Abstract
The ability to determine the global location of transcription factor binding sites in vivo is important for a comprehensive understanding of gene regulation in human cells. We have developed a technology, called serial analysis of binding elements (SABE), involving subtractive hybridization of chromatin immunoprecipitation-enriched DNA fragments followed by the generation and analysis of concatamerized sequence tags. We applied the SABE technology to search for p53 target genes in the human genome, and have identified several previously described p53 targets in addition to numerous potentially novel targets, including the DNA mismatch repair genes MLH1 and PMS2. Both of these genes were determined to be responsive to DNA damage and p53 activation in normal human fibroblasts, and have p53-response elements within their first intron. These two genes may serve as a sensor in DNA repair mechanisms and a critical determinant for the decision between cell-cycle arrest and apoptosis. These results also demonstrate the potential for use of SABE as a broadly applicable means to globally identify regulatory elements for human transcription factors in vivo.
Keywords: chromatin immunoprecipitation, DNA binding, transcription factor
A major challenge in the postgenome era is to elucidate global transcriptional regulatory networks (1). Transcription factors control gene expression through binding-specific regulatory sequences on DNA and recruiting chromatin modifying complexes and the general transcription machinery to initiate RNA synthesis (2). Alterations in gene expression required to coordinate various biological processes such as the cell cycle and normal development, and pathological states such as tumorigenesis are, in part, a consequence of changes in the DNA-binding status of various transcription factors, and, consequently, sensitive technologies to accurately and efficiently identify bona fide regulatory elements for specific transcription factors in vivo, under a variety of physiological conditions, will be needed to elucidate human gene regulatory networks. Computational analysis can provide predictions of regulatory elements within genome sequences (3-5). However, sites identified in silico may not necessarily represent regulatory elements in vivo. Many regulatory decisions for gene expression involve cooperative interactions between transcription factors bound to multiple adjacent weak elements (2). Accessibility of cis elements on DNA for some factors in vivo is also affected by nucleosomal organization in chromatin (6). Global localization of cis elements for sequence-specific factors in vivo can be performed in yeast by using chromatin immunoprecipitation (ChIP) and hybridization of intergenic sequence arrays (7, 8). A comparable strategy for globally analyzing binding of factors to the human genome is impracticable because of the enormous size and complexity, and because regulatory elements are often found at vast distances either upstream or downstream from the core promoter. Nonetheless, limited analysis of human transcription factor-binding sites by using hybridization of high-density arrays with probe prepared from ChIP DNA have been performed with promoters of interest (9), with CpG microarrays (10) or with selected chromosomes (11). Here, we describe a technology called serial analysis of binding elements (SABE) for globally identifying binding sites of mammalian transcription factors in vivo. SABE involves specific ChIP (12), enrichment of ChIP DNA by using representational difference analysis (13), and generation of sequence tags similar to serial analysis of gene expression (SAGE) (14). By using this approach, we have identified target genes for the tumor suppressor protein p53.
p53 is a sequence-specific DNA-binding protein that regulates transcription of genes, causing cell-cycle arrest or apoptosis in response to DNA damage (15, 16). The p53 gene is commonly mutated in human cancers, and the vast majority of mutations occur in the region required for DNA binding, thereby disabling its function in causing arrest or death of cells with damaged genetic information. p53 is regulated at multiple levels to control its interaction with DNA. In normal cycling cells, p53 is maintained in the cytoplasm by several mechanisms where it is rapidly turned over. In response to DNA damage, or other cellular stresses, p53 becomes modified by phosphorylation and acetylation, which promotes its accumulation and retention in the nucleus, stimulates DNA binding, and regulates recruitment of coactivator complexes. The mechanisms by which p53 controls the decision to cause cellular arrest or to undergo apoptosis is currently a question of considerable interest and likely involves differential regulation of specific classes of genes representing cell-cycle regulators to cause growth arrest or cell death pathway effectors. Various estimates have predicted 200-400 (17) or >1,600 (11) p53-binding sites in the human genome, but only a fraction of these have been identified, including 48 sites on chromosomes 21 and 22 by using a strategy with probe prepared from ChIP DNA (11). In addition to its role in regulating cell-cycle arrest and apoptosis, p53 is also known to be involved in regulating DNA repair mechanisms through the induction of P53R2, a ribonucleotide reductase subunit, and may also directly participate in repair by promoting annealing of single-stranded DNAs and rejoining double-stranded breaks. In this report, we identify two mismatch DNA repair genes, MLH1 and PMS2, as targets for p53 in normal fibroblasts. These results demonstrate a broader role for p53 in regulation of transcriptional responses to DNA damage than was previously understood, and suggest a possible link between induction of DNA-damage response and the decision to undergo cell-cycle arrest or apoptosis.
Experimental Procedures
Plasmids, Cell Lines, and Antibodies. For details of plasmid constructions, see Supporting Text, which is published as supporting information on the PNAS web site. A stable Jurkat cell line expressing p53-3XFLAG was produced by transfection with p3FLAG-53, and cloning by limiting dilution after 1 month of selection in 800 μg/ml G418 (Sigma). The resulting J53 cell line was shown to express p53-3XFLAG by immunoblotting. Minimal reporter genes bearing MLH1 and PMS2 p53-response elements were generated by cloning fragments generated from conventional ChIP analysis. Briefly, PCR products were inserted into pCR2.1 by using the TA cloning system (Invitrogen), and the sequences were confirmed before subcloning using KpnI-XhoI (mutL homolog 1; MLH1) or BamHI-XbaI (yeast postmeiotic segregation increased 2; PMS2) into the same sites (MLH1) or NheI-BglII sites (PMS2) of pTAL-Luc (Clontech). Antibodies against MLH1, PMS2, and actin were purchased from Santa Cruz Biotechnology.
