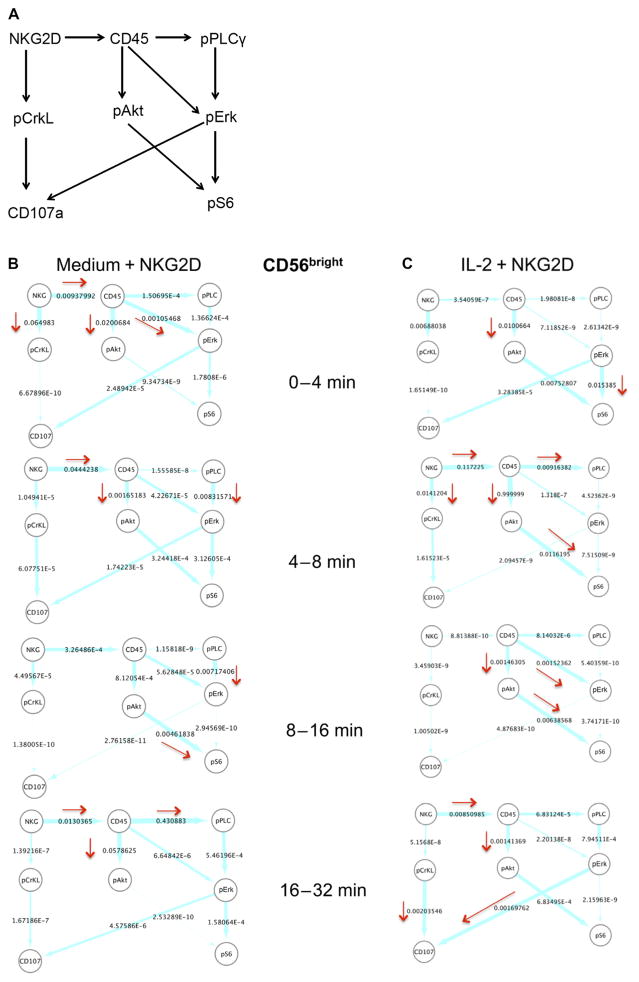
We considered an interaction network (Fig. 4A) involving eight different proteins that were analyzed in the mass cytometry experiments. The eight proteins, including NKG2D, CD45, pPLCγ2, pCrkL, pAkt, pErk, pS6, and CD107a, were chosen on the basis of previous results described in the literature regarding NKG2D-induced signaling and the available markers included in the mass cytometry experiment. We considered effective interactions, modeled as first-order chemical reactions, between different protein pairs (Fig. 4A). The presence of an effective interaction between a protein pair was based on the published literature.
Fig. 4. In silico characterization of the NKG2D-stimulated signaling kinetics in CD56bright NK cells.
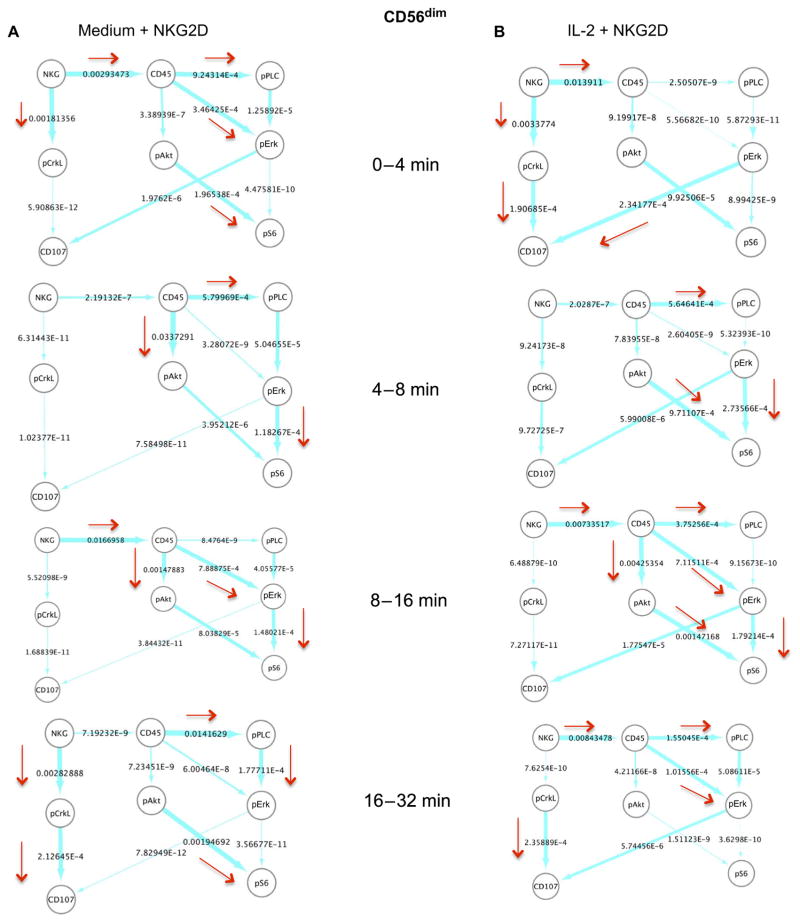
(A) Effective signaling model used to characterize signaling kinetics. The arrows indicate the presence of first-order reactions between the indicated protein pairs. The direction of the arrow shows the causality in the interaction; for example, A→B denotes that species B is generated or activated by A with a rate proportional to the abundance of A. (B and C) Instantaneous average fluxes in the network shown in (A). The average fluxes were calculated from the mass cytometry data obtained at two successive time points (as indicated) for CD56bright NK cells stimulated with anti-NKG2D after first being treated with medium (B) or IL-2 (C). The flux (described by Eq. 8) in the time interval from t1 to t2 (t2 > t1) was calculated at the earlier time point, t1. The fluxes were rescaled by the largest flux calculated for all data sets analyzed. The thickness of an arrow is proportional to the value (shown next to the arrows) of the associated flux on a logarithmic scale. The directions associated with relatively larger values (≥10−3) of the fluxes are emphasized with red arrows. NKG2D and pPLCγ2 are denoted as NKG and pPLC, respectively, for brevity. The flux analysis was performed on the mass cytometry data collected at 0, 4, 8, 16, and 32 min. For each time point, the sample size was about 250 single cells.
CD45 molecules are present in activating synapses but are excluded from inhibitory NK cell synapses that are formed upon ligand binding (22). Thus, a change in the spatial localization of CD45 upon NKR engagement could influence the indirect interactions mediated by Src family kinases (SFKs) between the NKG2D-DAP10 receptor complex and CD45. The first-order chemical reaction NKG2D→CD45 (Fig. 4A) considers the possibility of such an interaction between CD45 and NKG2D upon binding of the anti-NKG2D antibody. Upon receptor cross-linking with this antibody, the NKG2D-associated adaptor protein DAP10 is tyrosine-phosphorylated by potentially five different SFKs that are found in NK cells (1, 23). The phosphatase CD45 activates SFKs by dephosphorylating a tyrosine residue at an inhibitory C-terminal site (24, 25). Phosphorylation of DAP10 results in the recruitment of the adaptor protein Grb2, which, in turn, leads to the activation of Vav1, PLCγ2, and the guanosine triphosphatase Rac1, resulting in the activation of the mitogen-activated protein kinase (MAPK) Erk (1, 10, 26). These interactions are represented by the first-order chemical reactions CD45→PLCγ2→pErk (Fig. 4A). Alternatively, PI3K produces phosphatidylinositol (3,4,5)-triphosphate (PIP3) from the plasma membrane lipid phosphatidylinositol (4,5)-biphosphate, which leads to the activation of Rac1 and Erk (8). These signaling events are broadly represented by the first-order chemical reaction CD45→pErk (Fig. 4A). The generation of PIP3 by PI3K also leads to the phosphorylation of Akt (27) and the subsequent phosphorylation of ribosomal protein S6 through the mammalian target of rapamycin (mTOR) signaling pathway (21, 28). These events are described by the first-order chemical reactions CD45→pAkt→pS6 (Fig. 4A). Activated Erk stimulates the relocalization of the lysosomal-associated membrane protein CD107a to the cell surface (29), where it acts as a marker for cytokine secretion and the release of cytolytic granules (30). This event is described by the first-order chemical reaction pErk→CD107a (Fig. 4A). S6 is activated by pErk in T cells (21). Thus, we considered the possibility of this activation event in NK cells with the first-order chemical reaction pErk→pS6. We also included pCrkL, a member of the Crk family of adaptor proteins. CrkL regulates NK activation by influencing NK cell–target cell adhesion and NK cell polarity (9). In our mass cytometry experiments, pCrkL was increased transiently in abundance in primary human NK cells after NKG2D stimulation, which is consistent with previous findings (9). However, the role of pCrkL in NKG2D-mediated NK cell activation is unclear. We considered an association between pCrkL and the relocalization of CD107a upon NKG2D stimulation with the first-order chemical reactions NKG2D→pCrkL→CD107a (Fig. 4A). Next, we evaluated the strengths of the interactions and the corresponding fluxes between the proteins as the kinetics progressed in time. We chose to include the early times (0 to 32 min) in our analysis of NKG2D-mediated signaling kinetics because most NKG2D-mediated signaling events are thought to occur within this time frame after NKG2D stimulation (9, 31, 32).
