Abstract
Experimental efforts to understand how the brain represents, stores and processes information require high-fidelity recordings of multiple different forms of neural activity within functional circuits. Thus, creating improved technologies for large-scale recordings of neural activity in the live brain is a crucial goal in neuroscience. Over the past two decades, the combination of optical microscopy and genetically encoded fluorescent indicators has become a widespread means of recording neural activity in nonmammalian and mammalian nervous systems, transforming brain research in the process. In this review, we describe and assess different classes of fluorescent protein indicators of neural activity. We first discuss general considerations in optical imaging and then present salient characteristics of representative indicators. Our focus is on how indicator characteristics relate to their use in living animals and on likely areas of future progress.
A central goal of neuroscience is to understand how nervous systems encode and process information at circuit and cellular levels. The development of genetically encoded optical indicators of neuronal activity has enabled progress toward this goal to an extent that was unimagined two decades ago. Visualization of neuronal activity using calcium indicators has become a standard research approach in animal models, and interest in other types of activity sensors is increasing. However, the recent proliferation of different indicator types and variations can make it difficult for newcomers to the field to determine which ones best suit their needs. Previous reviews have focused on one category of indicator1–3 or instrumentation4, but have not attempted to consider all optical indicators under a common framework. Here we discuss multiple classes of genetically encoded activity indicators using common criteria and explain shared principles underlying their operation. We hope the reader will gain a mechanistic basis for understanding and evaluating activity indicators that will be useful in interpreting the literature and designing new experiments.
Advantages of genetically encoded activity indicators
The popularity of genetically encoded optical indicators is due to a set of compelling advantages that derive from either genetic encoding or optical reporting, or both. Genetic encoding enables reporters to be constructed from proteins that respond to a variety of neuronal events. Optical reporters have been constructed for vesicle release, changes in neurotransmitter concentrations, transmembrane voltage, and intracellular calcium dynamics, providing information that electrodes and functional magnetic resonance imaging cannot. Genetic encoding also allows selective sampling of neuronal subsets, including genetically defined neuronal subtypes; sparse random subsets of cells; or cells with specific patterns of anatomical connectivity, for example, via axonal or trans-synaptic labeling5,6. Finally, genetically encoded sensors can be stably expressed to study how neuronal dynamics evolve over time in individual animals during the course of learning, life experience, brain development, or disease progression7–9.
Other important advantages stem from the virtues of optical imaging. Optical imaging enables thousands of neurons to be simultaneously observed in vivo9–14, more than currently feasible with electrodes, and thus enables sophisticated analyses of ensemble neural activity9,10,15,16. Imaging can be less biased than electrodes, as some neuron types resist extracellular electrical recordings owing to unfavorable cell morphology, weak electrical dipoles, or the organization of the extracellular tissue17. Because of sampling bias for active cells, extracellular recordings typically overestimate rates of action potentials (APs), or spikes17, while estimates of spiking rates from optical reporters align more closely with those from intracellular recordings. Voltage and calcium indicators can reveal spatiotemporal activity patterns within neurons such as dendritic integration, voltage propagation5,18, or dendritic spiking19. A final advantage is minimization of neuronal damage. While microscopy requires optical access to the tissue of interest, there are many animal preparations that allow minimally invasive optical access or placement of optical probes up to millimeters away from the imaged cells. By comparison, electrodes must be located within 50 µm of the cell under study20. In mammals, methods such as thinned-skull, cranial-window, and microendoscope preparations allow the immediate vicinity of cells under study to be left unperturbed21,22, while certain model organisms such as zebrafish are translucent and can be imaged intact. This avoidance of local perturbation also aids long-term imaging.
Considerations when using genetically encoded indicators
Nevertheless, the proper use of genetically encoded optical indicators requires addressing certain technical issues. These relate to obtaining adequate and specific indicator expression and setting appropriate imaging parameters based on the expected signals. First, for long-term expression, indicators are expressed by viral infection or transgenesis, both of which require empirical optimization. There are many viral classes and serotypes, which must be tested for transduction efficiency and toxicity in the cells of interest. The alternative method of in utero electroporation can have unpredictable yields. Both viruses and in utero electroporation impose limitations on construct size, restricting the selection of promoters that can be used. Transgenic animals may not express indicators at high enough levels for the desired usage or can show expression in undesired cell types. Proper cell-type-restricted expression may require the crossing of two or three lines of transgenic animals, which can be time-consuming and costly.
Second, even under ideal conditions, optical imaging is limited by the inherent quantum mechanical randomness of photon emission and detection. This limits both the fidelity of detecting neural events and the accuracy of estimating event timing4,23. To characterize detection fidelity of single APs, the metric d′ from signal detection theory is useful23. The calculation of d′ takes into account several factors that influence event detection under practical situations, including indicator brightness, response amplitude and kinetics, as well as background fluorescence and fluctuations. Thus, d′ better predicts spike detection fidelity than older metrics such as fractional fluorescence change (ΔF/F) or signal-to-noise ratio, which omit consideration of indicator brightness, kinetics, and/or response waveforms. For example, indicator A could have maximum brightness lower than indicator B, but if it has disproportionately lower minimum brightness, it would have higher ΔF/F. Yet indicator B would be more useful for event detection in most cases. In the extreme example, an indicator could be engineered with infinite ΔF/F by having a completely dark baseline without improving induced brightness. This thought experiment makes apparent that brightness, not just fractional change, is useful to consider.
Calculating d′ values for specific indicators, labeling conditions, and optical instrumentation allows researchers to assess the feasibility of different experimental configurations and the statistical confidence of the resulting activity traces23. This is especially important when comparing the signaling fidelities attained with different microscopy modalities. For instance, laser-scanning modalities such as two-photon or confocal microscopy typically sample individual pixels for only ~0.1–2 µs per image frame but provide optical sectioning, which greatly reduces the impact of background fluorescence. By comparison, even with 100-Hz acquisition frame rates, wide-field epifluorescence samples individual pixels for 10 ms. However, wide-field imaging lacks optical sectioning and hence in densely labeled tissue background fluorescence can sharply lower image contrast. Notwithstanding the different appearances of images attained by these different microscopy modalities, signal detection analysis reveals that in many cases of practical interest the d′ values for spike detection using calcium imaging are often about the same, whereas naive analyses appealing to ΔF/F values can lead to incorrect conclusions4,23.
Lastly, an important consideration with genetically encoded indicators is that recorded signals are influenced by the indicator’s response characteristics. An indicator’s key attributes include the fluorescence response curve and response kinetics. As events measured with genetically encoded indicators can play out over time intervals from milliseconds to minutes, it is important to select an indicator with sufficient responsiveness and speed to report the underlying neural events at an acceptable d′ value and with suitable temporal resolution for the question at hand. An important related consideration is whether the experiment requires sensitive event detection (for example, of APs), precise event timing (for example, to address questions about synchronous neural firing), accurate reporting of temporal waveforms (such as fluctuations in membrane voltage or intracellular calcium), or combinations thereof. Another consideration is that different illumination and acquisition parameters influence achievable imaging durations, which are limited by photobleaching of optical probes. For example, calcium imaging can typically be performed over ~1 h sessions and repeated daily9. Voltage imaging sessions might last ~1 h for studies of aggregate neural activity before photobleaching reaches 50%, but only ~15 min for studies of single-cell activity24,25, owing to the faster frame rates and correspondingly higher illumination intensities needed.
INDICATORS
Recent reviews have covered strategies for indicator expression17,26, instrumentation for in vivo imaging4,26, and analysis of cell activity traces4. Hence we will focus instead on comparisons of the primary classes of indicators. For each class, we will discuss the general mechanisms of sensing, compare performance characteristics of specific sensors, and relate these to practical applications. We will discuss indicators following the natural physiological order of synaptic transmission and excitation: vesicle fusion, neurotransmitter release, voltage change, and calcium entry.
Vesicular release indicators
Synaptic vesicle release is a fundamental step in interneuronal signaling that is regulated by neuronal activity and regulatory processes27. Visualization of vesicle release also potentially offers a way of tracking the activity of specific neuronal outputs independent of postsynaptic processing.
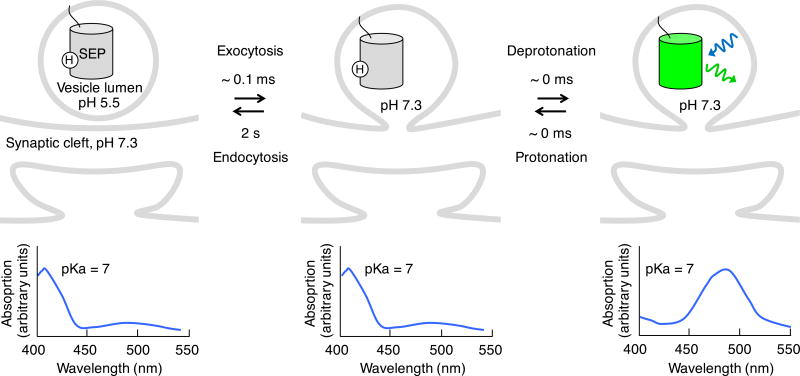
Synaptic vesicle release can be detected using vesicularly localized genetically encoded pH indicators (which we will call GEPIs). The pH of the vesicle lumen is typically ~5.5, whereas that of the extracellular environment is 7.0–7.5 (Fig. 1)2. Thus the contents of synaptic vesicles experience a change in pH from 5.5 to 7.0–7.5 upon membrane fusion. GEPIs are fluorescent proteins that demonstrate pH-dependent brightness at a specific wavelength. They were first developed from the naturally pH-sensitive GFP. In the wild-type GFP chromophore, a phenolic oxygen exists in a pH-dependent equilibrium between protonated and deprotonated states, which absorb maximally near 400 and 488 nm respectively28. A GEPI named ecliptic pHluorin was evolved from wild-type GFP to lack 488-nm excitability at pH 5.5 (hence becoming dark, or “ecliptic,” at low pH) while becoming fluorescent at higher pH values, with a pKa of 7.1 (refs. 29,30). However, ecliptic pHluorin is still mostly protonated at neutral pH, like wild-type GFP29. Addition of mutations from EGFP, which is completely deprotonated at neutral pH22, produced superecliptic pHluorin (SEP), with more complete deprotonation at neutral pH30,31 (Table 1).
Figure 1.
Genetically encoded pH indicators (GEPIs). Superecliptic pHluorin (SEP) is shown as an example. pH-dependent fluorescent proteins in the low pH of synaptic vesicles have a protonated chromophore (above) and absorb primarily at ~400 nm (below). Fusion of the synaptic vesicle induces near-instantaneous loss of the proton from the chromophore, shifting its absorbance peak to ~490 nm and allowing excitation by 488-nm light (blue sinusoidal arrow), with resulting green emission (green sinusoidal arrow). Times shown are half-rise and half-decay times. Half-reuptake time is from ref. 35.
Table 1.
Selected genetically encoded pH indicators (GEPIs)
| GEPI | ΔF/F, pH 5.5 to pH 7.4 |
Brightness at pH 5.5 (mM−1 cm−1)a |
Brightness at pH 7.4 (mM−1 cm−1)b |
pKa | Refs. |
|---|---|---|---|---|---|
| EGFP | 5.0 | 6.4 | 32 | 6.0 | 30,124 |
| Ecliptic pHluorin (EP) | 50 | 0.070 | 3.5c | 7.1 | 29,30 |
| Superecliptic pHluorin (SEP) | 50 | 0.44 | 22d | 7.2 | 30 |
| pHTomato | 2.0 | 24 | 48 | 7.8 | 31,125 |
| td-mOrange2 | 3.5 | 10 | 35 | 6.5 | 39,126,127 |
| pHoran4 | 17 | 3.2 | 55 | 7.5 | 31 |
| pHuji | 20 | 0.34 | 6.8 | 7.7 | 31 |
Brightness at pH 5.5 was calculated from brightness at pH 7.4 (below) and reported ΔF/F.