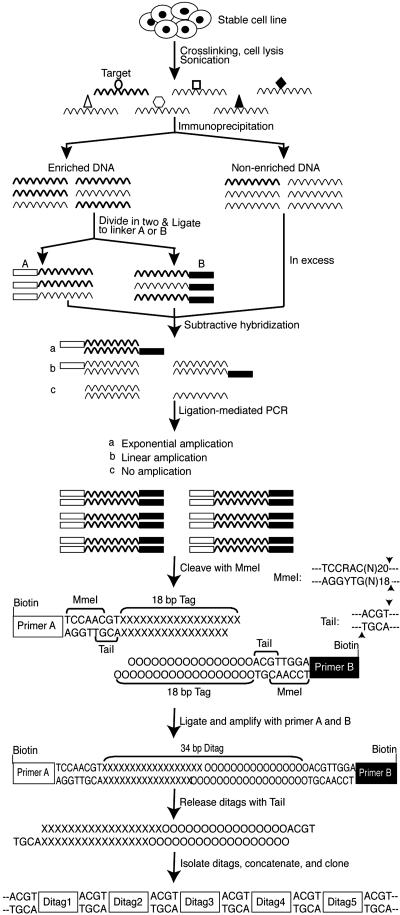
SABE. A schematic diagram of the strategy to generate SABE tag libraries is shown in Fig. 1, and a detailed description of the procedures are described in Supporting Text. Concatamerized SABE tags were ligated into AatII-digested pJC-Z2 and transformed into DH5α competent cells. Kanamycin-resistant colonies were analyzed by colony-PCR with the M13 forward and reverse primers, and clones bearing concatamers of >300 nt were sequenced with the T7 promoter primer. Ditags are 34 bp long and are separated by the TaiI recognition sequence (ACGT). Tag sequences were used to manually search human genomic D NA by using ENSEMBL (which can be accessed at www.ensembl.org/Homo_sapiens/) to identify the genomic location. Of 234 SABE tag sequences, 146 (62%) were uniquely represented in the human genome (indicated in Table 3, which is published as supporting information on the PNAS web site). Putative p53 responsive elements were identified from 1,000-bp DNA flanking the tag by using motif (which can be accessed at http://motif.genome.jp).
Fig. 1.
Schematic summary of SABE. Mammalian cells were crosslinked by treatment with formaldehyde, and specific protein-DNA complexes were isolated by IP. DNA is ligated to linkers and specific DNA selectively amplified after representational difference subtractive hybridization. After digestion with MmeI, ditags are produced by ligation, which are then released by digestion with TaiI. The released ditags are separated from the biotinylated linker and primer DNA by using streptavidin magnetic beads, concatamerized by ligation, and cloned. Ditag sequences are 34 bp long and are separated by the TaiI recognition sequence (ACGT).
Conventional ChIP Analysis. HS27 human diploid fibroblasts were treated with 25 μM cisplatin (Sigma-Aldrich) for 6 h before crosslinking and IP with anti-p53 antibody conjugated to agarose (p53-AC, Santa Cruz Biotechnology). Purified ChIP DNA was dissolved in 30 μl of TE, and 2 μl each of ChIP and input DNA control amplified by PCR using 30 reaction cycles and analysis on 2% agarose gels. Primers used for analysis of chromatin immunoprecipitated DNA are detailed in Supporting Text.
Preparation of Nuclear Extracts and EMSAs. Cisplatin-treated HS27 cells (107) were collected by low-speed centrifugation, washed three times with cold PBS, suspended in 1 ml of buffer A [10 mM Hepes (pH 7.9)/1.5 mM MgCl2/10 mM KCl/0.5 mM DTT] and passed 10 times through a 27 1/2 gauge needle. Nuclei were collected by spinning for 8 sec at 10,000 × g, washed once in buffer A, and suspended in 200 μl of cold buffer C (20 mM Hepes (pH 7.9)/25% glycerol/420 mM KCl/1.5 mM MgCl2/0.2 mM EDTA/0.5 mM DTT/0.5 mM PMSF), and incubated on ice for 15 min. Cold buffer D (200 μl) [20 mM Hepes (pH 7.9)/20% glycerol/0.2 mM EDTA/0.5 mM DTT/0.5 mM PMSF] was added, and the suspension was centrifuged for 15 min at 12000 × g at 4°C. The supernatant was transferred to a new tube and stored at -70°C until use. EMSA reactions were performed by preincubating 5 μg of nuclear extract/2 μg of poly dI-dC/2 μg of BSA/100 pmol of unlabeled competitor oligonucleotide or 0.2 μg antibody where indicated, in binding buffer [10 mM Hepes (pH 7.9)/100 mM KCl/5 mM MgCl2/and 5% glycerol] on ice for 30 min. Oligonucleotide probe (2 pmol), labeled by end filling with T4 DNA polymerase, was then added, and the reactions were incubated at room temperature for 1 h before resolving the complexes on 4% nondenaturing acrylamide gels. The oligonucleotide probe for MLH1 was 5′-GGC AGA GGC ATG TAC AGC GCA TGC CCA CAA-3′ (p53 consensus is underlined).