The calculation of the fluxes (based on Eq. 8) in the CD56bright NK cells showed that at early times after NKG2D stimulation (that is, 0 to 4 min), the signaling kinetics in the IL-2–treated cells occurred with a larger magnitude of flux in the pathway pErk→pS6 compared to that in their medium-treated counterparts (Fig. 4, B and C). This implies that between 0 and 4 min, the rate of change in the pErk abundance to produce pS6 (or the propensity of the reaction pErk→pS6) was greater than other reaction propensities (for example, the propensity for CD45→pErk in the IL-2–treated cells) considered in Fig. 4A. It is possible that the phosphorylation of S6 was induced by the high abundances of pErk present before the NKG2D stimulation in the IL-2–treated CD56bright NK cells before the NKG2D stimulation (Fig. 4C). At later time points after the NKG2D stimulation of the IL-2–treated CD56bright NK cells, fluxes with larger magnitudes in the signaling kinetics occurred in those pathways, in which CD45 was considered to stimulate S6 activation (CD45→pAkt→pS6) between 4 and 16 min, PLCγ2 activation (CD45→pPLCγ2) between 4 and 8 min, and Erk activation (CD45→pErk) between 8 and 16 min (Fig. 4C). Activated Erk stimulated the relocalization of CD107a to the cell surface (pErk→CD107a) between 16 and 32 min after NKG2D stimulation in the IL-2–treated CD56bright NK cells (Fig. 4C). In addition, pCrkL also appeared to contribute to CD107a relocalization (pCrkL→CD107a) in the same time interval (Fig. 4C). We found that NKG2D stimulation led to fluxes with greater magnitudes to CD45 (NKG2D→CD45) between the 4- to 8-min and 16- to 32-min time intervals in the IL-2–treated CD56bright NK cells (Fig. 4C). Overall, the calculation of fluxes suggested the possible involvement of CD45 with most of the major changes in the signaling pathways that led to the activation of S6 and Erk and, eventually, CD107a relocalization. In comparison, in the medium-treated CD56bright NK cells, CD45 was predicted to contribute to the activation of Erk (CD45→pErk) between 0 and 4 min and Akt (CD45→pAkt) between 0 and 8 min during the early stages of NKG2D-mediated signaling (Fig. 4B). In the medium-treated CD56bright NK cells, pErk did not stimulate large changes in CD107a localization, and pAkt stimulated S6 activation substantially only during the period 8 to 16 min after NKG2D stimulation (Fig. 4B). Changes to NKG2D→CD45 fluxes were still observed in the medium-treated NK cells at early (0 to 8 min) and late (16 to 32 min) times after NKG2D stimulation (Fig. 4B). Thus, in the absence of IL-2, the involvement of CD45 in the activation of Erk and S6 was substantially reduced, which could be a result of the lower abundances of CD45 in the medium-treated NK cells compared to those in their IL-2–treated counterparts (Fig. 1).
We also analyzed fluxes (Eq. 8) in the CD56dim NK cells (Fig. 5, A and B). Similar to the IL-2–treated CD56bright NK cells, NKG2D-mediated activation of Erk, Akt, and S6 in the CD56dim NK cells was induced by CD45 as the signaling progressed after NKG2D stimulation (Fig. 5B). However, the changes in fluxes in the IL-2–treated CD56dim NK cells were less substantial (less than fivefold) than those in the CD56bright NK cells (Figs. 4C and 5B). In addition, the relocalization of CD107a in response to Erk activation was about 1000-fold less between 16 and 32 min in the IL-2–treated CD56dim NK cells than in the IL-2–treated CD56bright NK cells (Figs. 4C and 5B). Therefore, although CD45 stimulated most of the larger changes in pErk, pAkt, and pS6 abundance and in CD107a relocalization, the effect of CD45 in the CD56dim NK cells was much weaker compared to that in the CD56bright NK cells (Figs. 4C and 5B). The signaling kinetics of the medium-treated CD56dim NK cells showed more similarity to those of the IL-2–treated CD56dim NK cells (Fig. 5, A and B) compared to their CD56bright counterparts (Fig. 4, B and C). In the medium-treated CD56dim NK cells, CD45 induced changes in the activation of PLCγ2, Akt, and Erk during different time intervals. However, the flux from pErk to CD107a relocalization was substantially less (<100-fold) in the medium-treated CD56dim NK cells (Fig. 5A) compared to that in the IL-2–treated CD56dim NK cells (Fig. 5B). Overall, the extent of the effect of IL-2 on the CD56dim NK cells was not as large as that in the CD56bright NK cells, and pErk in the CD56dim NK cells was less effective at stimulating CD107a relocalization than it was in the CD56bright NK cells (Figs. 4C and 5B). We obtained qualitatively similar results from our data-driven analysis of another NK cell donor (fig. S8).
Fig. 5. In silico characterization of the NKG2D-stimulated signaling kinetics in CD56dim NK cells.
(A and B) Instantaneous average fluxes (derived from Eq. 8) in the network shown in Fig. 4A. The average fluxes are calculated from the mass cytometry data obtained at two successive time points (as indicated) for CD56dim NK cells stimulated with anti-NKG2D after first being treated with medium (A) or IL-2 (B). The flux (as calculated from Eq. 8) in the time interval from t1 to t2 (t2 > t1) was calculated at the earlier time point t1. The fluxes were rescaled by the largest flux calculated for all data sets analyzed. The thickness of an arrow is proportional to the value (shown next to the arrows) of the flux on a logarithmic scale. The directions associated with relatively larger values (≥10−4) of the fluxes are emphasized with red arrows. NKG2D and pPLCγ2 are denoted as NKG and pPLC, respectively, for brevity. The flux analysis was performed on the mass cytometry data collected at 0, 4, 8, 16, and 32 min. For each time point, the sample size was about 4000 cells.
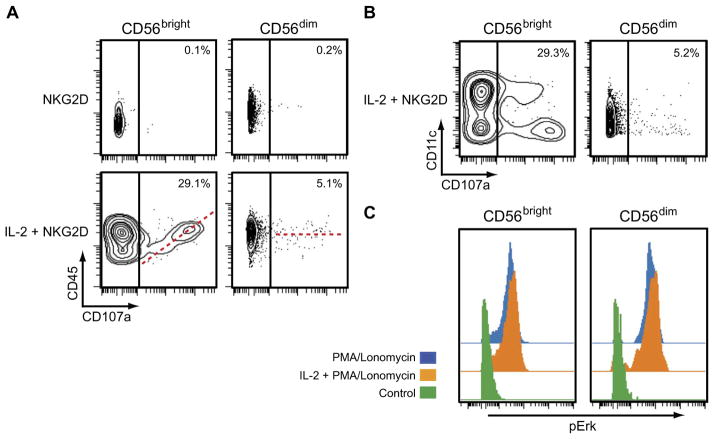
The in silico analysis of fluxes between the protein pairs predicted the involvement of CD45 in stimulating large changes in the signaling pathways in the IL-2–treated CD56bright NK cells (Fig. 4C). IL-2 stimulated almost a twofold increase in the average abundance of CD45 in the CD56bright NK cells compared to that in the IL-2–treated CD56dim NK cells, which led to enhanced Src activation and a resulting increase in Erk activation and CD107a relocalization after NKG2D stimulation. Thus, these results suggest that pretreatment with IL-2 promotes the robust NKG2D-mediated activation of the CD56bright NK cells by increasing the abundance of CD45. From this analysis, one could predict that those IL-2–treated CD56bright NK cells that had a CD45 abundance similar to that of medium-treated CD56bright NK cells would display substantially reduced amounts of CD107a on the cell surface after NKG2D stimulation. We tested this prediction by gating the IL-2–treated CD56bright NK cells based on low or high CD45 abundance late after NKG2D stimulation (t = 256 min), a time at which increased amounts of CD107a were detected at the cell surface (Fig. 1). We found that IL-2–pretreated CD56bright NK cells with less CD45 had reduced amounts of CD107a on the cell surface (Fig. 6A). This property of IL-2–treated CD56bright NK cells was reproducible in cells from donors #2 and #3 (figs. S9 and S10), as well as in all of the additional donors that were tested (fig. S11). Note that data from this late time point were not used to calculate the fluxes that were used to generate the predictions.