Brightness at pH 7.4 was directly measured as product of peak extinction coefficient and quantum yield of mature protein measured at pH 7.4, unless otherwise indicated.
Brightness at pH 7.4 was estimated from relative brightness of EP upon 488-nm excitation at pH 7.4 vs. maximal observed brightness (at pH 8.5), with maximal brightness assumed to be similar to that of the parental wild-type GFP, which has a similar excitation spectrum shape. Brightness of wild-type GFP at 488 nm was derived from the published absorbance spectrum, extinction coefficient, and quantum yield124. Brightness of EP in cells relative to EGFP and SEP may be disproportionally low owing to absence of the F64L folding mutation.
Brightness at pH 7.4 was estimated from relative brightness of SEP upon 488-nm excitation at pH 7.4 vs. maximal observed brightness (at pH 9.5), with maximal brightness assumed to be similar to that of the parental EGFP, which has a similar excitation spectrum shape. Published measurements were used for the brightness of EGFP at 488 nm (ref. 124). Values were rounded to two significant digits.
SEP fused to the luminal side of the vesicular transmembrane proteins VAMP, synaptophysin, or VGLUT26 have been used to visualize vesicle fusion. Calculations confirm their ability to detect single-AP events in vitro. Each synaptic vesicle may contain up to ~10 GEPI molecules32, and an AP at a hippocampal or cortical synapse evokes the release of less than one vesicle on average33. The maximum number of GEPI molecules exocytosed following a single AP is thus ~10. With indicator pKa values near neutral pH, ~5 of these ~10 molecules will be excitable. To detect these molecules, their signal must be discernible above autofluorescence. Cellular autofluorescence elicited by ~480-nm excitation is equivalent in brightness to 1,800 GFP molecules per µm2 of membrane34, corresponding to 110 or 290 molecules in typical one- and two-photon diffraction-limited spots of 0.06 µm2 or 0.16 um2, respectively. Thus the ~5 GEPI molecules responding in a single AP would produce a ΔF/F of 4.5% or 1.7%, respectively, over autofluorescence in a diffraction-limited spot. In cell cultures, single fusion events have approached the predicted 4.5% ΔF/F by one-photon microscopy, with detection aided by synchronization with stimuli and by total internal reflectance microscopy to eliminate out-of-focus background35. However, in vivo, increased autofluorescence and scattering will reduce the obtainable ΔF/F while decreasing signal photons acquired, adversely affecting d′ values23. Thus GEPIs are not likely to be useful for imaging single APs in vivo.
However, SEP can visualize responses integrated over large numbers of synapses or APs. SEP was the first genetically encoded sensor used in vivo to visualize population-level neuronal activity, detecting odorant responses in fly olfactory glomeruli36. It then became the first indicator to allow a similar feat in mammals, visualizing odorant representations in mouse olfactory glomeruli. Here observed ΔF/F were as high as 100% because there were multiple SEP-labeled synapses within each imaged pixel and multiple excitation events per time interval. SEP also detected evoked release at the mouse neuromuscular junction in response to AP trains37. In these experiments, relatively long exposures of 100–1,000 ms were used. As vesicular proteins have long residence times after vesicle fusion (~1 s), GEPIs effectively integrate multiple release events in these time intervals37,38.
Orange-red GEPIs may eventually help improve synaptic vesicle activity detection in vivo, as autofluorescence and scattering are reduced at longer wavelengths. The orange-red fluorescent proteins pHTomato, pHoran4, and pHuji were engineered to have pKa values near 7.5, like SEP (Table 1). However, their relative fluorescence at pH 5.5 is still higher than that of SEP owing to lower Hill coefficients31. In cultured neurons, pHuji reported some fusion events by brightening, but half of all vesicles showed high baseline fluorescence for unknown reasons39. Thus further improvement of orange-red GEPIs is still needed.
Neurotransmitter indicators
GEPIs report vesicle release, but GEPIs are not necessarily specific for the neurotransmitter content of those vesicles. There are dozens of neurotransmitters and modulators in the nervous system, and they have a range of effects on postsynaptic neuronal activity. Thus, imaging the release of specific neurotransmitters is an important step in dissecting neuronal circuitry.
Immediately upon vesicular fusion, neurotransmitters are released into the synaptic cleft. Here they can reach concentrations in the high micromolar to low millimolar range before being removed by neurotransmitter transporters with rapid kinetics (often <1 ms)40,41. The high concentrations and fast kinetics of neurotransmitters make them attractive targets for genetically encoded sensing. Further, unlike with vesicular exocytosis, voltage, or calcium, no small molecule fluorescent indicators exist for neurotransmitters42. In principle, genetically encoded transmitter indicators (GETIs) can be expressed on either pre- or postsynaptic cells, allowing visualization of neurotransmission from specific presynaptic or to specific postsynaptic cell types42.
The first GETI to visualize glutamate, the primary excitatory neurotransmitter in the mammalian CNS, was FLIPE, comprising CFP and YFP fused to the glutamate-binding domain of the bacterial glutamate transporter GltI. FLIPE shows ~8% emission ratio changes to 10 µM glutamate when expressed on hippocampal neurons43 (due to ~4% increases in CFP and ~4% decreases in YFP; Table 2). Purified FLIPE perfused in cortical slices detects evoked glutamate release over large regions, despite not being localized to neuronal surfaces44. An indicator of a similar design with optimized linkers, super glutamate-sensing fluorescent reporter (SuperGluSnFR), detects glutamate with up to 44% ratio changes and responds to single APs with 2% ratio changes when expressed on cultured hippocampal neurons (Table 2).
Table 2.
Selected genetically encoded transmitter indicators (GETIs)
| GETI | Maximum ΔF/F in vitro/ on neuronsa |
Glu-free brightness in vitro (mM−1 cm−1)b |
Glu-bound brightness in vitro (mM−1 cm−1)b |
Kd in vitro/ on neurons (µM) |
kon (M−1 s−1) |
koff (s−1) |
Refs. |
|---|---|---|---|---|---|---|---|
| FLIPE | +0.1/+0.04 (ECFP) | 7.8c | 8.0 | 0.6/ND | 100 × 106 | 60 | 43 |
| −0.1/−0.04 (Venus) | 4.0 | 3.7 | |||||
| SuperGluSnFR | +0.14/0.19 (ECFP) | 9.4d | 9.6 | 2.5/2.5 | 30 × 106 | 75 | 128 |
| −0.13/−0.17 (Citrine) | 1.9 | 1.5 | |||||
|
| |||||||
| iGluSnFR | +4.5/+1.0 | 5.3d | 29d | 110/4.9 | ND | ND | 46 |
Fluorescence change from zero to saturating glutamate in vitro at 25 °C at the emission peak of each channel. This number is empirically measured and, for the acceptor fluorophore in FRET sensors, is influenced by cross-excitation and bleed-through. As event detection is often optimized by single-channel imaging of FRET indicators23,83, the two channels are shown separately.
Estimated molar brightness produced by each fluorophore. For FRET sensors, glutamate-free/saturated brightness for the donor channel is calculated as the product of donor peak extinction coefficient and donor quantum yield multiplied by 1 – E, where E is glutamate-free/saturated FRET efficiency. Glutamate-free/saturated brightness for the acceptor channel is calculated as the product of donor peak extinction coefficient and acceptor quantum yield multiplied by E. Specific values for these parameters are noted below. These numbers do not account for cross-excitation and bleed-through of the other channel, but provide an estimate of the contribution of each fluorophore to indicator brightness.
Peak ECFP extinction coefficient is 28 mM−1 cm−1 at 433 nm, ECFP quantum yield is 0.37 (ref. 129), Venus quantum yield is 0.57 (ref. 130), and glutamate-free/saturated E of 0.25/0.23 is derived from published emission spectra in HBSS. dCitrine quantum yield is 0.76 (ref. 130), and glutamate-free/saturated E of 0.09/0.07 is derived from published emission spectra.
Brightness in the glutamate-bound state at pH 7.4 was first estimated from published data showing 90% of maximal brightness at pH 7.4, assuming maximal brightness is similar to that of EGFP. Glutamate-free brightness in vitro was then obtained by dividing by ΔF/F + 1. NA, not applicable. ND, not determined. Measurements were performed at room temperature. A maximum of two significant digits are used; some values measured from published graphs are less precise.
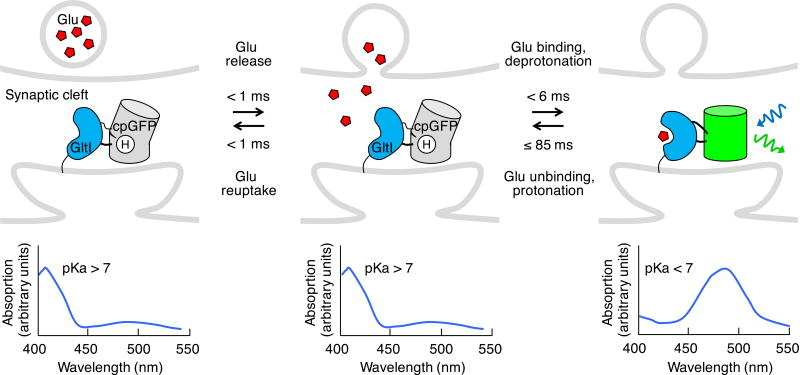
Subsequently, the more responsive GETI iGluSnFR was engineered using a circularly permuted GFP (cpGFP) and GltI (ref. 46). In circular permutation, a protein is recoded so that new N and C termini are located at previously adjacent internal positions and the original termini are connected by a linker. The concept of conformational modulation of fluorescence of a cpGFP was first introduced in the calcium indicator Camgaroo45 (discussed below) using GFP circularly permuted at amino acids 144 and 145 (cpGFP145), near the chromophore phenolic oxygen. iGluSnFR was obtained by screening Glt1 with cpGFP145 inserted at various locations for glutamate-responsive fluorescence. iGluSnFR brightened upon glutamate binding when excited at 488 nm, with a response in neuronal cultures of ~100% ΔF/F upon glutamate increase from 0 to 10 µM (Table 2). As glutamate shifts the excitation curve to increase excitability at 488 nm (ref. 46), the mechanism of iGluSnFR has some similarity to that of SEP, although with iGluSnFR it is chromophore pKa rather than environmental pH that changes (Fig. 2). As discussed later, modulation of the pKa of a cpGFP will be a common mechanism in many activity indicators.
Figure 2.
Genetically encoded transmitter indicators (GETIs). iGluSnFR reports glutamate with increased fluorescence (above). A glutamate-induced conformational change in the glutamate-binding domain from a bacterial glutamate transporter (Glt1) induces loss of the proton from the chromophore, shifting its absorbance peak to ~490 nm (below) and allowing excitation by 488-nm light (blue sinusoidal arrow), with resulting green emission (green sinusoidal arrow). Binding time was measured in vitro for an increase in glutamate concentration from 0 to 4.6 µM (ref. 46). The indicated unbinding time is an upper limit deduced from live cell experiments.
The sensitivity of iGluSnFR for synaptic activity surpasses that of GEPIs. iGluSnFR responds to single APs with mean fluorescence changes (averaged over entire fields of view) of 10% in cultured neurons46. The sensor’s baseline brightness is far brighter than that of SEP, and thus the 10% increase represents a larger number of signal photons. In the signal detection framework, a given response yields a larger d′ value when initiated from a brighter baseline with smaller fractional fluctuations due to shot noise. In addition, specific regions in the image field, namely synaptic regions, should show a much larger fluorescence change, as glutamate concentrations will be highest in synaptic clefts. The expected peak glutamate concentration of >0.1 mM following a single release in the synaptic cleft would be expected to saturate the sensor population there, yielding a 100% response. Comparisons in cell culture indeed showed iGluSnFR to be more sensitive than SEP in detecting synaptic release47.