Luciferase and RT-PCR Assays. HeLa cells were cotransfected with 1 μg of p53RE-luciferase reporter and the indicated amount of p53 expression vector by using SuperFect transfection reagent (Qiagen). The total DNA in each transfection was equalized by addition of empty pcDNA3.1 vector. Transfection efficiency was normalized by cotransfection of 1 μg of pCMV-β-gal (Promega) internal control plasmid. Lysates were prepared 24 h after transfection, and activity was measured with the luciferase assay system (Promega) and luminescent β-gal detection kit II (Clontech), by using a microplate luminometer (Turner Designs). Results are the average of data from a minimum of three separate transfections.
For RT-PCR, RNA was isolated from cisplatin-treated HS27 cells by using the Qiagen RNeasy kit, and mRNA was isolated using the Oligotex mRNA purification kit (Qiagen). RT-PCR was performed by using Ready-To-Go RT-PCR beads (Amersham) with primers specific for MLH1 (5′-GAG ACA GTG GTG AAC CGC AT-3′ and 5′-CTT GAT TGC CAG CAC ATG GT-3′), which produces a 403-bp product, or PMS2 (5′-AGA ACC TGC TAA GGC CAT CA-3′ and 5′-TAA GCC TTC GAA GTT TTC TTC TT-3′), which generates a 223-bp product.
Results
SABE. The strategy for identifying target genes for sequence-specific DNA-binding factors using SABE is illustrated in Fig. 1. DNA-protein complexes are crosslinked in vivo using formaldehyde, the cells are lysed, and DNA is sheared by sonication to produce fragments of ≈300 bp (see Fig. 6A, which is published as supporting information on the PNAS web site). Protein-DNA complexes are then immunoprecipitated by using an antibody specific for the factor of interest. IP provides only a partial enrichment of specific DNAs, and, consequently, the signal-to-noise ratio is too low to make direct analysis of target genes practical (data not shown). To address this problem, we used a modification of representational difference analysis (13) that enables amplification of specific ChIP DNA by subtractive hybridization against reference (nonenriched) DNA. Briefly, ChIP-enriched DNA is made blunt and ligated in separate reactions to two linkers A and B (Fig. 1). The ligated DNA is hybridized to 10-fold molar excess of input control DNA and then amplified by PCR with primers specific for the linkers. After amplification, nonspecific DNA sequences will be underrepresented in the product mixture relative to specific DNA fragments.
To analyze the enriched immunoprecipitated DNA fragments, we used a strategy modified from the SAGE technique (14). The linkers A and B were designed with overlapping recognition sites for the type III endonuclease MmeI (New England Biolabs) and TaiI (Fermentas) (Fig. 1). Additionally, to facilitate separation of the linkers from the final tag DNAs, the linkers and primers included a 5′ biotin moiety (see Supporting Text for sequences). DNA fragments from the amplification are digested with MmeI, and the 46-bp fragments, including 28 bp of the linker plus 18 bp of flanking tag sequence, are purified on 12% acrylamide gels (Fig. 6B). Because MmeI leaves a 2-bp 3′ overhang, to maximize information content of the tags, the digested fragments were ligated directly to form ditags, rather than trimming to create blunt ends (Fig. 1). The ligated ditags are amplified with primers A and B and then released by digestion with TaiI. TaiI was selected because it maximally overlaps with the MmeI site and is more efficient than NlaIII, the anchoring enzyme used in SAGE (18). After digestion, the ditags can be separated from the biotin-tagged linker and primer fragments by using streptavidin Dynabeads, purified by electrophoresis (Fig. 6C), ligated to form concatamers (Fig. 7A, which is published as supporting information on the PNAS web site), and directly cloned into a vector containing an AatII site (GACGT↓C). Clones containing concatamers of 300-1,200 bp are analyzed by sequencing (Fig. 7B). Ditags can be identified in the sequencing data because each is 34 bp long separated by a TaiI sequence (ACGT). The final tag generated by this strategy is 18 bp long, including a 2-bp overlap generated by the MmeI digestion (Fig. 1). Tag sequences are used to search the human genome database to identify its genomic location. Putative binding sites for the factor of interest can then be identified by analyzing flanking DNA on genes of particular interest for consensus sequences, with the rationale that the SABE tag must reside within a segment no greater than the length of the original sheared immunoprecipitated DNA fragment (Fig. 6A).
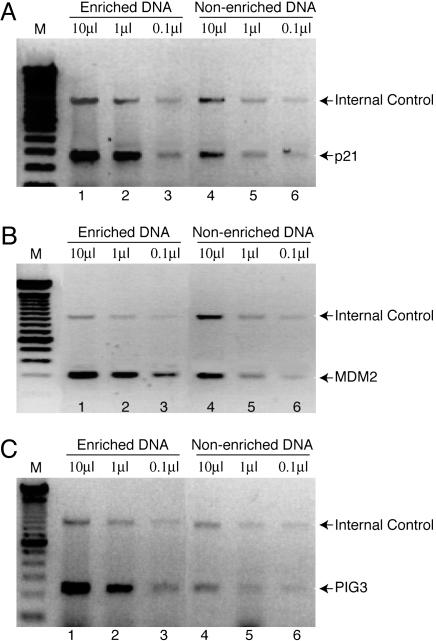
Identification of p53 Putative Target Genes by Using SABE. To enable identification of p53 target genes, we constructed a Jurkat T cell line that stably expresses p53 protein fused to a triple FLAG tag (p53-3XFLAG). Before subtractive hybridization, we analyzed DNA immunoprecipitated from crosslinked cells by PCR with primers specific for known p53 targets, p21/WAF, MDM2, and PIG3 to confirm that the FLAG-tagged protein was capable of binding to its natural cis elements in vivo. We found that ChIP DNA isolated by IP with anti-FLAG antibodies was enriched for all three known p53 targets relative to an internal control represented by USF1 (Fig. 2), which confirms that the tagged protein is functional for specific DNA binding. However, the fact that the immunoprecipitated (enriched) sample does produce a signal with the randomly chosen control primers illustrates the fact that DNA recovered in this way contains a significant amount of nonspecific template. This finding necessitates the representational difference subtractive strategy illustrated in Fig. 1.