Fig. 6. Testing of model predictions.
(A) Mass cytometry analysis of CD45 and CD107a in CD56bright and CD56dim NK cells (obtained from donor #1) at 256 min after the cells were stimulated with anti-NKG2D antibody having first been treated with medium (NKG2D) or IL-2 (IL-2 + NKG2D). As predicted by the model, IL-2–treated CD56bright NK cells with greater amounts of CD45 exhibited increased relocalization of CD107a to the cell surface. IL-2–treated CD56dim NK cells with the greater amounts of CD45 showed a bimodal distribution of CD107a abundances on the cell surface. The dashed red line denotes the correlation between the abundances of CD45 and CD107a. Data are from a single donor and are representative of five independent donors. (B) Mass cytometry analysis of CD11c and CD107a abundances (NK cells obtained from donor #1) at 256 min after IL-2–pretreated CD56bright and CD56dim NK cells were stimulated with anti-NKG2D antibody. As predicted by the model, CD11c abundance played no role in determining the extent of NKG2D-stimulated CD107a relocalization in either NK cell subset. Data are from a single donor and are representative of five independent donors. (C) Mass cytometry analysis of the distribution of pErk in CD56bright and CD56dim NK cells at 16 min after incubation with medium, PMA alone, or PMA and ionomycin. The NK cells were first pretreated with either IL-2 (orange) or medium (blue). Unstimulated control samples are shown in green. Data are from a single donor and are representative of three independent donors.
When the CD56bright NK cells were gated on CD11c, whose abundance was also increased after IL-2 treatment, but which is not known to play a role in NKG2D signaling, we did not detect any correlation between CD11c with the cell surface abundance of CD107a after NKG2D stimulation (Fig. 6B). This was also observed in NK cells from donors #2 and #3 (figs. S9B and S10B). In addition, no correlation was observed between CD45 abundance and IFN-γ production in the IL-2–treated CD56bright NK cells after NKG2D stimulation (fig. S11C). The in silico analysis of the CD56bright NK cells also showed that CD45 stimulated the activation of Erk in the IL-2–treated cells (Fig. 4C). Thus, our model predicts that both IL-2– and medium-treated cells would have similar amounts of pErk if the CD45-mediated Erk activation pathway was bypassed. Stimulation of cells with phorbol 12-myristate 13-acetate (PMA) and ionomycin leads to an increase in the abundance of intracellular calcium and the activation of Erk but bypasses the need for SFKs (25, 33). We found that PMA and ionomycin stimulated similar increases in pErk abundance in both IL-2– and medium-treated NK cells for both the CD56bright and CD56dim subsets (Fig. 6C). Furthermore, we did not observe any obvious correlation between CD45 abundance and CD107a cell surface abundance in the IL-2–treated CD56bright NK cells after treatment with PMA and ionomycin (fig. S11B), suggesting that the role of CD45 in CD56bright NK cells is upstream of calcium release and Erk activation.
Because our analysis showed that CD45 was involved to a similar extent in the IL-2– and medium-treated CD56dim NK cells, the model predicts that, unlike in the CD56bright cells, the increased abundance of CD45 in the IL-2–treated CD56dim NK cells alone is not a major factor in the increased activation of these cells. This prediction was confirmed by our analysis of the behavior of the IL-2–treated CD56dim NK cells at 256 min after NKG2D stimulation, in which the increased CD107a abundance on the cell surface occurred in cells with reduced amounts of CD45 (Fig. 6A). This property was also displayed by NK cells isolated from donors #2 and #3 (figs. S9A and S10A). The CD56dim NK cell population can be further divided into subpopulations based on the cell surface marker CD57, which defines terminally differentiated NK cells (34) and the expression of inhibitory receptors, such as NKG2A. Because the CD57− NKG2A+ subset of CD56dim NK cells exhibits enhanced responsiveness to IL-2 compared to that of their CD57− NKG2A− and CD57+ counterparts (34), we examined whether there was a CD45-mediated dependency, similar to the CD56bright NK cells, of CD107a mobilization to the cell surface after NKG2D stimulation in the CD57− NKG2A+ CD56dim NK cell subset. However, we observed no correlation between CD45 and CD107a in the CD57− NKG2A+ CD56dim NK cell subset (figs. S9A and S10A). Together, these results suggest that the correlation between CD45 and CD107a abundance is specific for the IL-2–treated CD56bright NK cell population. Because the relocalization of CD107a to the cell surface correlates with both the extent of cytokine secretion and the release of cytolytic granules, we used flow cytometry to examine any correlation between CD45 abundance and IFN-γ production by CD56bright and CD56dim NK cells after stimulation of NKG2D. Whereas we observed a correlation between CD45 and CD107a abundance in the IL-2–treated CD56bright NK cells (similar to that observed from our mass cytometry analysis), we did not observe any correlation between CD45 abundance and IFN-γ production in these cells (fig. S11C). Thus, these results suggest that CD45 may be linked to cytolytic granule secretion, rather than cytokine production, by the IL-2–treated CD56bright NK cells in response to NKG2D stimulation.
DISCUSSION
We analyzed the synergy between the IL-2 and NKG2D signaling pathways in NK cells residing at two different stages of development (immature CD56bright cells and mature CD56dim NK cells) through a combination of single-cell, mass cytometry experiments and a data-driven in silico framework. Measurement of the abundances of 37 different proteins by mass cytometry revealed that the pre-treatment of NK cells with IL-2 before stimulation through NKG2D generated moderate to large changes in the abundances of several proteins, including CD45 and NKG2D, with many of these changes depending on the maturation state of the NK cells. The differences in the protein amounts between the IL-2–treated CD56dim and the IL-2–treated CD56bright NK cells before the NKG2D stimulation could be the result of the type of IL-2 receptors found on these cells: CD56bright NK cells express the high-affinity, heterotrimeric IL-2Rαβγ receptors, whereas the CD56dim NK cells express the low-affinity, heterodimeric IL-2Rβγ receptors (4). Stimulation of the IL-2– or medium-treated NK cells by plate-bound agonistic anti-NKG2D antibodies resulted in increased mobilization of CD107a at the cell surface in the IL-2–treated cells compared to that in the medium-treated cells.
We developed a data-driven in silico framework to analyze cytokine-NKR signaling synergy with the time-stamped snapshot single-cell mass cytometry data. The data-driven method was developed to address the difficulties in establishing canonical mechanistic signaling models for NKG2D signaling kinetics because of the poor characterization of many associated signaling pathways. Furthermore, we needed to develop an in silico framework that was able to separate the contributions of signaling that occurred in response to pretreatment with IL-2 or medium and NKG2D stimulation in the single-cell data because the current methods (21) designed to quantify the strengths of protein-protein interactions in cytometry data are unable to do so. The data-driven scheme described interactions between measured proteins by a set of coupled, first-order reactions in which the rates of the reactions were estimated in a time interval with successive time-stamped mass cytometry data. Thus, the reaction rates can vary between different time intervals, which provides a description of the signaling kinetics in terms of piecewise first-order reactions. The piecewise description models the kinetics with first-order reactions, where the rate constants can be different during different time intervals.