The sensitivity of iGluSnFR has been validated in living mice. iGluSnFR successfully detected activity-dependent activation of a single dendritic bouton with 50% fluorescence rises, corresponding to an average glutamate concentration at the Kd of the sensor, 4.9 µM (ref. 46). An initial rise of 20% in fluorescence occurred within 2 ms. As 100% responses are expected within the synaptic cleft, the kinetics and magnitude of the initial part of the response, averaged across one surface of the dendritic spine, would be consistent with detection of glutamate from a single vesicle. In this case, the total response increased over the next 20 ms and lasted for 100 ms, suggesting the likelihood of additional rounds of neurotransmitter release, and so single-vesicle detection remains to be unambiguously demonstrated. iGluSnFR has also been used to produce a low-resolution map of activity in the entire mouse brain48, similarly to how SEP was used in the olfactory bulb. In the future, tethering of iGluSnFR to a synaptic molecule could confine expression to synaptic clefts and further enhance detected responses.
In addition to glutamate, GETIs for other neurotransmitters would clearly be useful. The existence of bacterial binding proteins for acetylcholine, GABA, and glycine49 suggests that indicators can be engineered for these neurotransmitters.
Voltage indicators
While GETIs report the release of neurotransmitters, the primary effect of neurotransmitters is to change the membrane potential in postsynaptic targets. To detect this subsequent type of neural activity, indicators of transmembrane voltage are needed. Transmembrane voltage is also the primary means of millisecond-scale computation within individual neurons and of rapid transmission of signals across long distances. Voltage changes in neurons display a rich repertoire of behaviors across a variety of time scales. Transmembrane voltage at any given time is influenced by depolarizing and hyperpolarizing synaptic inputs and by internal biochemical states50, and it can demonstrate coordinated oscillatory activity in neuronal populations. Synaptic activity can lead to transient depolarizations without a spike, or it can lead to voltage-gated channel activation and spike generation, localized within dendrites, to the soma and axon, or throughout the cell51. Axonal spikes can occur singly or in trains, and spike bursts can occur with frequencies above 100 Hz (ref. 52). Voltage indicators that can visualize these various types of events, in single cells and in populations, have long been desired. Organic voltage-sensitive dyes typically have fast kinetics, but they are often highly phototoxic, allow neither genetically targeted delivery nor long-term imaging studies of single cells, and have been incapable of reporting single spikes in the live mammalian brain53.
Of all activity indicator types, voltage indicators show the largest variety of designs and mechanisms3,54 (Fig. 3), perhaps because no single voltage indicator design has yet met all performance requirements satisfactorily. The first genetically encoded voltage indicator (GEVI), Flash55, was described in 1997 within a few months of the first genetically encoded calcium indicator (GECI), Cameleon56. Flash comprised a single GFP domain inserted into the Shaker potassium channel. However, Flash, as well as subsequent GEVIs comprising a GFP inserted into a sodium channel and a fluorescence resonance energy transfer (FRET) pair attached to the four-helix transmembrane voltage-sensing domain (VSD) of Shaker, failed to express at the membranes of mammalian cells57. GEVIs that function well in mammalian neurons were finally developed starting with the VSFP2 family in 2003, using the VSD of a voltage-sensing phosphatase58.
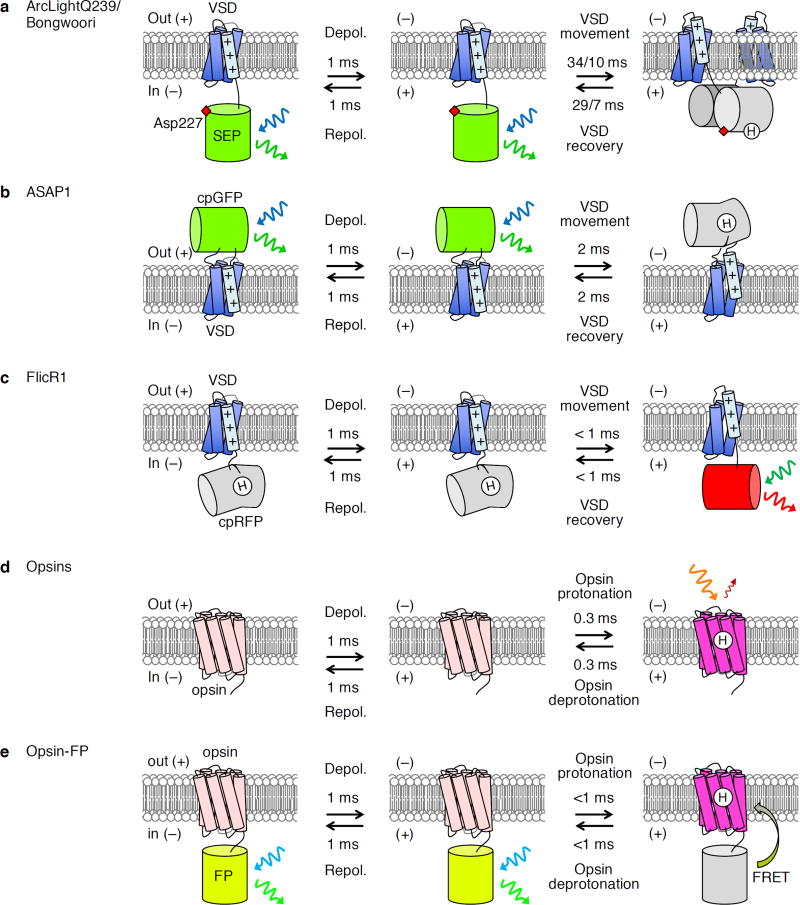
Figure 3.
Genetically encoded voltage indicators (GEVIs). (a) ArcLight-family GEVIs respond to depolarization by reduced green fluorescence (rightward green sinusoidal arrow) from superecliptic pHluorin (SEP) upon blue-light excitation (blue sinusoidal arrow). The mechanism is not fully known but is believed to involve voltage-dependent dimerization leading to protonation of the SEP chromophore. Kinetics are shown for ArcLightQ239 and Bongwoori as measured at 33 °C (ref. 123), with the slash (/) separating ArcLightQ239 and Bongwoori values. Among the ArcLight variants, these have the largest amplitude and fastest signaling kinetics, respectively. (b) GEVIs of the ASAP family also report depolarization by dimming of a circularly permuted GFP (cpGFP). The mechanism presumably involves coupling of VSD movement to chromophore protonation, similar to that in iGluSnFR and single-fluorophore GECIs. Kinetics were measured at 22 °C (ref. 25). (c) FlicR reports depolarization with increased red fluorescence (red sinusoidal arrow) from a circularly permuted red fluorescent protein (cpRFP) upon green excitation (leftward green sinusoidal arrow), presumably as a result of chromophore deprotonation. Kinetics shown were measured at 37 °C (ref. 59). (d) Opsins report depolarization with increased red fluorescence (red sinusoidal arrow) upon excitation by ~600-nm light (orange sinusoidal arrow), but this emission is weak (quantum yield < 0.01). Kinetics are shown for QuasAr2 measured at 34 °C (ref. 65). (e) Opsin–fluorescent protein fusions report depolarization with a dimming of fluorescence, due to absorbance shift in the opsin leading to increased FRET. In the case of Ace2N-mNeonGreen, emission is yellow-green (yellow-green sinusoidal arrow) and excitation is cyan (cyan sinusoidal arrow). Kinetics are shown for Ace2N-mNeonGreen measured at 22 °C (ref. 5).
Fluorescent-protein-based GEVIs with diverse architectures and response characteristics have since been developed, including the Butterfly family, the VSFP3 family, ElectricPk, the ArcLight family, the accelerated sensor of action potentials (ASAP) family, and fluorescent indicator for voltage imaging red 1 (FlicR1) (ref. 3). Except in FlicR1, VSD movement upon depolarization induces dimming of a fused fluorescent protein that, exception in ASAPs, is attached at the C terminus of the VSD (Table 3). VSFP2- and Butterfly-family GEVIs also contain a second fluorescent protein to serve as a FRET acceptor, and FRET changes occur on top of direct modulation of the fluorescent protein attached to the VSD. FlicR1 has a single fluorescent protein domain attached to the VSD C terminus and, unlike other GEVIs, brightens upon depolarization59. In the case of ElectricPk, ASAPs, and FlicR1, the fluorescent protein is circularly permuted, and voltage-responsiveness likely derives from conformational changes in the sensing domain affecting chromophore protonation, as in iGluSnFR (ref. 54). It is less well understood how voltage affects fluorescence in other GEVIs, although inducible fluorescent protein dimerization has been proposed for ArcLight60.
Table 3.
Selected genetically encoded voltage indicators (GEVIs)
| GEVI | ΔF/F −70 to +30 mVa |
Brightness at −70 mV (mM−1 cm−1)b |
Brightness at +30 mV (mM−1 cm−1)b |
Activationc τfast (ms), fast fraction, τslow (ms) |
Inactivationd τfast (ms), fast fraction, τslow (ms) |
ΔF/F per APe |
Refs. |
|---|---|---|---|---|---|---|---|
| VSFP2.3 | −0.06 (Cerulean) | 8.6f | ND | 3.0, 27%, 69 | 92, 100% | −0.005 | 131−133 |
| +0.03 (Citrine) | 10f | ND | +0.003 | ||||
| Butterfly1.2 | −0.05* (Citrine) | 36g | ND | 1.0, 41%, 12* | 90, 100%* | −0.007e* | 24 |
| +0.03* (mKate2) | 12g | ND | +0.003e* | ||||
| Mermaid2 | −0.24* (seCFP2) | NDh | ND | 0.92, 79%, 13* | 10, 100%* | −0.066e* | 69 |
| +0.12* (YFP) | NDh | ND | +0.033e* | ||||
|
| |||||||
| ArcLight Q239 | −0.35* | 22i | 14 | 9.0, 50%, 48* | 17, 79%, 60* | −0.022 | 134 |
| −0.32 | 22 | 15 | 104, 61%, 283 | 28, 39%, 271 | −0.082 | 68 | |
| ASAP1 | −0.18 | 4.2j | 3.4 | 2.0, 60%, 72 | 2.0, 44%, 51 | −0.048 | 18,25 |
| −0.29 | 4.2 | 3.0 | 3.0, 67%, 29 | 2.3, 88%, 39 | −0.178 | 68 | |
| ASAP2f | −0.22 | 3.2i | 2.5 | 2.8, 81%, 135 | 2.4, 71%, 155 | −0.090 | 18 |
| Archer1 | +0.80 | 0.028k | 0.050 | ~3, ND, ND | ~5, ND, ND | +0.3 | 63 |
| Mac-Citrine | −0.20 | ≤59l | ≤22 | 2.8, 71%, 74 | 5.4, 77%, 67 | −0.048 | 67 |
| Ace2N-mNeon | −0.18 | ≤90l | ≤74 | 0.36, 74%, 4.2 | 0.42, 64%, 5.2 | −0.12 | 5 |
| Ace2N-4aa-mNeon | −0.09 | ≤90l | ≤82 | 0.37, 58%, 5.5 | 0.50, 60%, 5.9 | −0.050 | 5 |
GEVIs with reported single-AP responses in neuronal culture and usage in live animals are included.
Fluorescence change from −70 to +30 mV in long-lasting voltage steps in HEK293 cells at the emission peak of each channel. This number is empirically measured and, for the acceptor fluorophore in FRET sensors, is influenced by cross-excitation and bleed-through. As event detection is often optimized by single-channel imaging of FRET indicators23,83, the two channels are shown separately.