Fig. 2.
Demonstration of known p53 targets in ChIP-enriched DNA. J53 cells expressing p53-3XFLAG were crosslinked, and DNA was sheared by sonication. The indicated volumes (marked at the top) of sheared input DNA before IP (lanes 4-6) or DNA enriched by IP with anti-FLAG antibody (lanes 1-3) were analyzed by PCR with specific primers corresponding to p53 responsive elements in p21/WAF (A), MDM2 (B), and PIG3 (C). Primers specific for the human USF1 gene were included as internal control.
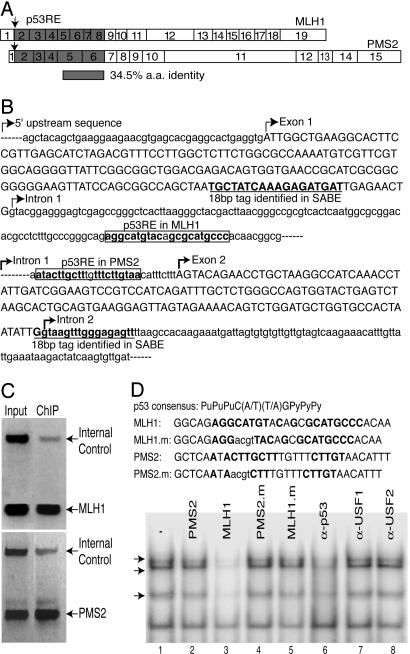
We then generated a SABE tag library from the p53-specific ChIP DNA, and manually determined the chromosomal location of 231 sequence tags by using blast (Table 3). From this analysis, we observed tags representing 17 previously known p53 target genes (Table 1), including MDM2, p21/WAF, 14-3-3∑, PIG3, and two recently identified p53 targets located in chromosomes 21 and 22 (11). Several of the previously known p53 target genes were represented by multiple SABE tags (Table 1). More than 80% of the tags were localized within the 5′ flanking or intron DNA sequences of putative target genes (Table 1). Interestingly, most of the potential target genes localized near SABE tags are predicted to be involved in tumor suppression, apoptosis, transcriptional regulation, growth factor signaling, or cell-cycle regulation (Table 3). Among these were two human mismatch repair (MMR) genes, MLH1 and PMS2, which encode proteins that are related within their N-terminal regions. Exons 2-8 of MLH1 encode a region that has 35% sequence identity as the protein segment encoded by exons 2-6 of PMS2 (Fig. 3A). Fig. 3B shows the structures of these potential target genes, and the location of the identified SABE tag in each. We searched 1 kb upstream and downstream of the tag in each gene for sequences matching the p53 consensus element and located potential binding sites within the first intron of each (Fig. 3B). The potential p53RE in MLH1 contains a nearly perfect match (19 of 20 nucleotides) to the defined consensus that is represented by two tandem repeats of the palindromic sequence PuPu-PuC(A/T) (T/A)GPyPyPy, separated by a 0- to 13-bp spacer (Fig. 3 B and D) (17). The putative p53 consensus found in intron 1 of PMS2 is more divergent, with only 16 of 20 nucleotides matching the consensus. We note that even with the degenerate p53 consensus sequence, the likelihood of observing a p53-binding site in a random DNA sequences is low. Hoh et al. (19) examined 30,000 reference sequences with base frequencies comparable to the human genome, and found that <1,500 (5%) exceeded a theoretical cutoff score.
Table 1. Summary of p53 target genes identified by unique SABE tags.
| Chromosome | 5′* | I† | E‡ | 3′§ | N¶ | Previously known p53 targets∥ |
|---|---|---|---|---|---|---|
| 1 | 3 | 8 | 0 | 1 | 1 | 14-3-3∑(2)** |
| 2 | 3 | 5 | 0 | 0 | 3 | TGFA, TP5313 (PIG3) |
| 3 | 0 | 7 | 0 | 0 | 0 | MLH1†† |
| 4 | 3 | 4 | 0 | 2 | 3 | |
| 5 | 2 | 3 | 0 | 1 | 0 | CSPG2 |
| 6 | 8 | 3 | 0 | 2 | 1 | CDKN1A (p21/WAF) (2)** |
| 7 | 3 | 6 | 0 | 1 | 0 | IGFBP3, EGFR, ING3, PMS2†† |
| 8 | 3 | 4 | 0 | 1 | 2 | |
| 9 | 3 | 4 | 0 | 1 | 0 | |
| 10 | 2 | 7 | 0 | 0 | 1 | UNC5B, TNFRSF6 |
| 11 | 5 | 1 | 0 | 2 | 0 | CASP1, NOX4 |
| 12 | 2 | 2 | 0 | 2 | 0 | MDM2 |
| 13 | 0 | 2 | 0 | 0 | 0 | |
| 14 | 4 | 0 | 0 | 0 | 0 | |
| 15 | 1 | 0 | 0 | 0 | 1 | |
| 16 | 1 | 1 | 0 | 0 | 0 | |
| 17 | 2 | 5 | 0 | 0 | 0 | |
| 18 | 1 | 1 | 0 | 0 | 0 | |
| 19 | 1 | 1 | 0 | 0 | 0 | |
| 20 | 0 | 1 | 0 | 0 | 0 | PCNA |
| 21 | 2 | 0 | 0 | 0 | 1 | RUNX1 |
| 22 | 1 | 1 | 0 | 1 | 0 | PACSIN2 |
| MT | 1 | 0 | 0 | 0 | 0 | |
| X | 2 | 0 | 0 | 0 | 0 | EGFL6 |
| Totals | 53 | 66 | 0 | 14 | 13 |
Number of tags localized to the 5′ upstream region.