The in silico framework enabled us to separate the changes in the protein abundances that were derived from the pretreatment with IL-2 or medium and from the NKG2D-mediated stimulation and enabled us to directly compare the signaling kinetics between the CD56bright and CD56dim NK cell subsets initiated by the NKG2D stimulation under these different pretreatment conditions. Using the estimated rates, we calculated average fluxes between signaling proteins at each time point. The fluxes between protein pairs provided a dynamic and mechanistic characterization of the complex time dependence of the average abundances and the covariances in the mass cytometry data. The magnitudes of the fluxes enabled us to identify proteins that play important roles in regulating the signaling kinetics. Our calculations of the fluxes showed donor-to-donor differences in these magnitudes (Fig. 4 and fig. S8). The estimation of the magnitudes of the fluxes in our in silico framework depended on the kinetics of changes in the average protein abundances and the pairwise correlations between the proteins, and because these quantities varied between different donors (see Figs. 1 and 2 and figs. S1 to S3), the magnitudes of the fluxes also showed variations. However, some of the general features of the kinetics, for example, the involvement of CD45 in stimulating Erk activation and CD107a relocalization in IL-2–treated CD56bright cells, were maintained across the different donors (Fig. 4 and fig. S8). A mechanistic understanding of which features of the signaling kinetics are insensitive to donor-to-donor variations will require further investigation.
The strong correlation between CD107a and CD45 abundance predicted by the in silico model, in which increased CD107a relocalization correlated with increased CD45 abundance, was observed in the IL-2–treated CD56bright NK cells from all of the donors tested. At the same time, this correlation was not observed in the IL-2–treated CD56dim NK cells from any of the donors or when we gated on a CD57− NKG2A+ subpopulation of the IL-2–treated CD56dim NK cells. CD45 is a phos-phatase that is present in all nucleated hematopoietic cells, and it activates SFKs by dephosphorylating their inhibitory tyrosine residues (24, 25). However, when present in greater abundance, CD45 can also diminish SFK activation by dephosphorylating a tyrosine residue in the kinase domain (35). The IL-2–treated CD56dim NK cells contained fewer NKG2D molecules (~1.5-fold less average abundance) but more CD45 molecules (~1.5-fold higher average abundance) than did the IL-2–treated CD56bright cells. It is possible that this makes the relationship between CD45 abundance and CD107a relocalization in the IL-2–treated CD56dim NK cells more complicated, which could lead to weaker signaling activity. Previous studies with Ptprc−/− (CD45-null) mice showed that NK cells lacking CD45 produced lower amounts of cytokines and showed a defect in CD107a relocalization compared to NK cells from wild-type mice when stimulated through immunoreceptor tyrosine-based activation motif (ITAM)–associated activating NKRs, such as CD16, Ly49H, and NKG2D (25). The effect of CD45 in the activation of immature and mature mouse NK cells was not examined in the CD45-deficient mice (36). We observed no correlation between CD45 abundance and IFN-γ production in the IL-2–treated CD56bright NK cells in response to the stimulation of NKG2D. However, it is difficult to compare NKG2D signaling in mouse and human NK cells because human NKG2D associates only with the adaptor DAP10, whereas mouse NKG2D associates with either DAP10 or DAP12 in activated mouse NK cells. DAP10 contains a YINM sequence and transmits signals through pathways that are different from those used by the ITAM-bearing adaptor DAP12 (37).
The flux analysis based on the mass cytometry data points to several differences between different protein pairs. In the IL-2–treated CD56bright NK cells, the calculation of fluxes showed large rates of change in CD45 abundances to produce pAkt (through CD45→pAkt) at all of the time points examined, whereas the rates of change in CD45 abundances to generate pErk (through CD45→pErk) became larger transiently at a specific time interval (16 to 32 min). Such differences could possibly arise because of the presence of multiple phosphorylation sites in Erk or other MAPKs. Multisite activations can give rise to bistable behavior in MAPK activation, resulting in a switch-like activation of the MAPK (for example, Erk) when the stimulation crosses a threshold value (38). In addition, CD45 and pErk (CD45→pSrc→pVav→Rac→PAK1→MEK→Erk) are separated by a relatively larger number of signaling events compared to those between CD45 and pAkt activation (CD45→pSrc→PI3K→PIP3→pAkt). Therefore, a signal generated at the receptor-ligand interaction could take some time to surpass the threshold needed for Erk activation, whereas pAkt might be more easily activated.
Another observation is of the relatively larger fluxes between NKG2D→CrkL→CD107a at 16 to 32 min in the medium-treated CD56dim NK cells, which did not exhibit any CD107a relocalization. The value of the flux for pCrkL→CD107a in these cells was almost 10-fold less than the CrkL→CD107a and Erk→CD107a fluxes for the IL-2–treated CD56bright cells, which exhibited increased CD107a relocalization. The smaller values of the flux in the medium-treated CD56dim NK cells could be responsible for the negligible relocalization of CD107a in these cells. However, the magnitude of the CrkL→CD107a flux at 16 to 32 min in the medium-treated CD56dim cells was similar to that in their IL-2–treated counterparts, which exhibited CD107a relocalization to a small extent. It is therefore possible that there are additional pathways, which were not analyzed here, that are responsible for CD107a relocalization in the IL-2–treated CD56dim cells.
The flux calculations also showed transient fluxes between NKG2D and CD45, which could have arisen due to indirect interactions regulated by multiple intermediate complexes that were not measured in the experiments. For example, the activation of SFKs by CD45 results in the phosphorylation of tyrosine residues in NKG2D-DAP10, which leads to Erk activation and NKG2D internalization (39). Furthermore, the spatial localization of CD45 at the cell surface of NK cells can change depending on the type of synapse (activating or inhibitory) that is formed (22). CD45 is evenly distributed at the target cell–NK cell interface when cells of the YT human NK cell line form activating synapses. Similar to what occurs in the immunological synapse formed by T cells (40), CD45 is excluded from the inhibitory synapses in the YT cells (22). The presence of wide regions (size, >30 nm) alternating with narrow regions (size, ~14 nm) in the synaptic cleft in the activating NK cell synapses possibly enables large CD45 molecules to be present in the synapse (22). Thus, the transient fluxes between NKG2D and CD45 observed in this study could also be influenced by changes in the spatial localization of CD45 in the NK cell synapses.
The combined approach that we developed to use single-cell mass cytometry data and data-driven in silico modeling could be applied to analyze signaling mechanisms in a wide range of systems, especially when synergistic interactions between different signaling networks take place or when the contribution of the pretreatment condition regulating the receptor-induced signaling kinetics is not negligible. However, the in silico framework works successfully only under two specific conditions: (i) when a reasonable amount of previous knowledge regarding the interactions between the measured proteins is available and (ii) when the effects of intrinsic noise fluctuations are not substantial. In the absence of any information about the potential interactions between the measured species, any estimation of the fluxes would involve minimization of a cost function in large dimensions without any good initial guess. The solution of such problems can be difficult, particularly for reaction networks in which many reaction parameters (also known as sloppy modes) vary across a large range without affecting the cost function substantially (41). Therefore, the analysis of a signaling system can begin with calculations involving molecular species that have known interactions, and then small sets of proteins with unknown interactions in the scheme can be included iteratively, where each iteration is followed up by test of predictions. Signaling events in single cells usually involve large amounts of proteins; thus, cell-to-cell variations in total protein content (or extrinsic noise fluctuations) dominate over intrinsic noise fluctuations (42), which can become important for proteins with low copy numbers (43). Intrinsic noise fluctuations can be important during receptor stimulation when the numbers of ligands or receptors pertaining to a single cell are very small. In such cases, considering stochastic kinetics for the first-order reactions (44) could provide a way to incorporate these fluctuations into the model.