Estimated molar brightness produced by each fluorophore. For FRET sensors, brightness for the donor channel at −70 mV is calculated as the product of donor peak extinction coefficient and donor quantum yield multiplied by 1 − E, where E is FRET efficiency at −70 mV. Brightness for the acceptor channel at −70 mV is calculated as the product of donor peak extinction coefficient and acceptor quantum yield multiplied by E. Values for specific parameters are noted below. Values do not account for any cross-excitation or bleed-through, but provide an estimate of the contribution of each fluorophore to indicator brightness. Brightness values for FRET indicators at +30 mV are not shown, as emission spectra at +30 mV have not been published. However, as the measured ΔF/F in each channel is small, brightness will not change much at +30 mV.
During steps from −70 to +30 mV.
During steps from +30 to −70 mV.
In mouse or rat cortical or hippocampal neurons.
Peak Cerulean extinction coefficient is 28 mM−1 cm−1 at 433 nm, Cerulean quantum yield is 0.51 (ref. 135), Citrine quantum yield is 0.76 (ref. 130), and E at −70 mV is 0.40 as derived from the published emission spectrum as previously described131,132.
Citrine extinction coefficient is 77 mM−1 cm−1 (ref. 130), mKate2 quantum yield is 0.40 (ref. 136), and E at −70 mV is 0.39 as derived from the published emission spectrum.
Not determined, as relevant emission spectra were not published.
Based on ArcLight reaching maximal fluorescence at −70 mV with the brightness of SEP from Table 1.
Calculated from the relative brightness of the indicator at −70 mV vs. a similar construct with sfGFP instead of cpsfGFP, and the measured brightness of sfGFP.
Calculated as 5 × the previously measured brightness of wild-type Archaerhodopsin62.
FRET efficiency at −70 mV is unknown, so brightness is indicated as less than or equal to the brightness of the fluorescent protein alone. Measurements were performed at room temperature unless otherwise indicated. Up to two significant digits are used; some values measured from published graphs are less precise.
Measurements were performed at 33–35 °C. ND, not determined.
Opsins represent another class of voltage-sensing protein domains. Nonconducting mutants of opsin-family pumps or channels contain as a chromophore a Schiff base of retinal in a voltage-sensitive protonation–deprotonation equilibrium. Depolarization shifts the equilibrium from deprotonation toward protonation and the main absorbance peak from blue (~400 nm) to orange (~600 nm) wavelengths61, which can be detected as a rise in fluorescence upon yellow light excitation62. This fluorescence is weak (quantum yields of < 0.001, compared to >0.1 for fluorescent proteins), but the chromophore is also unusually robust to photobleaching62. Opsin-only indicators such as Archaerhodopsin and QuasAr variants thus can report voltage in cultured neurons and in cultured slices using their intrinsic fluorescence62–65. However, the illumination intensities required in tissue (~12 W/mm2)65 could induce thermal or photochemical damage in living animals. More compatible with in vivo usage are fusions of fluorescent proteins with opsins to form a FRET pair. FRET efficiency increases upon depolarization, leading to a decrease in brightness from the fluorescent protein FRET donor66–68. As the fluorescent protein is bright, this change can be detected with more moderate illumination intensities. The diverse mechanisms and in vitro performance of both VSD- and opsin-based GEVIs have recently been reviewed54, and performance parameters of representative GEVIs are also summarized in Table 3.
Several VSD-based GEVIs have been used successfully in living animals to detect population voltage dynamics and stimulus-evoked single-cell voltage changes. VSFP2.3, VSFP-Butterfly1.2, and Mermaid2, which feature fast activation and slow inactivation kinetics (<3 ms and >10 ms) at 22 °C, were used to detect aggregate voltage dynamics of ~200 ms duration in populations of neurons in mice, enabling the visualization of activity spreading through cortical regions after sensory stimulation3,69. Evoked responses of single cells were detectable by averaging 10 s of trials70. ArcLight was used to detect APs and subthreshold depolarizations in single cells in flies71, but distinguishing between the two was difficult because of the slow kinetics of ArcLight for both activation and inactivation (10 ms and 28 ms respectively at 22 °C)25. Although not as bright as ArcLight5, the faster ASAP1 (~2 ms for activation and inactivation at 22 °C) can discern subthreshold depolarizations from APs in cultured neurons and brain slices25, owing to larger responses to APs of 5–18% in cultured neurons and 5% in slices25,68. ASAP1 and its derivative ASAP2f were used to characterize voltage responses in different neuronal compartments in response to visual stimuli under two-photon illumination in flies, demonstrating that voltage–calcium relationships differ between neurons18.
Opsins have been used in animals to visualize spiking and subcellular voltage dynamics. Unfused opsins can report neuronal activity in worms, where autofluorescence and scatter are minor concerns63. In mice, Mac-Citrine detected voltage changes from the ~10 ms dendritic Ca2+ spikes of cerebellar Purkinje neurons with 1% ΔF/F (ref. 67). Ace2N-4aa-mNeon was the first GEVI to detect single APs in live mammals, reporting the relatively fast (2-ms) APs of cortical neurons with 5% ΔF/F (ref. 5). Ace2N-4aa-mNeon was also capable of reporting subthreshold hyperpolarizations, dendritic activity, and fast spike trains up to ~70 Hz with 0.2-ms accuracy of spike timing. With multitrial averaging, Ace2N-4aa-mNeon also reported the propagation of voltage within individual dendrites and axons5.
It is important to note that electrical signals by neurons are transformed by the kinetics and the linearity of GEVIs to produce the observed fluorescence responses. For example, GEVIs with activation and inactivation kinetics slower than the 2-ms duration of APs show less sensitivity for APs than for slower membrane voltage changes. Thus, faster sensors will produce a less distorted representation of voltage. GEVI improvements have reached a point where, for single AP detection, response amplitudes can be similar to those of commonly used calcium indicators68,72. However, the signals created by fast sensors such as ASAP1, Mac-Citrine, and Ace-mNeonGreen do not persist appreciably beyond the 2-ms durations of APs. Thus AP detection requires fast sampling (>300 Hz). These sampling rates, 10–20 times higher than typically used for calcium imaging, in turn also necessitate high excitation intensities to achieve reasonable rates of event detection, causing faster bleaching and limiting experiment duration.
As no existing GEVI combines all desirable features, GEVI engineering continues to be an active area of research. A study of several opsin-based GEVIs found reduced response amplitudes under two-photon illumination with 80 MHz pulsing compared to that under one-photon illumination73. This could be related to an incompatibility between laser pulse rates and opsin photocycle kinetics. Only one of the photostates of opsins is fluorescent and voltage-sensitive; if this state decays faster than the time between laser pulses, then the effective voltage sensitivity would be reduced66. For two-photon imaging, an opsin with a prolonged voltage-sensitive state may thus be useful. Improving membrane localization of FRET opsins and FlicR1 would also be desirable, so that more of the cellular fluorescence is voltage-responsive. When detection of APs and subthreshold voltage changes is desired but submillisecond timing is not necessary, a GEVI exhibiting fast activation but slightly slower inactivation could be useful. Finally, further improvements in response amplitude, maximal brightness, and optimization of kinetics is always useful. Here, the development of Ace2N-mNeon exemplifies how identification of brightness as a limiting factor in detectability motivated the testing of brighter fluorescent proteins5.
Calcium indicators
Calcium imaging is in many ways the most mature modality for optical imaging of neural activity, allowing studies of neural ensemble dynamics and coding9,10,15,16, dendritic processing19, and synaptic function72, including studies in chronic preparations allowing long-term time-lapse imaging7–9.
Calcium is a second messenger for neurotransmitter reception and membrane depolarization. Cytoplasmic calcium is maintained at 50–100 nM at rest, but neuronal activity induces influx via multiple routes1. Ionotropic receptors for excitatory neurotransmitters pass calcium; for example, NMDA-type glutamate receptors mediate a calcium rise in synaptic spines to ~1 µM (ref. 74). Dendritic voltage-gated calcium channels allow calcium entry during local dendritic spiking51. Finally, voltage-gated calcium channels allow calcium entry throughout the neuron during AP propagation. In cortical neurons, this results in an intracellular calcium rise of ~150 nM within 10 ms that then persists with a half decay time of 50–70 ms (refs. 75,76). Early work using calcium dyes loaded via a patch pipette demonstrated that calcium imaging could indeed report single synaptic responses and AP firing in vivo75.
The primary advantages and limitations of calcium imaging derive from the magnitude and kinetics of calcium entry and extrusion. Calcium entry transforms the transient membrane-localized events of neurotransmitter receptor opening or action potential generation into a volume-filling and more prolonged biochemical change. This amplification, combined with the brightness of optimized indicators, makes calcium indicator responses larger in terms of photonic output and thus easier to detect. For example, a single AP can be initiated and completed within 3–5 ms, before the calcium transient even reaches its peak75,76. The slower kinetics of the resulting calcium transients allow them to be detected at sampling intervals of 30–60 ms (ref. 72), an order of magnitude longer than the APs themselves. Major limitations of GECI imaging are that they do not report neurotransmitter receptor activation or AP firing with temporal precision, nor do they report membrane hyperpolarizations77 or subthreshold voltage changes well. Timing of sparse APs can be indirectly inferred by deconvolution of the indicator response and calcium transient kinetics78,79, but this generally requires prior knowledge of indicator kinetics and calcium kinetics in the neuron type being imaged. Calculation of spike rates from GECI data is also difficult, with the best algorithms achieving only 40–60% accuracy for higher frequency events80.
The development, mechanisms, and uses of GECIs have recently been reviewed extensively1,26. We will thus only briefly discuss GECI mechanisms and some of their more distinctive applications. The first GECI, Cameleon, composed of calmodulin (CaM) and M13 domains between ECFP and EYFP, reported calcium with increased FRET56. This basic architecture was retained in YC2.60, YC3.60, D3cpv, and TnXL81 and then simplified in Twitch-family GECIs, which contain a single troponin domain between ECFP and EYFP82. FRET sensors exhibit high basal brightness in at least one channel (Table 4). In low-photon-count situations, analyzing only the brighter or more responsive of the fluorophores in a FRET GECI rather than the emission ratio improves detection, and thus the ratiometric nature of the measurement is not usually advantageous for large-scale imaging in vivo23,83, but it is useful for calibrated measurements of calcium concentrations84. While responses to single APs are insufficient to rise above noise in the smaller neurons of the fly85, YC2.60, YC3.60, and D3cpv can reveal single APs in cortical neurons in living mice86,87. However, discrimination of closely spaced APs is limited by observed half-decay times of >200 ms (refs. 86,87).
Table 4.
Selected genetically encoded voltage indicators (GECIs)
| GECI | Maximum ΔF/F in vitroa |
Ca2+-free brightness (mM−1 cm−1)b |
Ca2+-saturated brightness (mM−1 cm−1)b |
Kd
in vitro (nM)c |
ΔF/F per AP in tissued |
Half-decay rate in tissue (ms)e |
Refs. |
|---|---|---|---|---|---|---|---|
| YC3.60 | −0.66 (ECFP) | 8.8f | 3.1 | 780 | −0.01 | 410 | 137,138 |
| +0.77 (cpVenus) | 2.4f | 11 | +0.02 | ||||
| YC3.60 | −0.66 (ECFP) | 8.8g | 3.1 | 140 | −0.01f | 470 | 139,140 |
| 3GS | +0.77 (cpVenus) | 2.4g | 11 | +0.01f | |||
| D3cpV | −0.46 (ECFP) | 7.3h | 3.6 | 530 | −0.03f | 9,500 | 141,142 |
| +1.1 (cpVenus) | 4.8h | 10 | +0.02f | ||||
| TN-XXL | −0.5 (ECFP) | 9.6i | 5.4 | 800 | −0.01f | 1,600 | 142,143 |
| +1.0 (cpCitrine) | 1.5i | 10 | +0.02f | ||||
| Twitch-2B | −0.77 (mCerulean3) | 22j | 5.8 | 200 | −0.12f | 2,100 | 82,142 |
| +0.87 (cpVenus) | 0.83j | 12 | +0.12f | ||||
|
| |||||||
| GCaMP3 | +12 | 1.8 | 23 | 540 | +0.14 | 650 | 114,142 |
| GCaMP5k | +9.4 | ND | ND | 190 | +0.04 | 270 | 72,144 |
| GCaMP6f | +52 | 0.70 | 37 | 380 | +0.22 | 140 | 72,91 |
| GCaMP6s | +63 | 0.66 | 42 | 140 | +0.25 | 550 | 72 |
| R-CaMP2 | +4.8 | 2.3 (1.6)k | 11 | 69 | +0.60 | 150 | 106 |
| jRGECO1a | +11 | 1.0 (0.74)k | 12 | 150 | +0.19 | 200 | 7 |
| jRCaMP1b | +6.2 | 4.0 (4.0)k | 29 | 712 | NDl | ND | 7 |
GECIs with reported single-AP responses in live animals or acute rodent slices are included. Some values are estimates based on published graphs.