Number of tags localized to the intron.
Number of tags localized to the exon.
Number of tags localized to the 3′ region.
Not located near the annotated gene.
Previously known target genes represented by SABE tags.
Genes for which two tags were identified.
Demonstrated as p53-responsive in this study.
Fig. 3.
MLH1 and PMS2 identified as p53 target genes. (A) The proteins encoded by MLH1 and PMS2 share sequence identity within their N-terminal regions, representing exons 2-8 and exons 2-6, respectively. (B) Sequences of the MLH1 (Upper) and PMS2 (Lower) regions flanking the identified SABE tag (underlined in bold). Putative p53-response elements (p53RE) are boxed. (C) p53 binds to the first intron of MLH1 and PMS2 in vivo. Normal HS27 human fibroblasts were induced with cisplatin, and ChIP was performed by using anti-p53 antibodies and PCR with primers specific for MLH1 (Upper) and PMS2 (Lower). (D) p53 binds to the MLH1 p53RE in vitro. Labeled MLH1 p53RE oligonucleotide (Upper) was used as probe for EMSA with nuclear extracts prepared from cisplatin-treated HS27 cells. Competitor oligonucleotides were added at 50-fold molar excess as indicated (top, lanes 2-5), or antibodies against p53 (lane 6), USF1 (lane 7), or USF2 (lane 8). Arrows indicate specific p53-DNA complexes.
MLH1 and PMS2 Are Directly Regulated by p53. To confirm that p53 is capable of binding to the potential p53-response elements in MLH1 and PMS2, we performed conventional ChIP with normal human fibroblasts. Expression of p53 was induced by treatment with cisplatin, and crosslinked complexes were immunoprecipitated with specific antibodies. Under these conditions, we observed p53 bound to the first intron of both MLH1 and PMS2, demonstrating that p53 binds to these regions in vivo, but not to an internal control represented by the USF1 gene (Fig. 3C). We then examined whether p53 could bind to the putative p53 elements from MLH1 and PMS2 in vitro by using EMSA. We found that labeled p53RE probe from MLH1 formed several complexes with proteins present in nuclear extracts prepared from cisplatin-treated HS27 fibroblasts (Fig. 3D, lane 1). Several of the slower migrating complexes (indicated with arrows) could be eliminated by inclusion of excess WT unlabeled MLH1 p53RE competitor (lane 3) but not competitor containing a mutation of the p53 consensus sequence (lane 5). Additionally, formation of these complexes was inhibited by inclusion of specific antibodies against p53 (lane 6), demonstrating that p53 can bind to the MLH1 p53RE in vitro. The multiple complexes observed in EMSA with the p53RE may represent a monomer, dimer, and tetramer of p53 as observed previously (20). Binding of p53 complexes could not be competed with an oligonucleotide containing the putative PMS2 p53RE (Fig. 3D, lane 2), and we did not observe efficient binding to this oligonucleotide when used as probe in EMSA (data not shown). This finding suggests that the putative PMS2 p53RE is significantly weaker for binding of p53 in vitro.
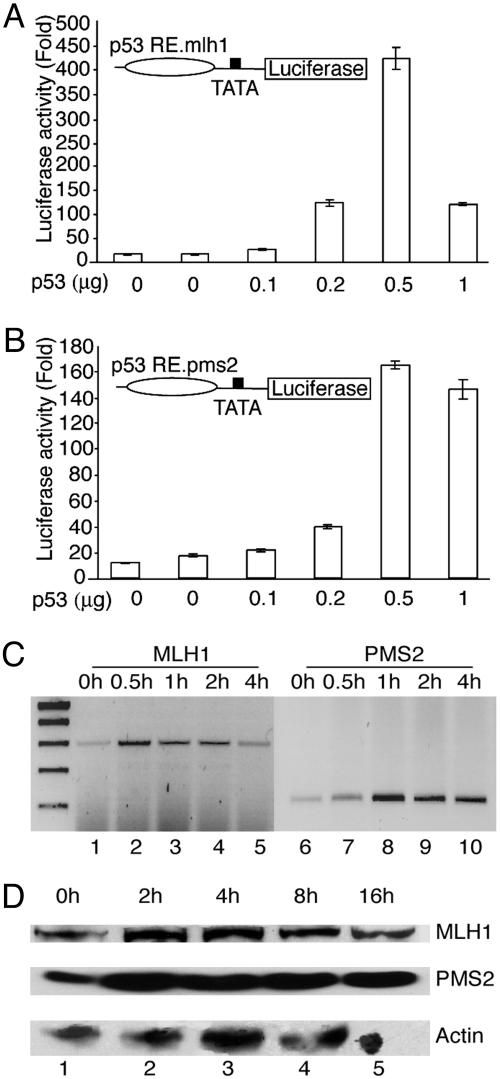
To determine whether p53 can activate transcription from the putative response elements in vivo, we constructed reporter genes bearing the elements upstream of a minimal TATA box promoter in the vector pTAL-luciferase. HeLa cells were cotransfected with the reporter genes and a p53 expression vector. In these experiments, we observed dose-dependent activation of both reporters in response to p53 expression (Fig. 4 A and B); however, the reporter bearing an upstream MLH1 p53RE was ≈4-fold more responsive to p53 than the PMS2 reporter. This finding is consistent with that shown above where the MLH1 p53RE binds p53 more efficiently in vitro than does PMS2.