MATERIALS AND METHODS
Development of the in silico framework in terms of first-order chemical reactions
The mass cytometry data showed that both the CD56bright and CD56dim NK cells exhibited moderate amounts of basal activation, which changed substantially for several proteins when the NK cells were treated with IL-2 (see table S1). Upon stimulation of the cells with anti-NKG2D antibody, the ensuing signaling kinetics changed single-cell protein abundances, which produced time-dependent changes in the average values and covariances in the protein abundances. Therefore, the measured single-cell abundances after NKG2D stimulation contained contributions arising from the unstimulated condition as well as from the changes induced by NKG2D signaling (Fig. 3). Because individual cells were not tracked in the mass cytometry measurements, it is difficult to separate the changes in single-cell protein abundances that arise from these two sources. We considered a model system in which the receptor-induced (for example, NKG2D) signaling kinetics in individual cells were described by a set of first-order reactions involving the measured protein species. We considered deterministic mass-action kinetics; thus, we captured the cell-to-cell variations in protein abundances that arose because of differences in signaling induced by IL-2 or medium before NKG2D stimulation. However, these kinetics do not take into account the intrinsic noise fluctuations in the chemical reactions, which can become relevant for molecular species that are present at low abundances (43). Such fluctuations play a minor role in signaling kinetics, presumably due to the presence of large copy numbers of signaling proteins (42). Consider a model system of N number of single cells, with each cell (indexed by α) containing n different molecular species (indexed by i) occurring with copy numbers or abundances, { }. The interactions between the molecular species can be described by a set of coupled first-order biochemical reactions. The single-cell abundances then follow a deterministic mass-action linear kinetics:
| (1) |
in which Mji describes the rate of the reaction j→i. The strength of the interaction between species i and j is given by the magnitudes of the rates of the forward (j→i) and the reverse (i→j) reactions, Mji and Mij, respectively. The flux fji = Mji xj(t) for i≠j [or fij = Mij xi(t)] gives the rate of change of species i (or j) by j (or i) through the reaction j→i (or i→j). Thus, when fji(t) > fij(t), it implies that at time t, species j is causing the abundance of species i to change by a greater amount than vice versa. This provides a precise notion of causality in the interactions in the signaling pathway. Note that the M matrix does not depend on the cell index, which implies that the signaling reactions occur at the same rate in each cell. We considered cell-to-cell variations of species abundances at the prestimulus (t = 0) state due to the extrinsic noise fluctuations in total species abundances, tonic (basal) signaling, and treatment conditions (for example, cells treated before NKG2D stimulation with either medium or cytokine, such as IL-2). Equation 1 can be solved analytically to calculate single-cell species abundances at any time t:
| (2) |
In the mass cytometry data sets, we are provided with measurements that pertain to the abundances {xi(α)(t)} at a particular time t in single cells indexed by α. The challenge is to use the time-stamped snapshot data at multiple time points to infer the reaction rates or {Mji}. The difficulties for achieving that are as follows: First, the n × n M matrix in general contains n2 independent elements; therefore, we need at least n2 linear equations involving these matrices to solve for all the elements of M. Suppose that we are provided with data sets at two different time points, t1 and t2 (>t1). The population averages and covariances of the proteins at any time t can be easily computed from the data; that is:
| (3A) |
| (3B) |
These quantities at the time points t1 and t2 are related by:
| (4A) |
| (4B) |
Note that Eqs. 4A and 4B provide n + n(n + 1)/2 = n2/2 + 3n/2 (<n2) linear equations for determining n2 elements of M from the data; thus, the system is underdetermined. The nonuniqueness in the estimation of M using Eqs. 4A and 4B can be further characterized with the following relationship:
| (5) |
where Q is any orthogonal matrix (QQT = 1) with real elements, and J(t2)1/2 is the square root of the matrix J(t2).
Second, the fact that elements of M must be calculated uniquely from the average values and covariances makes it difficult to separate the changes that occurred during NKG2D-stimulated signaling from those that occurred as a result of the pretreatment with IL-2 or medium. For example, the covariance between species [J(t)] at a particular time
| (6) |
is affected by the signaling kinetics (∝eMt) and by the covariances at the prestimulus level [J(t = 0)]. It is not possible to separate eMt from J(t = 0) by just calculating J(t) or by considering functions of J(t). Several methods, based on the calculation of mutual information for joint probability distributions (45) or conditional joint probability distributions (21), aim to infer the strength of interactions with covariances of the species abundances; however, those estimates can be biased by covariances between the species at the prestimulus level. Thus, even in this relatively simple and idealized situation, it can be challenging to infer the strength of interactions from time-stamped, mass cytometry measurements. We addressed this difficulty by estimating the elements of the M matrix with a simulated annealing technique. We first created a cost function
| (7) |
which was minimized in the simulated annealing calculation. The average abundances and covariances computed at times t1 and t2 from the mass cytometry data were used in Eq. 7 to estimate the M matrix (M̂). When the data were generated by the signaling kinetics given by Eq. 1, the correct M matrix yielded χ2 = 0. The average effective flux at time t was calculated using the i→j flux is given by:
| (8) |
where the first term is the flux from i→j and the second term is the flux from j→i. When fi→j > 0, the effective flux from i→j is represented with an arrow from i to j, and when fi→j < 0, we showed the effective flux with an arrow from j to i.
Estimation of the M matrix using simulated annealing
We started our annealing scheme with an initial matrix M, in which the Mii’s for the reaction network (Fig. 4A) were set to 1, whereas the rest of the off-diagonal elements were set to zero. The diagonal elements were chosen such that all of the columns of the matrix were added up to zero. Using this M matrix, we calculated an initial cost χ2initial, which was given by Eq. 7. The initial temperature (σ2T)initial was set to 10χ2initial. After each annealing step, the temperature was lowered according to an exponential annealing scheme, (σ2T)p+1 = β × (σ2T)p, where p is the annealing index and β (<1) was chosen such that (σ2T)end = 10−6. We used a total of 6000 annealing steps. For every annealing step p, we ran 106 Monte Carlo (MC) updates. For each MC update, we chose an element Mji by drawing the row index i and the column index j randomly from a uniform distribution U(1,n), where n is the total number of proteins considered in the model. When i = j, we proposed a new Mpropose(i, j) = Mcurrent(i, j) + Δ (2ξ − 1), in which Δ is the maximum step size and ξ is a random number between (0,1). We have performed simulations for Δ = 1 and 0.01, and both produced the same final result. If i ≠ j, then we first checked whether Mcurrent(i, j) + Δ (2ξ − 1) > 1 or < 0. If these conditions were met, we set Mpropose(i, j) = Mcurrent(i, j); otherwise, we set Mpropose(i, j) = Mcurrent(i, j) + Δ (2ξ − 1). This condition ensured that the off-diagonal elements of the matrix M lay between 0 and 1. Note that the diagonal elements of the proposed matrix M could, in principal, take any (positive or negative) real value. We then calculated the cost function given by Eq. 7. The proposed M matrices whose columns did not add up to zero paid a penalty equal to the third term in Eq. 7. A choice of χ2 given by Eq. 7 biased our search for networks in which the total number of molecules was conserved at all times. We accepted the proposed moves using the standard Metropolis-Hastings algorithm (46), in which the acceptance probability η is given by:
The method was validated with synthetic data (see fig. S4 for details). We set Δt to 1 min in Eq. 7 for all of the simulations. The flux values shown in Figs. 4 and 5 were normalized by a constant factor (maximum flux magnitude occurred in all the data analyzed) such that the flux magnitudes were always less than or equal to unity.
Enrichment of primary human NK cells
Blood was obtained from the Blood Centers of the Pacific under an Institutional Review Board–approved protocol (IRB #10-00265). Peripheral blood mononuclear cells (PBMCs) were then isolated by density gradient centrifugation with Ficoll-Paque PLUS (GE Healthcare Bio-Sciences AB). NK cells were purified from the PBMCs with an NK Cell Isolation kit (Miltenyi Biotec GmbH) and were >90% pure. The enriched primary human NK cells were cultured in RPMI 1640 (Corning Cellgro, Mediatech Inc.) containing 10% fetal bovine serum (Thermo Fisher Scientific), 2 mM L-glutamine [University of California, San Francisco (UCSF) cell culture facility], 1× nonessential amino acids (UCSF cell culture facility), sodium pyruvate (0.11 mg/ml; UCSF cell culture facility), 100 IU penicillin, and streptomycin (100 μg/ml; Corning Cellgro) (R10 medium) and were pretreated with either medium or human IL-2 (200 U/ml; provided by Prometheus Laboratories Inc.) for 24 hours at 37°C and 5% CO2.