Fluorescence change from zero to saturating calcium in vitro at 25 °C at the emission peak of each channel. This number is empirically measured and, for the acceptor fluorophore in FRET sensors, is influenced by cross-excitation and bleed-through. As event detection is often optimized by single-channel imaging of FRET indicators23,83, the two channels are shown separately.
Estimated molar brightness produced by each fluorophore. For FRET sensors, brightness for the donor channel is calculated as the product of donor peak extinction coefficient and donor quantum yield multiplied by 1 – E, where E is measured FRET efficiency in calcium-free or calcium-saturated conditions. Brightness for the acceptor channel is calculated as the product of donor peak extinction coefficient and acceptor quantum yield multiplied by E. Values for specific parameters are noted below. Values do not account for any cross-excitation or bleed-through, but provide an estimate of the contribution of each fluorophore to indicator brightness. For single-protein sensors, calcium-free brightness is calculated from calcium-saturated fluorescence and maximum ΔF/F.
Measured at 25 °C.
In mouse brain or mouse acute slices at 35 °C.
In vitro measurements at 37 °C; neuronal measurements in mouse brain or mouse acute slices at 35 °C in response to a single AP.
Peak ECFP extinction coefficient is 28 mM−1 cm−1 at 433 nm, ECFP quantum yield is 0.37 (ref. 129), cpVenus quantum yield is assumed to be 0.57 as with Venus130, and calcium-free/saturated E of 0.15/0.70 is derived from published emission spectra as previously described131,132.
FRET emission spectra are not published, but E values are inferred to be similar to that of YC3.60 on the basis of similar ratio changes.
Calcium-free/saturated E of 0.30/0.65 is derived from published emission spectra.
Citrine quantum yield is 0.76 (ref. 130), calcium-free/saturated E of 0.07/0.48 is derived from calcium-free/saturated emission spectrum (A.J. Lam and M.Z.L., unpublished data).
Peak mCerulean3 extinction coefficient is 29 mM−1 cm−1 at 433 nm, Cerulean quantum yield is 0.80 (ref. 135), and calcium-free/saturated E of 0.05/0.75 is derived from published emission spectra.
Values in parentheses are from direct measurement of extinction coefficient and quantum yield without calcium. ND, not determined.
Single-fluorophore GECIs have achieved larger responses than FRET-based GECIs. The first single-fluorophore GECI was Camgaroo, a YFP bearing a CaM domain between positions 144 and 145, near the chromophore45. At rest, the Camgaroo chromophore is mostly protonated and dark to excitation at 488 nm, while calcium binding increases its pKa, causing chromophore deprotonation and increased excitation at 488 nm. Camgaroo was the first indicator in which conformational changes in a domain attached near the chromophore modulate its brightness45. The sensors pericam and G-CaMP (later condensed to GCaMP) extended this concept, fusing CaM to the C terminus and the CaM-binding peptide to the N terminus of cpGFP145 (refs. 88,89). In GCaMP, residues 145–148 were deleted as well. Notably, an important function of His148 in hydrogen-bonding to and thereby stabilizing the deprotonated chromophore is replaced by Arg377 in the CaM domain of GCaMP, but only in the calcium-bound state90. This finding explains the nearly complete chromophore protonation in calcium-free GCaMP90.
GCaMP-family GECIs have since been improved iteratively by several groups through many rounds of mutagenesis and selection (Table 4)72. Currently the most commonly used GECIs are those of the GCaMP6 series. GCaMP6f exhibits fluorescence changes of 2,820% across calcium concentrations of 0 to ~1 µM and half decay times of 71 ms in vitro at 37 °C (refs. 72,91). GCaMP6f reports single APs in mouse cortex with ~20% ΔF/F, superior to that of organic dyes, and a 142-ms half-decay time72. GCaMP6m, GCaMP6s, and GCaMP7 produce even larger responses to single APs, but at the cost of decays that are 93–190% longer than that of GCaMP6f72,92. For more temporally accurate calcium reporting, a GCaMP6f variant, GCaMP6fRS09, reduces half-decay times to 20 ms in vitro at 37 °C and 110 ms in mouse neurons in vivo, while maintaining 86% of the responsiveness of GCaMP6f91. A variant of GCaMP3, GCaMP3fast, is even faster, with an in vitro half-decay time of 3 ms at 37 °C, and has lower baseline fluorescence, although its performance in vivo has yet to be characterized93. These faster GECIs are as fast as organic calcium dyes in their off-rates, which are reaching practically useful limits, as single AP-induced calcium transients themselves have half-decay times of 50–60 ms (without indicator buffering)75.
GECIs have demonstrated uses in detecting AP firing and certain subcellular events in vivo. One of their first applications in vivo was to visualize odorant responses in flies, where GCaMP1 was found to have higher signal-to-noise ratio than SEP94. GCaMPs have since been used to visualize activity in entire worm and fish brains, implicating specific neurons in decision-making or learning95–98. GCaMP reporting of APs in the same neuronal population over time has revealed recruitment of neurons into functional circuits during learning in the mouse8,9,99. More applications to learning are discussed in a recent review17. GCaMPs have also been used to localize activity to specific postsynaptic and presynaptic compartments100. For example, GCaMP6s revealed that different spines on the same visual cortex neuron respond to visual stimuli of different orientations72. Using GCaMP6s, local dendritic spiking was found to correlate with synaptic potentiation in cortical neurons101 and the acquisition of place specificity in hippocampal neurons19. GCaMP6s was used to detect orientation selectivity of individual axonal boutons in projections from visual cortex102. Postsynaptic GCaMP5 was used to localize single release events at the fly neuromuscular junction, revealing that spontaneous and evoked release events occur at different boutons103. The use of GECIs instead of GEPIs to localize synaptic vesicle exocytosis illustrates the relative ease of detecting GECI responses, which benefit from the amplified nature of the calcium signal.
An active area of research is the engineering of GECIs with optical characteristics other than green fluorescence. Red GECIs should improve signal over autofluorescence in vivo and also allow a blue excitation channel to be used for exciting other indicators or optogenetic actuators. Red GECIs have been created from circularly permuted RFPs with a similar design to pericams, similar to GCaMP but without a deletion of essential amino acids104,105. Recent variants jRGECO1a and R-CaMP2 report single spikes in mouse cortex with similar (jRGECO1a) or higher (R-CaMP2) ΔF/F to that of GCaMP6f (Table 4), and with similar kinetics, and they show less attenuation by tissue depth7,106. However, they are dimmer and show photoswitching by blue light7. jRCaMP1b has slower on- and off-kinetics, but is brighter and does not photoswitch7. However, all red GECIs exhibit increased green fluorescence and reduced responsiveness after long-term expression7.
To allow optical selection and tracing of neurons for calcium imaging, photoactivatable GECIs named sPA-GCaMP6f and sPA-GCaMP6s were engineered by combining features of superfolder GFP, photoactivatable GFP (PA-GFP), and GCaMP6f or GCaMP6s107. sPA-GCaMP6f is dark to 488-nm excitation in either calcium-free or calcium-bound states until 400-nm photoactivation, after which approximately half of the chromophore population is deprotonated and bright without calcium. Calcium addition to ~1 µM induces a further ~120% increase in 488-nm absorption. In mouse cortical neurons, sPA-GCaMP6f and sPA-GCaMP6s allowed optical selection of cells for calcium reporting. They reported single APs with similar kinetics as the parental GCaMP6 variants but one-fifth the amplitude. sPA-GCaMPs can be useful for visualizing responses in synapses that belong to a cell body of interest, or vice versa. A different photoactivatable GECI, PA-TnXL, was created by replacing the CFP-YFP FRET pair in TnXL with a PA-GFP as the donor and a dim YFP variant as the acceptor108. After 400-nm photoactivation, 488-nm excitation of PA-TnXL reports calcium with 800% fluorescence increases in vitro, but has not yet been characterized in vivo.
Photoconvertible GECIs that change wavelengths upon illumination could allow optical selection of neurons for activity tracking, or optical marking of neurons that exhibit interesting activity patterns for later follow-up. One such GECI, GR-GECO, arose from a recreation of the pericam design in the photoconvertible fluorescent protein mMaple. GR-GECO permanently converts from green to red fluorescence upon 400-nm illumination while maintaining >500% calcium responses in both forms109. A recent study revealed that many GCaMPs are inherently photoconvertible to red emission by 400-nm light, with the red species retaining up to 41% of the responsiveness of the original green form110. These findings add to earlier observations of green-to-red photoconversion in GFP derivatives111, although the photoconversion may be less efficient than in fluorescent proteins evolved for photoconvertibility. Finally, a unique GECI, CAMPARI, undergoes green-to-red photoconversion by 400-nm light only when bound to calcium112. While CAMPARI responds dynamically to calcium in both green and red forms (undergoing protonation and thereby dimming in high calcium, the opposite behavior of other GECIs), its unique ability is in retroactively reporting calcium activity in time windows defined by light. For example, CAMPARI was used to permanently mark neurons activated by sensory stimulation or during specific behavior112.
How can GECIs be further improved? Current GECIs already show peak brightness similar to that of unmodified fluorescent proteins7,72,106, so it is unlikely brightness can be further increased. Increasing ΔF/F by reducing baseline brightness is of limited utility, as baseline fluorescence of GCaMP6-family GECIs is already indistinguishable from autofluorescence at lower expression levels113. A relatively unexplored avenue for GECI improvement, however, is acceleration of calcium binding. The fastest known half-rise time in the GCaMP series is 600 ms for GCaMP3RS09, measured in vitro as the time to reach half of the equilibrium fluorescence intensity when calcium is stepped from 0 to 200 nM at 25 °C (ref. 114). As based on their relative kon association rate constants (which can be derived from measured Kd dissociation equilibrium and koff dissociation rate constants), analogous half-rise times for GCaMP6f and GCaMP6fRS09 are expected to be 1,800 and 800 ms, respectively. The slow rise kinetics of GECIs to 200 nM calcium may seem surprising, as measurements with 1 µM calcium produce half-rise times of several milliseconds89,114, but there is a strong dependency of rise kinetics on concentration114. These kinetics may also seem nonintuitive given that responses of faster GECIs peak about 200 ms following an AP72, but they are consistent with differences in amplitudes of GECI transients following APs and steady-state GECI responses in vitro. For instance, GCaMP6f demonstrates peak ΔF/F of 20% following an AP, during which calcium concentration changes from ~50 to ~200 nM, but has a steady-state response of ~700% ΔF/F from 50 to 200 nM calcium91. The peak ΔF/F following an AP only reaches 20% and not 700% because calcium concentrations start to fall before binding equilibrium of calcium to GECI is reached. Thus all parameters of the GECI fluorescence transient following an AP—peak time, peak amplitude, and half-decay time—are products of the kinetics and amplitudes of both intracellular calcium transients and GECI responses (Fig. 4). The importance of on-kinetics is further demonstrated by R-CaMP2, which shows faster activation kinetics than other GECIs and has larger responses to single APs than some red GECIs with larger total ΔF/F (refs. 7,106). Thus, if binding kinetics can be accelerated, larger indicator responses to APs may be achievable for other GECI families as well.