Fig. 4.
MLH1 and PMS2 are targets for activation of p53 in vivo. (A and B) HeLa cells were cotransfected with 1 μg of MLH1 p53RE-Luc (A) or PMS2 p53RE-Luc (B) reporter genes and various amounts of p53 expression vector (indicated at the bottom). Luciferase activity was normalized by cotransfection of an internal control, pCMV-β-gal, and activity is represented proportional to the vector control. All determinations are from a minimum of three separate transfections. (C) MLH1 (lanes 1-5) and PMS2 (lanes 6-10) transcripts were measured by RT-PCR from unstimulated HS27 fibroblasts (lanes 1 and 6) or cells treated with cisplatin for the indicated time (top). The amplified products representing MLH1 and PMS2 transcripts were 303 and 223 bp, respectively. (D) Untreated HS27 fibroblasts (lane 1) or cells treated with cisplatin for the indicated times (top, lanes 2-5) were analyzed by immunoblotting with antibodies against MLH1 (Top), PMS2 (Middle), or actin (Bottom).
We also examined responsiveness of these genes to DNA damage in normal HS27 fibroblasts by using RT-PCR, and found that both transcripts were induced in response to cisplatin treatment (Fig. 4C). Additionally, the steady-state levels of both MLH1 and PMS2 protein were observed to be elevated within 2 h of treatment with cisplatin (Fig. 4D). These results indicate that MLH1 and PMS2 are DNA-damage-inducible genes in normal human fibroblasts, which is consistent with the presence of p53-response elements within their first intron.
p53 Binds Multiple Members of the PMS2 Gene Family. Human PMS2 is a homologue of the Saccharomyces cerevisiae PMS2 gene, and is one of 10 members of a highly homologous gene family (21) (see Table 2). The PMS2 gene encodes a 2.7-kb transcript consisting of 15 exons encompassing 16 kb of chromosome 7 (21). We designed primers to examine p53-specific ChIPs for potential interaction with these additional family members. DNA recovered by ChIP from Jurkat cells expressing p53-3XFLAG was amplified with primers recognizing all of the PMS2 family members (see Supporting Text) and cloned directly by using the TA cloning kit (Invitrogen). We analyzed 115 clones from this experiment to determine the relative proportion of each family member (Table 2). We observed that the majority of clones had sequences representative of PMS2L2a and PMS2L2b, which have identical sequences but are present at separate loci. A significant number of clones were also recovered that represent PMS2 and PMS2L5a and b, which also have identical sequences, but relatively few clones were isolated for the other family members (Table 2). These results suggest that of these family members, PMS2, PMS2L2, and PMS2L5, are likely the predominant targets for regulation by p53.
Table 2. Potential p53RE in PMS2 gene family members.
| Gene | Genome location (chromosome 7) | Location of putative p53RE | Clones from ChIP DNA, % |
|---|---|---|---|
| PMS2 | 57.89M | Intron 1 | 21 (18.3) |
| PMS2L2a, b | 74.54M, 74.56M | 5′ Upstream | 48 (41.7) |
| PMS2L5a, b | 71.92M, 73.72M | Intron 1 | 23 (20.0) |
| PMS2L9 | 74.75M | Intron 5 | 6 (5.2) |
| PMS2L4 | 66.17M | Intron 1 | 4 (3.5) |
| PMS2L5c | 99.54M | Intron 1 | 5 (4.3) |
| PMS2L11 | 76.25M | Intron 4 | 2 (1.7) |
| Pseudogene (no transcript) | 76.28M | Intron 1 | 6 (5.2) |
| Total | 115 (100) |
Discussion
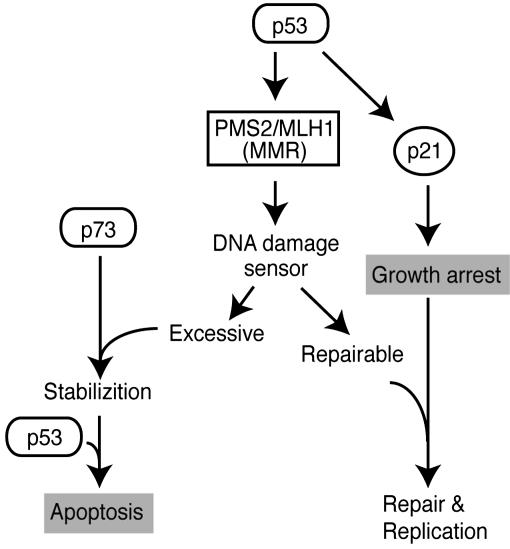
Activation of p53 in response to DNA damage induces either cell-cycle arrest, allowing the cell to repair DNA and recover before further replication, or initiation of programmed cell death (apoptosis), if the damage to DNA is excessive (16, 22). A key unresolved question is how a cell establishes the point at which DNA damage is excessive, and how this determination triggers a decision to undergo apoptosis rather than attempt DNA repair. MMR systems are conserved mechanisms which involve a group of proteins related in function to the bacterial MutS and MutL homologs, which recognize mismatched bases in double-stranded DNA, and initiate the repair process, respectively (23). The MLH1 protein forms a heterodimer with PMS2 and recruits additional enzymes necessary to correct mismatches generated by replication errors (24, 25). MMR proteins are also involved in activation of the cell-cycle checkpoint and induction of apoptosis when DNA damage overwhelms a critical threshold (reviewed in ref. 26). The cellular response to DNA damage requires activation of MLH1, which can cooperate with