In vitro stimulation of primary human NK cells
Nunc MaxiSorp enzyme-linked immunosorbent assay (ELISA) plates (Thermo Fisher Scientific) were washed twice in phosphate-buffered saline (PBS) and then were coated with anti-NKG2D mAb (5 μg/ml; clone 1D11, BioLegend) in PBS for 24 hours at 4°C. The ELISA plates were washed twice in PBS and blocked in complete culture medium for 10 min at room temperature before 150,000 enriched primary human NK cells were added to each well in the ELISA plate. The cells were left unstimulated (t = 0) or stimulated with anti-NKG2D mAb in R10 medium with or without IL-2 (200 U/ml) for 4, 8, 16, 32, 64, 128, or 256 min at 37°C and 5% CO2. For experiments in which the cells were stimulated with PMA and ionomycin, the cells were incubated for 16 min at 37°C and 5% CO2 with PMA (50 ng/ml; Sigma-Aldrich) and ionomycin (1 μg/ml; Sigma-Aldrich). For mass cytometry analysis, the cells were fixed with 1.5 % paraformaldehyde (PFA) in PBS for 10 min at room temperature immediately after stimulation.
Mass cytometry
Labeling of cells and the viability staining for mass cytometry were performed as previously described (47, 48), with a few modifications. Before the cells were stimulated, viability staining was performed by incubating the cells with 50 μM cisplatin (Sigma-Aldrich) in 1 ml of serum-free RPMI 1640 per 106 cells for 1 min at room temperature. An equal volume of R10 medium was added, and the cells were then incubated for an additional 5 min at room temperature. The cells were washed in R10 medium and resuspended at a concentration of 750,000 cells/ml. 151Eu-conjugated anti-CD107a mAb was added to the cells just before they were stimulated. Immediately after stimulation, the cells were fixed with 1.5% PFA in PBS for 10 min at room temperature. The cells were then washed twice with cell-staining medium (CSM) [PBS containing 0.5% bovine serum albumin (BSA) and 0.02% sodium azide] and bar-coded as previously described (49). Briefly, the cells were washed once with PBS and once with 0.02% saponin in PBS and then incubated with barcode reagents in 1 ml of 0.02% saponin in PBS at room temperature for 15 min. After the incubation, the cells were washed twice with CSM and then pooled for subsequent staining. All of the antibodies used for mass cytometry were validated and titrated before use. The bar-coded samples were surface-stained by incubation with a cocktail of metal-conjugated antibodies (listed in table S2) for 1 hour at room temperature with continuous shaking. Cells were washed twice with CSM, fixed with 1.5% PFA in PBS for 10 min at room temperature, and then permeabilized in cold methanol for 10 min at 4°C. Intracellular staining was performed by incubating the cells with a cocktail of metal-conjugated antibodies (listed in table S2) for 1 hour at room temperature with continuous shaking. Cells were washed twice with CSM and then incubated for 20 min in 1 ml of iridium DNA intercalator [diluted 1:5000 in PBS with 1.6% PFA (DVS Sciences)] for 20 min at room temperature or overnight at 4°C. Before the mass cytometry analysis was performed, the cells were washed once with CSM and twice with double-distilled water and then resuspended in double-distilled water containing the bead standard for normalization (50). Cells were resuspended in double-distilled water at about 1 million cells/ml and analyzed on a mass cytometer (Fluidigm). The data were normalized and debarcoded as previously described (49, 50). The protein abundance data were extracted from the NK cells by gating on caspase3−cPARP−CD45+CD235−CD61−CD33−CD20−CD3−CD56+ cells using the Cytobank platform.
Flow cytometry
The mAbs used for cell surface staining were as follows: fluorescein isothiocyanate–conjugated anti-human CD107a (H4A3, BioLegend), peridinin chlorophyll protein–Cy5.5–conjugated anti-human CD56 (HCD56, BioLegend), Alexa Fluor 700–conjugated anti-human CD3 (HIT3a, BioLegend), and allophycocyanin-conjugated anti-human CD45 (Hle-1, BD Biosciences). After stimulation, the cells were washed twice in staining buffer (PBS, 1% BSA, and 0.5 mM EDTA) and stained with the appropriate mAbs against cell surface markers for 20 min on ice. The cells were then washed twice in FACS staining buffer, fixed, and permeabilized using the Cytofix/Cytoperm kit (BD Biosciences) according to the manufacturer’s protocol. The cells were subsequently stained with Alexa Fluor 647–conjugated anti-human IFN-γ antibody for 20 min on ice. After the final wash, the samples were acquired on an LSRII flow cytometer (BD Biosciences) and analyzed with FlowJo software (Tree Star). Single live cells were gated on the basis of forward and side light scatter profiles, and NK cells were gated as CD3−CD56bright or CD3−CD56dim cells.
Statistical analysis
The parameters in the model were estimated as described in the beginning of this section.
Supplementary Material
Fig. S1. Kinetics of cell population–averaged protein abundances in mass cytometry measurements in IL-2– or medium-treated CD56bright and CD56dim primary human NK cells (from donor #1) stimulated by anti-NKG2D.
Fig. S2. Kinetics of cell population–averaged protein abundances in mass cytometry measurements in IL-2– or medium-treated CD56bright and CD56dim primary human NK cells (from donor #2) stimulated by anti-NKG2D.
Fig. S3. Kinetics of correlations between protein species in CD56bright and CD56dim NK cells from donor #2.
Fig. S4. Estimation of reaction rates with synthetic data generated from a biochemical network composed of coupled first-order chemical reactions.
Fig. S5. Estimation of reaction fluxes with synthetic data generated from the Ras activation network composed of coupled, nonlinear chemical reactions.
Fig. S6. In silico characterization of naïve CD4+ T cell signaling stimulated with cross-linking antibodies against CD3, CD28, and CD4.
Fig. S7. Kinetics of the average abundance of proteins when IL-2–pretreated cells obtained from donor #3 were stimulated by isotype-matched IgG or anti-NKG2D.
Fig. S8. In silico characterization of the NKG2D signaling kinetics in CD56bright and CD56dim NK cells from donor #2.
Fig. S9. Test of correlation between CD45 expression and CD107a mobilization to the cell surface of human NK cells from donor #2.
Fig. S10. Test of correlation between CD45 expression and CD107a mobilization to the cell surface of human NK cells from donor #3.
Fig. S11. Test of correlation between CD45 expression and CD107a mobilization to the cell surface of human NK cells from donors #4 to #7.
Fig. S12. Matrix plot showing the changes in average protein abundances in CD56bright and CD56dim NK cells in response to IL-2 treatment.
Table S1. Changes in average protein abundances in CD56bright and CD56dim NK cells in response to IL-2.
Table S2. Mass cytometry antibody panel.
Acknowledgments
We thank Prometheus Laboratories Inc. for providing human IL-2 and A. Snedden at the High Performance Computing Center at the Nationwide Children’s Research Institute for the help with graphics. J.D. and S.M. thank D. Wethington for help in generating some of the figures.
Funding: The work was partially supported by grant R56AI108880-01 from the National Institute of Allergy and Infectious Diseases to J.D. and L.L.L. L.L.L. is an American Cancer Society Professor and is supported by the Parker Institute for Cancer Immunotherapy and NIH grants AI066897 and AI068129. H.J. is supported by a Lundbeck Foundation Postdoctoral Fellowship. S.M. was supported, in part, by a grant from the Department of Biotechnology (BTPR12422/MED/31/287/2014, valid from November 2014 to 2017). In addition, the work in the Nolan Lab was supported by NIH grants U19AI057229, U19AI100627, R33CA183654, R33CA0183692, R01GM10983601, R01CA184968, R01CA19665701, R21CA183660, R01NS08953301, 5UH2AR067676, and R01HL120724; the Northrop-Grumman Corporation; Novartis grant CMEK162AUS06T; Pfizer grant 123214; Juno Therapeutics grant 122401; U.S. Department of Defense grants OC110674 and W81XWH-14-1-0 180; Gates Foundation grant OPP1113682; and U.S. Food and Drug Administration grant BAA-15-00121.