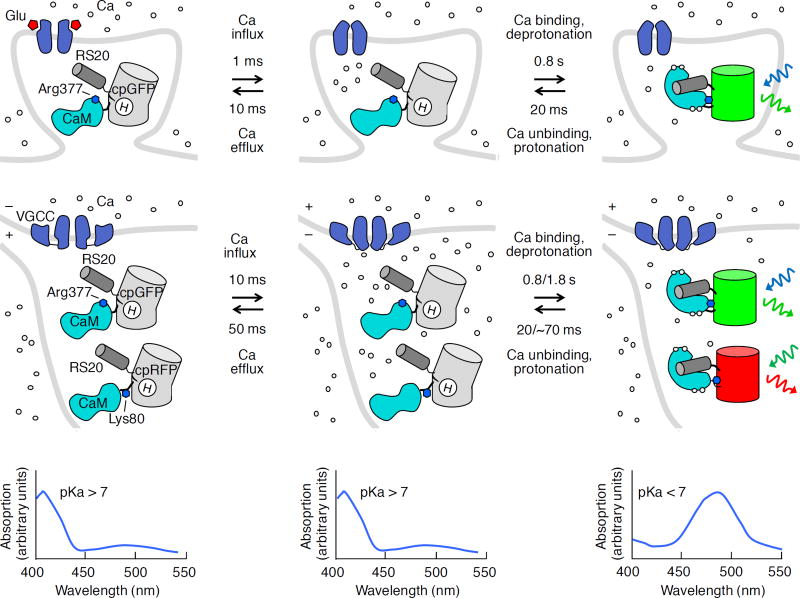
Figure 4.
Genetically encoded calcium indicators (GECIs). GECIs respond to calcium with increased fluorescence. Events following a glutamate release event (top row) and a single AP (middle row) that opens voltage-gated calcium channels (VGCC) are shown, with open channels in the first time point and closed channels before the second time point. Calcium-induced binding of calmodulin (CaM) to a peptide from smooth-muscle myosin light-chain kinase (RS20) causes repositioning of Arg377 in GCaMPs and Lys80 in R-GECOs, inducing loss of a proton from the chromophore and an absorbance shift (bottom row). GCaMPs can detect calcium transients induced by synaptic activation (top row) and action potentials (middle row), with increased green emission (rightward green sinusoidal arrow) upon blue excitation (blue sinusoidal arrow). R-GECOs can report APs with red emission (red sinusoidal arrow) upon green excitation (leftward green sinusoidal arrow). Spine calcium kinetics are from ref. 74. Dendrite and soma kinetics are from ref. 75. With repeated neurotransmitter release or prolonged depolarization, calcium rise and decay times will be longer. Times for calcium half-binding or half-unbinding are for GCaMP6f-RS09 and jRGECO1a, separated by a slash (/), as these are respectively the fastest green and red GECIs tested in neurons. GCaMP6f values were used for jRGECO1a, as they show similar in cellulo kinetics, but only GCaMP6f in vitro kinetics were measured. Half-binding times for GCaMP6f and GCaMP6f-RS09 were calculated by normalizing the half-binding time of GCaMP3-RS06 (ref. 91) after a 200-nM step at 25 °C (ref. 114) by the kon values of GCaMP6f and GCaMP6f-RS09 relative to GCaMP3-RS06 at 25 °C (ref. 91). Times at 37 °C may be similar, as GCaMPs show little temperature dependence in binding rates93. Unbinding times shown measured at 37 °C for GCaMP6fRS09 and GCaMP6f93. Note that observed rise times of fluorescence transients in cells will be mostly determined by calcium decay kinetics.
CONCLUSION
Genetically encoded calcium indicators have revolutionized systems neuroscience, allowing the study of neural encoding in neuronal subpopulations and whole brains, activity changes over time, and subcellular responses to synaptic activation. Genetically encoded sensors for other types of neuronal activity—vesicle fusion, neurotransmitters, and voltage—continue to be improved at a rapid pace. In particular, recent years have seen a resurgence in the development of voltage indicators, and examples of their use in reporting single-trial responses in vivo have recently appeared. It is clear that the uses of genetically encoded activity indicators will broaden in variety while also becoming a central part of efforts to understand the functions of specific circuits in the brain.
And yet, optical activity indicators are just one of many transformative optical methods developed over the past decade. These methods include optogenetic control of neuronal excitation and signaling115,116, long-term fluorescent labeling of activated neurons117,118, and post hoc imaging of optically cleared tissues119,120. A powerful aspect of optical methods is that they can be easily combined, so that the same cells are studied by multiple approaches across different phases of an experiment. For example, it is becoming increasingly feasible to combine neuronal activity visualization, optogenetic manipulations, and post-mortem examination of morphology, connectivity, and macromolecular distributions in the same cells121,122. In the future, studies that combine optical methods are likely to become common, with genetically encoded activity indicators enabling the initial step of observing neuronal activity in vivo.
Acknowledgments
We gratefully acknowledge research funding from the Defense Advanced Research Projects Agency (M.J.S. and M.Z.L.), the Rita Allen Foundation (M.Z.L.) and NIH BRAIN Initiative grant 1U01NS090600 (M.Z.L. and M.J.S.).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Grienberger C, Konnerth A. Imaging calcium in neurons. Neuron. 2012;73:862–885. doi: 10.1016/j.neuron.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 2.Kavalali ET, Jorgensen EM. Visualizing presynaptic function. Nat. Neurosci. 2014;17:10–16. doi: 10.1038/nn.3578. [DOI] [PubMed] [Google Scholar]
- 3.Knöpfel T, Gallero-Salas Y, Song C. Genetically encoded voltage indicators for large scale cortical imaging come of age. Curr. Opin. Chem. Biol. 2015;27:75–83. doi: 10.1016/j.cbpa.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 4.Hamel EJ, Grewe BF, Parker JG, Schnitzer MJ. Cellular level brain imaging in behaving mammals: an engineering approach. Neuron. 2015;86:140–159. doi: 10.1016/j.neuron.2015.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gong Y, et al. High-speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor. Science. 2015;350:1361–1366. doi: 10.1126/science.aab0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osakada F, et al. New rabies virus variants for monitoring and manipulating activity and gene expression in defined neural circuits. Neuron. 2011;71:617–631. doi: 10.1016/j.neuron.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dana H, et al. Sensitive red protein calcium indicators for imaging neural activity. Elife. 2016;5:e12727. doi: 10.7554/eLife.12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peters AJ, Chen SX, Komiyama T. Emergence of reproducible spatiotemporal activity during motor learning. Nature. 2014;510:263–267. doi: 10.1038/nature13235. [DOI] [PubMed] [Google Scholar]
- 9.Ziv Y, et al. Long-term dynamics of CA1 hippocampal place codes. Nat. Neurosci. 2013;16:264–266. doi: 10.1038/nn.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahrens MB, Orger MB, Robson DN, Li JM, Keller PJ. Whole-brain functional imaging at cellula resolution using light-sheet microscopy. Nat. Methods. 2013;10:413–420. doi: 10.1038/nmeth.2434. [DOI] [PubMed] [Google Scholar]
- 11.Lecoq J, et al. Visualizing mammalian brain area interactions by dual-axis two-photon calcium imaging. Nat. Neurosci. 2014;17:1825–1829. doi: 10.1038/nn.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sofroniew NJ, Flickinger D, King J, Svoboda K. A large field of view two-photon mesoscope with subcellular resolution for in vivo imaging. Elife. 2016;5:e14472. doi: 10.7554/eLife.14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stirman JN, Smith IT, Kudenov MW, Smith SL. Wide field-of-view, multi-region, two-photon imaging of neuronal activity in the mammalian brain. Nat. Biotechnol. 2016 doi: 10.1038/nbt.3594. http://dx.doi.org/10.1038/nbt.3594. [DOI] [PMC free article] [PubMed]
- 14.Tomer R, et al. SPED light sheet microscopy: fast mapping of biological system structure and function. Cell. 2015;163:1796–1806. doi: 10.1016/j.cell.2015.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dombeck DA, Harvey CD, Tian L, Looger LL, Tank DW. Functional imaging of hippocampal place cells at cellular resolution during virtual navigation. Nat. Neurosci. 2010;13:1433–1440. doi: 10.1038/nn.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Connor DH, Peron SP, Huber D, Svoboda K. Neural activity in barrel cortex underlying vibrissa-based object localization in mice. Neuron. 2010;67:1048–1061. doi: 10.1016/j.neuron.2010.08.026. [DOI] [PubMed] [Google Scholar]
- 17.Jercog P, Rogerson T, Schnitzer MJ. Large-scale fluorescence calcium-imaging methods for studies of long-term memory in behaving mammals. Cold Spring Harb. Perspect. Biol. 2016;8:a021824. doi: 10.1101/cshperspect.a021824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang HH, et al. Subcellular imaging of voltage and calcium signals reveals neural processing in vivo. Cell. 2016;166:245–257. doi: 10.1016/j.cell.2016.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheffield ME, Dombeck DA. Calcium transient prevalence across the dendritic arbour predicts place field properties. Nature. 2015;517:200–204. doi: 10.1038/nature13871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buzsáki G. Large-scale recording of neuronal ensembles. Nat. Neurosci. 2004;7:446–451. doi: 10.1038/nn1233. [DOI] [PubMed] [Google Scholar]
- 21.Betley JN, et al. Neurons for hunger and thirst transmit a negative-valence teaching signal. Nature. 2015;521:180–185. doi: 10.1038/nature14416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghosh KK, et al. Miniaturized integration of a fluorescence microscope. Nat. Methods. 2011;8:871–878. doi: 10.1038/nmeth.1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilt BA, Fitzgerald JE, Schnitzer MJ. Photon shot noise limits on optical detection of neuronal spikes and estimation of spike timing. Biophys. J. 2013;104:51–62. doi: 10.1016/j.bpj.2012.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akemann W, et al. Imaging neural circuit dynamics with a voltage-sensitive fluorescent protein. J. Neurophysiol. 2012;108:2323–2337. doi: 10.1152/jn.00452.2012. [DOI] [PubMed] [Google Scholar]
- 25.St-Pierre F, et al. High-fidelity optical reporting of neuronal electrical activity with an ultrafast fluorescent voltage sensor. Nat. Neurosci. 2014;17:884–889. doi: 10.1038/nn.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Broussard GJ, Liang R, Tian L. Monitoring activity in neural circuits with genetically encoded indicators. Front. Mol. Neurosci. 2014;7:97. doi: 10.3389/fnmol.2014.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Südhof TC. Neurotransmitter release: the last millisecond in the life of a synaptic vesicle. Neuron. 2013;80:675–690. doi: 10.1016/j.neuron.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsien RY. The green fluorescent protein. Annu. Rev. Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 29.Miesenböck G, De Angelis DA, Rothman JE. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature. 1998;394:192–195. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
- 30.Sankaranarayanan S, De Angelis D, Rothman JE, Ryan TA. The use of pHluorins for optical measurements of presynaptic activity. Biophys. J. 2000;79:2199–2208. doi: 10.1016/S0006-3495(00)76468-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen Y, Rosendale M, Campbell RE, Perrais D. pHuji, a pH-sensitive red fluorescent protein for imaging of exo- and endocytosis. J. Cell Biol. 2014;207:419–432. doi: 10.1083/jcb.201404107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mutch SA, et al. Protein quantification at the single vesicle level reveals that a subset of synaptic vesicle proteins are trafficked with high precision. J. Neurosci. 2011;31:1461–1470. doi: 10.1523/JNEUROSCI.3805-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alabi AA, Tsien RW. Synaptic vesicle pools and dynamics. Cold Spring Harb. Perspect. Biol. 2012;4:a013680. doi: 10.1101/cshperspect.a013680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sjulson L, Miesenböck G. Optical recording of action potentials and other discrete physiological events: a perspective from signal detection theory. Physiology (Bethesda) 2007;22:47–55. doi: 10.1152/physiol.00036.2006. [DOI] [PubMed] [Google Scholar]