the tumor-suppressor p53 gene to promote cell-cycle arrest and cell death (27, 28). It has recently been shown that activated PMS2 stabilizes p73 (29, 30), a member of the p53 family required for p53-dependent apoptosis in response to DNA damage. These lines of evidence imply a strong relationship between MMR and the role of p53 in regulation of the cell-cycle arrest/apoptosis decision process. Our identification of MLH1 and PMS2 as direct targets for p53 defines a signaling pathway that couples two important cellular guardian pathways, growth arrest and apoptosis (Fig. 5). MMR may serve as a DNA-damage sensor and a critical determinant for the decision between cell-cycle arrest and apoptosis. Therefore, we propose that upon induction of DNA damage, activated p53 induces cell-cycle arrest through induction of p21/WAF, and the expression of MLH1 and PMS2 as part of the response to initiate repair. When damage exceeds the point at which repair is possible, MLH1 and PMS2 as sensors of the extent of DNA damage may then function to trigger apoptosis by stabilizing p73, which is required for p53-dependent apoptosis (Fig. 5).
Fig. 5.
A model for the role of MLH1 and PMS2 in the growth arrest-apoptosis decision. MLH1 and PMS2 proteins are induced by p53 in response to DNA damage, and may function as a sensor of the extent of damage. In cells where damage exceeds a critical threshold, PMS2 may stabilize p73 by direct interaction, thereby causing induction of apoptosis.
The results shown here also demonstrate the usefulness of SABE for identification of target genes for transcription factors on the human genome. Our approach is similar in concept to the genome-wide mapping technique (GMAT) as recently described by Rho et al. (31), which was used to localize hyperacetylated histone H3 protein on the S. cerevisiae genome. However, to achieve the specificity and sensitivity necessary for localization of a sequence-specific DNA-binding factor on the human genome, we combined a ChIP-SAGE strategy with representational difference subtractive hybridization to specifically amplify ChIP DNAs relative to nonspecific DNA that is typically present in ChIP samples. An additional significant difference between our strategy and GMAT is that SABE produces completely random 18-mer sequence tags that are not anchored by digestion of the template with a restriction endonuclease. We believe that this will enhance the resolution at which specific binding elements could be localized on genomic DNA by sequencing significant numbers of tags. In our analysis, we have found six putative p53 target genes in chromosomes 21 and 22, and two of them, RUNX1 and PACSIN2, are located near the binding sites identified by Cawley et al. (11), using a ChIP microarray strategy (11). Considering our small sample size, we believe this result is significant, particularly because results with different microarray technologies generally only overlap by ≈40% (32). Additionally, only a portion of the potential targets (20 of 48) detected by p53 full-length antibody could be detected using a different p53 antibody (11). However, in contrast to results produced by ChIP microarray analysis for p53, the vast majority of the unique SABE tags we identified are localized near predicted genes (133 of 146), with >80% of the tags localized to a 5′ upstream region or intron (Table 1). These results demonstrate that SABE will provide a broadly applicable means for the genome-wide location analysis of DNA-binding transcription factors in a variety of physiological, developmental, and disease states in human cells in vivo.
Supplementary Material
Acknowledgments
We thank Chris Nelson, David Mitchell, Martin Hirst, and Tom Malcolm for comments on the manuscript. This work was supported by funds from the Canadian Institute for Health Research and the National Cancer Institute of Canada with funds from the Canadian Cancer Society. J.C. is a Postdoctoral Fellow of the Natural Sciences and Engineering Research Council of Canada and Michael Smith Foundation for Health Research.
Author contributions: J.C. and I.S. designed research; J.C. performed research; J.C. and I.S. analyzed data; and J.C. and I.S. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: SABE, serial analysis of binding elements; IP, immunoprecipitation; ChIP, chromatin IP; SAGE, serial analysis of gene expression; MMR, mismatch repair; MLH1, mutL homolog 1; PMS2, yeast postmeiotic segregation increased 2.