Footnotes
Author contributions: S.M., H.J., L.L.L., and J.D. designed the in silico and bench experiments. H.J. and S.-Y.C. performed the experiments. S.M. and J.D. developed the mathematical models and performed the computer simulations. D.S., W.C.R., and W.S. contributed to developing computational analysis and visualization tools. S.-Y.C. and G.P.N. provided help with the mass cytometry experiments. S.M., H.J., L.L.L., and J.D. analyzed the data and wrote the manuscript.
Competing interests: L.L.L. and the UCSF have licensed intellectual property rights regarding NKG2D for commercial applications.
Data and materials availability: The mass cytometry data have been made publicly available on the Cytobank webpage (https://community.cytobank.org).
REFERENCES AND NOTES
- 1.Lanier LL. Up on the tightrope: Natural killer cell activation and inhibition. Nat Immunol. 2008;9:495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Long EO. Negative signaling by inhibitory receptors: The NK cell paradigm. Immunol Rev. 2008;224:70–84. doi: 10.1111/j.1600-065X.2008.00660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryceson YT, March ME, Ljunggren HG, Long EO. Synergy among receptors on resting NK cells for the activation of natural cytotoxicity and cytokine secretion. Blood. 2006;107:159–166. doi: 10.1182/blood-2005-04-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooper MA, Fehniger TA, Caligiuri MA. The biology of human natural killer-cell subsets. Trends Immunol. 2001;22:633–640. doi: 10.1016/s1471-4906(01)02060-9. [DOI] [PubMed] [Google Scholar]
- 5.Lanier LL, Le AM, Civin CI, Loken MR, Phillips JH. The relationship of CD16 (Leu-11) and Leu-19 (NKH-1) antigen expression on human peripheral blood NK cells and cytotoxic T lymphocytes. J Immunol. 1986;136:4480–4486. [PubMed] [Google Scholar]
- 6.Cooper MA, Fehniger TA, Turner SC, Chen KS, Ghaheri BA, Ghayur T, Carson WE, Caligiuri MA. Human natural killer cells: A unique innate immunoregulatory role for the CD56bright subset. Blood. 2001;97:3146–3151. doi: 10.1182/blood.v97.10.3146. [DOI] [PubMed] [Google Scholar]
- 7.Long EO, Kim HS, Liu D, Peterson ME, Rajagopalan S. Controlling natural killer cell responses: Integration of signals for activation and inhibition. Annu Rev Immunol. 2013;31:227–258. doi: 10.1146/annurev-immunol-020711-075005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vivier E, Nunès JA, Vély F. Natural killer cell signaling pathways. Science. 2004;306:1517–1519. doi: 10.1126/science.1103478. [DOI] [PubMed] [Google Scholar]
- 9.Segovis CM, Schoon RA, Dick CJ, Nacusi LP, Leibson PJ, Billadeau DD. PI3K links NKG2D signaling to a CrkL pathway involved in natural killer cell adhesion, polarity, and granule secretion. J Immunol. 2009;182:6933–6942. doi: 10.4049/jimmunol.0803840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Upshaw JL, Arneson LN, Schoon RA, Dick CJ, Billadeau DD, Leibson PJ. NKG2D-mediated signaling requires a DAP10-bound Grb2-Vav1 intermediate and phosphatidylinositol-3-kinase in human natural killer cells. Nat Immunol. 2006;7:524–532. doi: 10.1038/ni1325. [DOI] [PubMed] [Google Scholar]
- 11.Upshaw JL, Leibson PJ. NKG2D-mediated activation of cytotoxic lymphocytes: Unique signaling pathways and distinct functional outcomes. Semin Immunol. 2006;18:167–175. doi: 10.1016/j.smim.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 12.Chiesa S, Mingueneau M, Fuseri N, Malissen B, Raulet DH, Malissen M, Vivier E, Tomasello E. Multiplicity and plasticity of natural killer cell signaling pathways. Blood. 2006;107:2364–2372. doi: 10.1182/blood-2005-08-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altan-Bonnet G, Germain RN. Modeling T cell antigen discrimination based on feedback control of digital ERK responses. PLOS Biol. 2005;3:e356. doi: 10.1371/journal.pbio.0030356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das J, Ho M, Zikherman J, Govern C, Yang M, Weiss A, Chakraborty AK, Roose JP. Digital signaling and hysteresis characterize Ras activation in lymphoid cells. Cell. 2009;136:337–351. doi: 10.1016/j.cell.2008.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mukherjee S, Rigaud S, Seok SC, Fu G, Prochenka A, Dworkin M, Gascoigne NRJ, Vieland VJ, Sauer K, Das J. In silico modeling of Itk activation kinetics in thymocytes suggests competing positive and negative IP4 mediated feedbacks increase robustness. PLOS ONE. 2013;8:e73937. doi: 10.1371/journal.pone.0073937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das J. Activation or tolerance of natural killer cells is modulated by ligand quality in a nonmonotonic manner. Biophys J. 2010;99:2028–2037. doi: 10.1016/j.bpj.2010.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watzl C, Sternberg-Simon M, Urlaub D, Mehr R. Understanding natural killer cell regulation by mathematical approaches. Front Immunol. 2012;3:359. doi: 10.3389/fimmu.2012.00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukherjee S, Seok SC, Vieland VJ, Das J. Cell responses only partially shape cell-to-cell variations in protein abundances in Escherichia coli chemotaxis. Proc Natl Acad Sci USA. 2013;110:18531–18536. doi: 10.1073/pnas.1311069110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swain PS, Elowitz MB, Siggia ED. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc Natl Acad Sci USA. 2002;99:12795–12800. doi: 10.1073/pnas.162041399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bendall SC, Nolan GP, Roederer M, Chattopadhyay PK. A deep profiler’s guide to cytometry. Trends Immunol. 2012;33:323–332. doi: 10.1016/j.it.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krishnaswamy S, Spitzer MH, Mingueneau M, Bendall SC, Litvin O, Stone E, Pe’er D, Nolan GP. Conditional density-based analysis of T cell signaling in single-cell data. Science. 2014;346:1250689. doi: 10.1126/science.1250689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCann FE, Vanherberghen B, Eleme K, Carlin LM, Newsam RJ, Goulding D, Davis DM. The size of the synaptic cleft and distinct distributions of filamentous actin, ezrin, CD43, and CD45 at activating and inhibitory human NK cell immune synapses. J Immunol. 2003;170:2862–2870. doi: 10.4049/jimmunol.170.6.2862. [DOI] [PubMed] [Google Scholar]
- 23.Billadeau DD, Upshaw JL, Schoon RA, Dick CJ, Leibson PJ. NKG2D-DAP10 triggers human NK cell–mediated killing via a Syk-independent regulatory pathway. Nat Immunol. 2003;4:557–564. doi: 10.1038/ni929. [DOI] [PubMed] [Google Scholar]
- 24.Hermiston ML, Xu Z, Weiss A. CD45: A critical regulator of signaling thresholds in immune cells. Annu Rev Immunol. 2003;21:107–137. doi: 10.1146/annurev.immunol.21.120601.140946. [DOI] [PubMed] [Google Scholar]
- 25.Hesslein DGT, Palacios EH, Sun JC, Beilke JN, Watson SR, Weiss A, Lanier LL. Differential requirements for CD45 in NK–cell function reveal distinct roles for Syk–family kinases. Blood. 2011;117:3087–3095. doi: 10.1182/blood-2010-06-292219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tassi I, Cella M, Presti R, Colucci A, Gilfillan S, Littman DR, Colonna M. NK cell-activating receptors require PKC-θ for sustained signaling, transcriptional activation, and IFN-γ secretion. Blood. 2008;112:4109–4116. doi: 10.1182/blood-2008-02-139527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ali AK, Nandagopal N, Lee SH. IL-15–PI3K–AKT–mTOR: A critical pathway in the life journey of natural killer cells. Front Immunol. 2015;6:355. doi: 10.3389/fimmu.2015.00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okkenhaug K. Signaling by the phosphoinositide 3-kinase family in immune cells. Annu Rev Immunol. 2013;31:675–704. doi: 10.1146/annurev-immunol-032712-095946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang K, Zhong B, Gilvary DL, Corliss BC, Hong-Geller E, Wei S, Djeu JY. Pivotal role of phosphoinositide-3 kinase in regulation of cytotoxicity in natural killer cells. Nat Immunol. 2000;1:419–425. doi: 10.1038/80859. [DOI] [PubMed] [Google Scholar]