- 35.Leitz J, Kavalali ET. Fast retrieval and autonomous regulation of single spontaneously recycling synaptic vesicles. Elife. 2014;3:e03658. doi: 10.7554/eLife.03658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ng M, et al. Transmission of olfactory information between three populations of neurons in the antennal lobe of the fly. Neuron. 2002;36:463–474. doi: 10.1016/s0896-6273(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 37.Tabares L, et al. Monitoring synaptic function at the neuromuscular junction of a mouse expressing synaptopHluorin. J. Neurosci. 2007;27:5422–5430. doi: 10.1523/JNEUROSCI.0670-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bozza T, McGann JP, Mombaerts P, Wachowiak M. In vivo imaging of neuronal activity by targeted expression of a genetically encoded probe in the mouse. Neuron. 2004;42:9–21. doi: 10.1016/s0896-6273(04)00144-8. [DOI] [PubMed] [Google Scholar]
- 39.Gandasi NR, et al. Survey of Red Fluorescence Proteins as Markers for Secretory Granule Exocytosis. PLoS One. 2015;10:e0127801. doi: 10.1371/journal.pone.0127801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diamond JS. Deriving the glutamate clearance time course from transporter currents in CA1 hippocampal astrocytes: transmitter uptake gets faster during development. J. Neurosci. 2005;25:2906–2916. doi: 10.1523/JNEUROSCI.5125-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.von Gersdorff H, Sakaba T, Berglund K, Tachibana M. Submillisecond kinetics of glutamate release from a sensory synapse. Neuron. 1998;21:1177–1188. doi: 10.1016/s0896-6273(00)80634-0. [DOI] [PubMed] [Google Scholar]
- 42.Liang R, Broussard GJ, Tian L. Imaging chemical neurotransmission with genetically encoded fluorescent sensors. ACS Chem. Neurosci. 2015;6:84–93. doi: 10.1021/cn500280k. [DOI] [PubMed] [Google Scholar]
- 43.Okumoto S, et al. Detection of glutamate release from neurons by genetically encoded surface-displayed FRET nanosensors. Proc. Natl. Acad. Sci. USA. 2005;102:8740–8745. doi: 10.1073/pnas.0503274102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dulla C, et al. Imaging of glutamate in brain slices using FRET sensors. J. Neurosci. Methods. 2008;168:306–319. doi: 10.1016/j.jneumeth.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baird GS, Zacharias DA, Tsien RY. Circular permutation and receptor insertion within green fluorescent proteins. Proc. Natl. Acad. Sci. USA. 1999;96:11241–11246. doi: 10.1073/pnas.96.20.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marvin JS, et al. An optimized fluorescent probe for visualizing glutamate neurotransmission. Nat. Methods. 2013;10:162–170. doi: 10.1038/nmeth.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hikima T, Garcia-Munoz M, Arbuthnott GW. Presynaptic D1 heteroreceptors and mGlu autoreceptors act at individual cortical release sites to modify glutamate release. Brain Res. 2016;1639:74–87. doi: 10.1016/j.brainres.2016.02.042. [DOI] [PubMed] [Google Scholar]
- 48.Xie Y, et al. Resolution of high-frequency mesoscale intracortical maps using the genetically encoded glutamate sensor iGluSnFR. J. Neurosci. 2016;36:1261–1272. doi: 10.1523/JNEUROSCI.2744-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berntsson RP, Smits SH, Schmitt L, Slotboom DJ, Poolman B. A structural classification of substrate-binding proteins. FEBS Lett. 2010;584:2606–2617. doi: 10.1016/j.febslet.2010.04.043. [DOI] [PubMed] [Google Scholar]
- 50.Sabatini BL, Regehr WG. Timing of synaptic transmission. Annu. Rev. Physiol. 1999;61:521–542. doi: 10.1146/annurev.physiol.61.1.521. [DOI] [PubMed] [Google Scholar]
- 51.Stuart GJ, Spruston N. Dendritic integration: 60 years of progress. Nat. Neurosci. 2015;18:1713–1721. doi: 10.1038/nn.4157. [DOI] [PubMed] [Google Scholar]
- 52.Bean BP. The action potential in mammalian central neurons. Nat. Rev. Neurosci. 2007;8:451–465. doi: 10.1038/nrn2148. [DOI] [PubMed] [Google Scholar]
- 53.Peterka DS, Takahashi H, Yuste R. Imaging voltage in neurons. Neuron. 2011;69:9–21. doi: 10.1016/j.neuron.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.St-Pierre F, Chavarha M, Lin MZ. Designs and sensing mechanisms of genetically encoded fluorescent voltage indicators. Curr. Opin. Chem. Biol. 2015;27:31–38. doi: 10.1016/j.cbpa.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siegel MS, Isacoff EY. A genetically encoded optical probe of membrane voltage. Neuron. 1997;19:735–741. doi: 10.1016/s0896-6273(00)80955-1. [DOI] [PubMed] [Google Scholar]
- 56.Miyawaki A, et al. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 57.Baker BJ, et al. Three fluorescent protein voltage sensors exhibit low plasma membrane expression in mammalian cells. J. Neurosci. Methods. 2007;161:32–38. doi: 10.1016/j.jneumeth.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 58.Dimitrov D, et al. Engineering and characterization of an enhanced fluorescent protein voltage sensor. PLoS One. 2007;2:e440. doi: 10.1371/journal.pone.0000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abdelfattah AS, et al. A bright and fast red fluorescent protein voltage indicator that reports neuronal activity in organotypic brain slices. J. Neurosci. 2016;36:2458–2472. doi: 10.1523/JNEUROSCI.3484-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jung A, Garcia JE, Kim E, Yoon BJ, Baker BJ. Linker length and fusion site composition improve the optical signal of genetically encoded fluorescent voltage sensors. Neurophotonics. 2015;2:021012. doi: 10.1117/1.NPh.2.2.021012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kolodner P, Lukashev EP, Ching YC, Rousseau DL. Electric-field-induced Schiff-base deprotonation in D85N mutant bacteriorhodopsin. Proc. Natl. Acad. Sci. USA. 1996;93:11618–11621. doi: 10.1073/pnas.93.21.11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kralj JM, Douglass AD, Hochbaum DR, Maclaurin D, Cohen AE. Optical recording of action potentials in mammalian neurons using a microbial rhodopsin. Nat. Methods. 2012;9:90–95. doi: 10.1038/nmeth.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Flytzanis NC, et al. Archaerhodopsin variants with enhanced voltage-sensitive fluorescence in mammalian and Caenorhabditis elegans neurons. Nat. Commun. 2014;5:4894. doi: 10.1038/ncomms5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gong Y, Li JZ, Schnitzer MJ. Enhanced archaerhodopsin fluorescent protein voltage indicators. PLoS One. 2013;8:e66959. doi: 10.1371/journal.pone.0066959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hochbaum DR, et al. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat. Methods. 2014;11:825–833. doi: 10.1038/nmeth.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bayraktar H, et al. Ultrasensitive measurements of microbial rhodopsin photocycles using photochromic FRET. Photochem. Photobiol. 2012;88:90–97. doi: 10.1111/j.1751-1097.2011.01011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gong Y, Wagner MJ, Zhong Li J, Schnitzer MJ. Imaging neural spiking in brain tissue using FRET-opsin protein voltage sensors. Nat. Commun. 2014;5:3674. doi: 10.1038/ncomms4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zou P, et al. Bright and fast multicoloured voltage reporters via electrochromic FRET. Nat. Commun. 2014;5:4625. doi: 10.1038/ncomms5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsutsui H, et al. Improved detection of electrical activity with a voltage probe based on a voltage-sensing phosphatase. J. Physiol. (Lond.) 2013;591:4427–4437. doi: 10.1113/jphysiol.2013.257048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Akemann W, et al. Two-photon voltage imaging using a genetically encoded voltage indicator. Sci. Rep. 2013;3:2231. doi: 10.1038/srep02231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cao G, et al. Genetically targeted optical electrophysiology in intact neural circuits. Cell. 2013;154:904–913. doi: 10.1016/j.cell.2013.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen TW, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brinks D, Klein AJ, Cohen AE. Two-photon lifetime imaging of voltage indicating proteins as a probe of absolute membrane voltage. Biophys. J. 2015;109:914–921. doi: 10.1016/j.bpj.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Higley MJ, Sabatini BL. Calcium signaling in dendritic spines. Cold Spring Harb. Perspect. Biol. 2012;4:a005686. doi: 10.1101/cshperspect.a005686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Helmchen F, Borst JG, Sakmann B. Calcium dynamics associated with a single action potential in a CNS presynaptic terminal. Biophys. J. 1997;72:1458–1471. doi: 10.1016/S0006-3495(97)78792-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koester HJ, Sakmann B. Calcium dynamics associated with action potentials in single nerve terminals of pyramidal cells in layer 2/3 of the young rat neocortex. J. Physiol. (Lond.) 2000;529:625–646. doi: 10.1111/j.1469-7793.2000.00625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gore BB, Soden ME, Zweifel LS. Visualization of plasticity in fear-evoked calcium signals in midbrain dopamine neurons. Learn. Mem. 2014;21:575–579. doi: 10.1101/lm.036079.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lütcke H, Gerhard F, Zenke F, Gerstner W, Helmchen F. Inference of neuronal network spike dynamics and topology from calcium imaging data. Front. Neural Circuits. 2013;7:201. doi: 10.3389/fncir.2013.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pnevmatikakis EA, et al. Simultaneous denoising, deconvolution, and demixing of calcium imaging data. Neuron. 2016;89:285–299. doi: 10.1016/j.neuron.2015.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Theis L, et al. Benchmarking spike rate inference in population calcium imaging. Neuron. 2016;90:471–482. doi: 10.1016/j.neuron.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Miyawaki A, Niino Y. Molecular spies for bioimaging—fluorescent protein-based probes. Mol. Cell. 2015;58:632–643. doi: 10.1016/j.molcel.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 82.Thestrup T, et al. Optimized ratiometric calcium sensors for functional in vivo imaging of neurons and T lymphocytes. Nat. Methods. 2014;11:175–182. doi: 10.1038/nmeth.2773. [DOI] [PubMed] [Google Scholar]
- 83.Hires SA, Tian L, Looger LL. Reporting neural activity with genetically encoded calcium indicators. Brain Cell Biol. 2008;36:69–86. doi: 10.1007/s11068-008-9029-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Grynkiewicz G, Poenie M, Tsien RY. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J. Biol. Chem. 1985;260:3440–3450. [PubMed] [Google Scholar]
- 85.Hendel T, et al. Fluorescence changes of genetic calcium indicators and OGB-1 correlated with neural activity and calcium in vivo and in vitro. J. Neurosci. 2008;28:7399–7411. doi: 10.1523/JNEUROSCI.1038-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wallace DJ, et al. Single-spike detection in vitro and in vivo with a genetic Ca2+ sensor. Nat. Methods. 2008;5:797–804. doi: 10.1038/nmeth.1242. [DOI] [PubMed] [Google Scholar]
- 87.Yamada Y, et al. Quantitative comparison of genetically encoded Ca indicators in cortical pyramidal cells and cerebellar Purkinje cells. Front. Cell. Neurosci. 2011;5:18. doi: 10.3389/fncel.2011.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nagai T, Sawano A, Park ES, Miyawaki A. Circularly permuted green fluorescent proteins engineered to sense Ca2+ Proc. Natl. Acad. Sci. USA. 2001;98:3197–3202. doi: 10.1073/pnas.051636098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nakai J, Ohkura M, Imoto K. A high signal-to-noise Ca2+ probe composed of a single green fluorescent protein. Nat. Biotechnol. 2001;19:137–141. doi: 10.1038/84397. [DOI] [PubMed] [Google Scholar]