References
- 1.Venter, J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J., Sutton, G. G., Smith, H. O., Yandell, M., Evans, C. A., Holt, R. A., et al. (2001) Science 291, 1304-1351. [DOI] [PubMed] [Google Scholar]
- 2.Ptashne, M. & Gann, A. (1997) Nature 386, 569-577. [DOI] [PubMed] [Google Scholar]
- 3.Hardison, R. C. (2000) Trends Genet. 16, 369-372. [DOI] [PubMed] [Google Scholar]
- 4.Lockhart, D. J. & Winzeler, E. A. (2000) Nature 405, 827-836. [DOI] [PubMed] [Google Scholar]
- 5.Heinemeyer, T., Wingender, E., Reuter, I., Hermjakob, H., Kel, A. E., Kel, O. V., Ignatieva, E. V., Ananko, E. A., Podkolodnaya, O. A., Kolpakov, F. A., et al. (1998) Nucleic Acids Res. 26, 362-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolffe, A. P. (2001) Essays Biochem. 37, 45-57. [DOI] [PubMed] [Google Scholar]
- 7.Iyer, V. R., Horak, C. E., Scafe, C. S., Botstein, D., Snyder, M. & Brown, P. O. (2001) Nature 409, 533-538. [DOI] [PubMed] [Google Scholar]
- 8.Ren, B., Robert, F., Wyrick, J. J., Aparicio, O., Jennings, E. G., Simon, I., Zeitlinger, J., Schreiber, J., Hannett, N., Kanin, E., et al. (2000) Science 290, 2306-2309. [DOI] [PubMed] [Google Scholar]
- 9.Ren, B., Cam, H., Takahashi, Y., Volkert, T., Terragni, J., Young, R. A. & Dynlacht, B. D. (2002) Genes Dev. 16, 245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weinmann, A. S., Yan, P. S., Oberley, M. J., Huang, T. H. & Farnham, P. J. (2002) Genes Dev. 16, 235-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cawley, S., Bekiranov, S., Ng, H. H., Kapranov, P., Sekinger, E. A., Kampa, D., Piccolboni, A., Sementchenko, V., Cheng, J., Williams, A. J., et al. (2004) Cell 116, 499-509. [DOI] [PubMed] [Google Scholar]
- 12.Solomon, M. J., Larsen, P. L. & Varshavsky, A. (1988) Cell 53, 937-947. [DOI] [PubMed] [Google Scholar]
- 13.Lisitsyn, N. & Wigler, M. (1993) Science 259, 946-951. [DOI] [PubMed] [Google Scholar]
- 14.Velculescu, V. E., Zhang, L., Vogelstein, B. & Kinzler, K. W. (1995) Science 270, 484-487. [DOI] [PubMed] [Google Scholar]
- 15.Levine, A. J. (1997) Cell 88, 323-331. [DOI] [PubMed] [Google Scholar]
- 16.Ryan, K. M., Phillips, A. C. & Vousden, K. H. (2001) Curr. Opin. Cell Biol. 13, 332-337. [DOI] [PubMed] [Google Scholar]
- 17.el-Deiry, W. S., Kern, S. E., Pietenpol, J. A., Kinzler, K. W. & Vogelstein, B. (1992) Nat. Genet. 1, 45-49. [DOI] [PubMed] [Google Scholar]
- 18.Saha, S., Sparks, A. B., Rago, C., Akmaev, V., Wang, C. J., Vogelstein, B., Kinzler, K. W. & Velculescu, V. E. (2002) Nat. Biotechnol. 20, 508-512. [DOI] [PubMed] [Google Scholar]
- 19.Hoh, J., Jin, S., Parrado, T., Edington, J., Levine, A. J. & Ott, J. (2002) Proc. Natl. Acad. Sci. USA 99, 8467-8472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McLure, K. G. & Lee, P. W. (1998) EMBO J. 17, 3342-3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicolaides, N. C., Carter, K. C., Shell, B. K., Papadopoulos, N., Vogelstein, B. & Kinzler, K. W. (1995) Genomics 30, 195-206. [DOI] [PubMed] [Google Scholar]
- 22.Vogelstein, B., Lane, D. & Levine, A. J. (2000) Nature 408, 307-310. [DOI] [PubMed] [Google Scholar]
- 23.Harfe, B. D. & Jinks-Robertson, S. (2000) Annu. Rev. Genet. 34, 359-399. [DOI] [PubMed] [Google Scholar]
- 24.Lipkin, S. M., Wang, V., Jacoby, R., Banerjee-Basu, S., Baxevanis, A. D., Lynch, H. T., Elliott, R. M. & Collins, F. S. (2000) Nat. Genet. 24, 27-35. [DOI] [PubMed] [Google Scholar]
- 25.Prolla, T. A., Pang, Q., Alani, E., Kolodner, R. D. & Liskay, R. M. (1994) Science 265, 1091-1093. [DOI] [PubMed] [Google Scholar]
- 26.Bellacosa, A. (2001) Cell Death Differ. 8, 1076-1092. [DOI] [PubMed] [Google Scholar]
- 27.Lakin, N. D. & Jackson, S. P. (1999) Oncogene 18, 7644-7655. [DOI] [PubMed] [Google Scholar]
- 28.Vikhanskaya, F., Colella, G., Valenti, M., Parodi, S., D'Incalci, M. & Broggini, M. (1999) Clin. Cancer Res. 5, 937-941. [PubMed] [Google Scholar]
- 29.Shimodaira, H., Yoshioka-Yamashita, A., Kolodner, R. D. & Wang, J. Y. (2003) Proc. Natl. Acad. Sci. USA 100, 2420-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flores, E. R., Tsai, K. Y., Crowley, D., Sengupta, S., Yang, A., McKeon, F. & Jacks, T. (2002) Nature 416, 560-564. [DOI] [PubMed] [Google Scholar]
- 31.Roh, T. Y., Ngau, W. C., Cui, K., Landsman, D. & Zhao, K. (2004) Nat. Biotechnol. 22, 1013-1016. [DOI] [PubMed] [Google Scholar]
- 32.Kuo, W. P., Jenssen, T. K., Butte, A. J., Ohno-Machado, L. & Kohane, I. S. (2002) Bioinformatics 18, 405-412. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.