- 30.Alter G, Malenfant JM, Altfeld M. CD107a as a functional marker for the identification of natural killer cell activity. J Immunol Methods. 2004;294:15–22. doi: 10.1016/j.jim.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 31.Cella M, Fujikawa K, Tassi I, Kim S, Latinis K, Nishi S, Yokoyama W, Colonna M, Swat W. Differential requirements for Vav proteins in DAP10- and ITAM-mediated NK cell cytotoxicity. J Exp Med. 2004;200:817–823. doi: 10.1084/jem.20031847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khurana D, Arneson LN, Schoon RA, Dick CJ, Leibson PJ. Differential regulation of human NK cell-mediated cytotoxicity by the tyrosine kinase Itk. J Immunol. 2007;178:3575–3582. doi: 10.4049/jimmunol.178.6.3575. [DOI] [PubMed] [Google Scholar]
- 33.Lee SH, Yun S, Lee J, Kim MJ, Piao ZH, Jeong M, Chung JW, Kim TD, Yoon SR, Greenberg PD, Choi I. RasGRP1 is required for human NK cell function. J Immunol. 2009;183:7931–7938. doi: 10.4049/jimmunol.0902012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Björkström NK, Riese P, Heuts F, Andersson S, Fauriat C, Ivarsson MA, Björklund AT, Flodström-Tullberg M, Michaelsson J, Rottenberg ME, Guzman CA, Ljunggren HG, Malmberg KJ. Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK-cell differentiation uncoupled from NK-cell education. Blood. 2010;116:3853–3864. doi: 10.1182/blood-2010-04-281675. [DOI] [PubMed] [Google Scholar]
- 35.McNeill L, Salmond RJ, Cooper JC, Carret CK, Cassady-Cain RL, Roche-Molina M, Tandon P, Holmes N, Alexander DR. The differential regulation of Lck kinase phosphorylation sites by CD45 is critical for T cell receptor signaling responses. Immunity. 2007;27:425–437. doi: 10.1016/j.immuni.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 36.Hesslein DGT, Takaki R, Hermiston ML, Weiss A, Lanier LL. Dysregulation of signaling pathways in CD45-deficient NK cells leads to differentially regulated cytotoxicity and cytokine production. Proc Natl Acad Sci USA. 2006;103:7012–7017. doi: 10.1073/pnas.0601851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lanier LL. DAP10- and DAP12-associated receptors in innate immunity. Immunol Rev. 2009;227:150–160. doi: 10.1111/j.1600-065X.2008.00720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Markevich NI, Hoek JB, Kholodenko BN. Signaling switches and bistability arising from multisite phosphorylation in protein kinase cascades. J Cell Biol. 2004;164:353–359. doi: 10.1083/jcb.200308060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quatrini L, Molfetta R, Zitti B, Peruzzi G, Fionda C, Capuano C, Galandrini R, Cippitelli M, Santoni A, Paolini R. Ubiquitin-dependent endocytosis of NKG2D-DAP10 receptor complexes activates signaling and functions in human NK cells. Sci Signal. 2015;8:ra108. doi: 10.1126/scisignal.aab2724. [DOI] [PubMed] [Google Scholar]
- 40.Leupin O, Zaru R, Laroche T, Müller S, Valitutti S. Exclusion of CD45 from the T-cell receptor signaling area in antigen-stimulated T lymphocytes. Curr Biol. 2000;10:277–280. doi: 10.1016/s0960-9822(00)00362-6. [DOI] [PubMed] [Google Scholar]
- 41.Transtrum MK, Machta BB, Brown KS, Daniels BC, Myers CR, Sethna JP. Perspective: Sloppiness and emergent theories in physics, biology, and beyond. J Chem Phys. 2015;143:010901. doi: 10.1063/1.4923066. [DOI] [PubMed] [Google Scholar]
- 42.Cotari JW, Voisinne G, Dar OE, Karabacak V, Altan-Bonnet G. Cell-to-cell variability analysis dissects the plasticity of signaling of common γ chain cytokines in T cells. Sci Signal. 2013;6:ra17. doi: 10.1126/scisignal.2003240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Kampen NG. Stochastic Processes in Physics and Chemistry. North-Holland Personal Library; 1992. p. 465. [Google Scholar]
- 44.Jahnke T, Huisinga W. Solving the chemical master equation for monomolecular reaction systems analytically. J Math Biol. 2007;54:1–26. doi: 10.1007/s00285-006-0034-x. [DOI] [PubMed] [Google Scholar]
- 45.Margolin AA, Nemenman I, Basso K, Wiggins C, Stolovitzky G, Dalla Favera R, Califano A. ARACNE: An algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7(Suppl 1):S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Laarhoven PJM, Aarts EHL. Simulated annealing: Theory and applications. In: Reidel D, editor. Mathematics and Its Applications. Kluwer Academic Publishers; 1987. p. 186. [Google Scholar]
- 47.Bendall SC, Simonds EF, Qiu P, Amir E-aD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, Balderas RS, Plevritis SK, Sachs K, Pe’er D, Tanner SD, Nolan GP. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fienberg HG, Simonds EF, Fantl WJ, Nolan GP, Bodenmiller B. A platinum-based covalent viability reagent for single-cell mass cytometry. Cytometry A. 2012;81A:467–475. doi: 10.1002/cyto.a.22067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zunder ER, Finck R, Behbehani GK, Amir E-aD, Krishnaswamy S, Gonzalez VD, Lorang CG, Bjornson Z, Spitzer MH, Bodenmiller B, Fantl WJ, Pe’er D, Nolan GP. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat Protoc. 2015;10:316–333. doi: 10.1038/nprot.2015.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Finck R, Simonds EF, Jager A, Krishnaswamy S, Sachs K, Fantl W, Pe’er D, Nolan GP, Bendall SC. Normalization of mass cytometry data with bead standards. Cytometry A. 2013;83A:483–494. doi: 10.1002/cyto.a.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Kinetics of cell population–averaged protein abundances in mass cytometry measurements in IL-2– or medium-treated CD56bright and CD56dim primary human NK cells (from donor #1) stimulated by anti-NKG2D.
Fig. S2. Kinetics of cell population–averaged protein abundances in mass cytometry measurements in IL-2– or medium-treated CD56bright and CD56dim primary human NK cells (from donor #2) stimulated by anti-NKG2D.
Fig. S3. Kinetics of correlations between protein species in CD56bright and CD56dim NK cells from donor #2.
Fig. S4. Estimation of reaction rates with synthetic data generated from a biochemical network composed of coupled first-order chemical reactions.
Fig. S5. Estimation of reaction fluxes with synthetic data generated from the Ras activation network composed of coupled, nonlinear chemical reactions.
Fig. S6. In silico characterization of naïve CD4+ T cell signaling stimulated with cross-linking antibodies against CD3, CD28, and CD4.
Fig. S7. Kinetics of the average abundance of proteins when IL-2–pretreated cells obtained from donor #3 were stimulated by isotype-matched IgG or anti-NKG2D.
Fig. S8. In silico characterization of the NKG2D signaling kinetics in CD56bright and CD56dim NK cells from donor #2.
Fig. S9. Test of correlation between CD45 expression and CD107a mobilization to the cell surface of human NK cells from donor #2.
Fig. S10. Test of correlation between CD45 expression and CD107a mobilization to the cell surface of human NK cells from donor #3.
Fig. S11. Test of correlation between CD45 expression and CD107a mobilization to the cell surface of human NK cells from donors #4 to #7.
Fig. S12. Matrix plot showing the changes in average protein abundances in CD56bright and CD56dim NK cells in response to IL-2 treatment.
Table S1. Changes in average protein abundances in CD56bright and CD56dim NK cells in response to IL-2.
Table S2. Mass cytometry antibody panel.