- 90.Wang Q, Shui B, Kotlikoff MI, Sondermann H. Structural basis for calcium sensing by GCaMP2. Structure. 2008;16:1817–1827. doi: 10.1016/j.str.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Badura A, Sun XR, Giovannucci A, Lynch LA, Wang SS. Fast calcium sensor proteins for monitoring neural activity. Neurophotonics. 2014;1:025008. doi: 10.1117/1.NPh.1.2.025008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Podor B, et al. Comparison of genetically encoded calcium indicators for monitoring action potentials in mammalian brain by two-photon excitation fluorescence microscopy. Neurophotonics. 2015;2:021014. doi: 10.1117/1.NPh.2.2.021014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Helassa N, et al. Fast-response calmodulin-based fluorescent indicators reveal rapid intracellular calcium dynamics. Sci. Rep. 2015;5:15978. doi: 10.1038/srep15978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang JW, Wong AM, Flores J, Vosshall LB, Axel R. Two-photon calcium imaging reveals an odor-evoked map of activity in the fly brain. Cell. 2003;112:271–282. doi: 10.1016/s0092-8674(03)00004-7. [DOI] [PubMed] [Google Scholar]
- 95.Ahrens MB, et al. Brain-wide neuronal dynamics during motor adaptation in zebrafish. Nature. 2012;485:471–477. doi: 10.1038/nature11057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dunn TW, et al. Brain-wide mapping of neural activity controlling zebrafish exploratory locomotion. Elife. 2016;5:e12741. doi: 10.7554/eLife.12741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nguyen JP, et al. Whole-brain calcium imaging with cellular resolution in freely behaving Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2016;113:E1074–E1081. doi: 10.1073/pnas.1507110112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Venkatachalam V, et al. Pan-neuronal imaging in roaming Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2016;113:E1082–E1088. doi: 10.1073/pnas.1507109113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lovett-Barron M, et al. Dendritic inhibition in the hippocampus supports fear learning. Science. 2014;343:857–863. doi: 10.1126/science.1247485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Siegel F, Lohmann C. Probing synaptic function in dendrites with calcium imaging. Exp. Neurol. 2013;242:27–32. doi: 10.1016/j.expneurol.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 101.Cichon J, Gan WB. Branch-specific dendritic Ca2+ spikes cause persistent synaptic plasticity. Nature. 2015;520:180–185. doi: 10.1038/nature14251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sun W, Tan Z, Mensh BD, Ji N. Thalamus provides layer 4 of primary visual cortex with orientation- and direction-tuned inputs. Nat. Neurosci. 2016;19:308–315. doi: 10.1038/nn.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Melom JE, Akbergenova Y, Gavornik JP, Littleton JT. Spontaneous and evoked release are independently regulated at individual active zones. J. Neurosci. 2013;33:17253–17263. doi: 10.1523/JNEUROSCI.3334-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Akerboom J, et al. Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Front. Mol. Neurosci. 2013;6:2. doi: 10.3389/fnmol.2013.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhao Y, et al. An expanded palette of genetically encoded Ca2+ indicators. Science. 2011;333:1888–1891. doi: 10.1126/science.1208592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Inoue M, et al. Rational design of a high-affinity, fast, red calcium indicator R-CaMP2. Nat. Methods. 2015;12:64–70. doi: 10.1038/nmeth.3185. [DOI] [PubMed] [Google Scholar]
- 107.Berlin S, et al. Photoactivatable genetically encoded calcium indicators for targeted neuronal imaging. Nat. Methods. 2015;12:852–858. doi: 10.1038/nmeth.3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Matsuda T, Horikawa K, Saito K, Nagai T. Highlighted Ca2+ imaging with a genetically encoded ‘caged’ indicator. Sci. Rep. 2013;3:1398. doi: 10.1038/srep01398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hoi H, Matsuda T, Nagai T, Campbell RE. Highlightable Ca2+ indicators for live cell imaging. J. Am. Chem. Soc. 2013;135:46–49. doi: 10.1021/ja310184a. [DOI] [PubMed] [Google Scholar]
- 110.Ai M, et al. Green-to-red photoconversion of GCaMP. PLoS One. 2015;10:e0138127. doi: 10.1371/journal.pone.0138127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sattarzadeh A, Saberianfar R, Zipfel WR, Menassa R, Hanson MR. Green to red photoconversion of GFP for protein tracking in vivo. Sci. Rep. 2015;5:11771. doi: 10.1038/srep11771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fosque BF, et al. Neural circuits.Labeling of active neural circuits in vivo with designed calcium integrators. Science. 2015;347:755–760. doi: 10.1126/science.1260922. [DOI] [PubMed] [Google Scholar]
- 113.Chu J, et al. A bright cyan-excitable orange fluorescent protein facilitates dual-emission microscopy and enhances bioluminescence imaging in vivo. Nat. Biotechnol. 2016;34:760–767. doi: 10.1038/nbt.3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sun XR, et al. Fast GCaMPs for improved tracking of neuronal activity. Nat. Commun. 2013;4:2170. doi: 10.1038/ncomms3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fenno L, Yizhar O, Deisseroth K. The development and application of optogenetics. Annu. Rev. Neurosci. 2011;34:389–412. doi: 10.1146/annurev-neuro-061010-113817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhou XX, Pan M, Lin MZ. Investigating neuronal function with optically controllable proteins. Front. Mol. Neurosci. 2015;8:37. doi: 10.3389/fnmol.2015.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jagadish S, Barnea G, Clandinin TR, Axel R. Identifying functional connections of the inner photoreceptors in Drosophila using Tango-Trace. Neuron. 2014;83:630–644. doi: 10.1016/j.neuron.2014.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kawashima T, Okuno H, Bito H. A new era for functional labeling of neurons: activity-dependent promoters have come of age. Front. Neural Circuits. 2014;8:37. doi: 10.3389/fncir.2014.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Seo J, Choe M, Kim SY. Clearing and labeling techniques for large-scale biological tissues. Mol. Cells. 2016;39:439–446. doi: 10.14348/molcells.2016.0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Susaki EA, Ueda HR. Whole-body and whole-organ clearing and imaging techniques with single-cell resolution: toward organism-level systems biology in mammals. Cell Chem. Biol. 2016;23:137–157. doi: 10.1016/j.chembiol.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 121.Grosenick L, Marshel JH, Deisseroth K. Closed-loop and activity-guided optogenetic control. Neuron. 2015;86:106–139. doi: 10.1016/j.neuron.2015.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jorgenson LA, et al. The BRAIN Initiative: developing technology to catalyse neuroscience discovery. Phil. Trans. R. Soc. Lond. B Biol. Sci. 2015;370:20140164. doi: 10.1098/rstb.2014.0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Piao HH, Rajakumar D, Kang BE, Kim EH, Baker BJ. Combinatorial mutagenesis of the voltage-sensing domain enables the optical resolution of action potentials firing at 60 Hz by a genetically encoded fluorescent sensor of membrane potential. J. Neurosci. 2015;35:372–385. doi: 10.1523/JNEUROSCI.3008-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Patterson GH, Knobel SM, Sharif WD, Kain SR, Piston DW. Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy. Biophys. J. 1997;73:2782–2790. doi: 10.1016/S0006-3495(97)78307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Li Y, Tsien RW. pHTomato, a red, genetically encoded indicator that enables multiplex interrogation of synaptic activity. Nat. Neurosci. 2012;15:1047–1053. doi: 10.1038/nn.3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li H, et al. Concurrent imaging of synaptic vesicle recycling and calcium dynamics. Front. Mol. Neurosci. 2011;4:34. doi: 10.3389/fnmol.2011.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shaner NC, et al. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat. Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hires SA, Zhu Y, Tsien RY. Optical measurement of synaptic glutamate spillover and reuptake by linker optimized glutamate-sensitive fluorescent reporters. Proc. Natl. Acad. Sci. USA. 2008;105:4411–4416. doi: 10.1073/pnas.0712008105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rizzo MA, Springer GH, Granada B, Piston DW. An improved cyan fluorescent protein variant useful for FRET. Nat. Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- 130.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat. Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 131.Lam AJ, et al. Improving FRET dynamic range with bright green and red fluorescent proteins. Nat. Methods. 2012;9:1005–1012. doi: 10.1038/nmeth.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lundby A, Akemann W, Knöpfel T. Biophysical characterization of the fluorescent protein voltage probe VSFP2.3 based on the voltage-sensing domain of Ci-VSP. Eur. Biophys. J. 2010;39:1625–1635. doi: 10.1007/s00249-010-0620-0. [DOI] [PubMed] [Google Scholar]
- 133.Mishina Y, Mutoh H, Knöpfel T. Transfer of Kv3.1 voltage sensor features to the isolated Ci-VSP voltage-sensing domain. Biophys. J. 2012;103:669–676. doi: 10.1016/j.bpj.2012.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jin L, et al. Single action potentials and subthreshold electrical events imaged in neurons with a fluorescent protein voltage probe. Neuron. 2012;75:779–785. doi: 10.1016/j.neuron.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cranfill PJ, et al. Quantitative assessment of fluorescent proteins. Nat. Methods. 2016;13:557–562. doi: 10.1038/nmeth.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shcherbo D, et al. Far-red fluorescent tags for protein imaging in living tissues. Biochem. J. 2009;418:567–574. doi: 10.1042/BJ20081949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lütcke H, et al. Optical recording of neuronal activity with a genetically-encoded calcium indicator in anesthetized and freely moving mice. Front. Neural Circuits. 2010;4:9. doi: 10.3389/fncir.2010.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Nagai T, Yamada S, Tominaga T, Ichikawa M, Miyawaki A. Expanded dynamic range of fluorescent indicators for Ca2+ by circularly permuted yellow fluorescent proteins. Proc. Natl. Acad. Sci. USA. 2004;101:10554–10559. doi: 10.1073/pnas.0400417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chen JL, Carta S, Soldado-Magraner J, Schneider BL, Helmchen F. Behaviour-dependent recruitment of long-range projection neurons in somatosensory cortex. Nature. 2013;499:336–340. doi: 10.1038/nature12236. [DOI] [PubMed] [Google Scholar]
- 140.Horikawa K, et al. Spontaneous network activity visualized by ultrasensitive Ca2+ indicators, yellow Cameleon-Nano. Nat. Methods. 2010;7:729–732. doi: 10.1038/nmeth.1488. [DOI] [PubMed] [Google Scholar]
- 141.Palmer AE, et al. Ca2+ indicators based on computationally redesigned calmodulin-peptide pairs. Chem. Biol. 2006;13:521–530. doi: 10.1016/j.chembiol.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 142.Tian L, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat. Methods. 2009;6:875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mues M, et al. Real-time in vivo analysis of T cell activation in the central nervous system using a genetically encoded calcium indicator. Nat. Med. 2013;19:778–783. doi: 10.1038/nm.3180. [DOI] [PubMed] [Google Scholar]
- 144.Akerboom J, et al. Optimization of a GCaMP calcium indicator for neural activity imaging. J. Neurosci. 2012;32:13819–13840. doi: 10.1523/JNEUROSCI.2601-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]