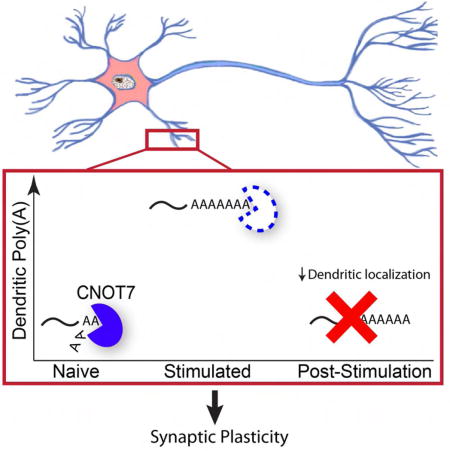
Summary
Translation of mRNAs in dendrites mediates synaptic plasticity, the probable cellular basis of learning and memory. Coordination of translational inhibitory and stimulatory mechanisms as well as dendritic transport of mRNA is necessary to ensure proper control of this local translation. Here, we find that the deadenylase CNOT7 dynamically regulates dendritic mRNA translation and transport as well as synaptic plasticity and higher cognitive function. In cultured hippocampal neurons, synaptic stimulation induces a rapid decrease in CNOT7 which in the short-term results in poly(A) tail lengthening of target mRNAs. However, at later times following stimulation, decreased poly(A) and dendritic localization of mRNA take place, similar to what is observed when CNOT7 is depleted over several days. In mice, CNOT7 is essential for hippocampal-dependent learning and memory. This study identifies CNOT7 as an important regulator of RNA transport and translation in dendrites as well as higher cognitive function.
Keywords: CNOT7, polyadenylation, deadenylation, translation, RNA transport
eTOC Blurb
McFleder et al. find that CNOT7, the major deadenylase in eukaryotes, has a specialized role in neurons. CNOT7 regulates the dendritic localization and translation of specific mRNAs, and mediates synaptic plasticity and learning and memory in mice.
Introduction
Experience-induced modifications of synapses are thought to serve as the molecular basis of learning and memory (Kandel, 2001). Synaptic plasticity provides long-lasting alterations in neuronal communication that allows memories to be retained for many years (Costa-Mattioli et al., 2009). Of the several forms of synaptic plasticity, at least three are dependent on protein synthesis: long-lasting neurotrophin-induced enhancement of synaptic efficacy (Kang H, 1996), metabotropic glutamate receptor long-term depression (mGluR-LTD) (Huber KM, 2000), and N-methyl-D-aspartate late-phase long-term potentiation (L-LTP) (Bradshaw et al., 2003, Miller et al., 2002). The necessity for new protein production in synaptic plasticity is independent of transcription and relies upon mRNAs and translation factors in dendrites (Bradshaw et al., 2003, Kang H, 1996, Martin and Kandel, 1996). Following synaptic stimulation, dendritic mRNAs are translated at postsynaptic sites where their protein products modify synapse structure and function (Sutton and Schuman, 2006). Based on sequence analysis of RNAs in the mammalian hippocampal neuropil, a region rich in axons and dendrites, there are >2,500 mRNAs localized to neurites (Cajigas et al., 2012). It is almost axiomatic that regulation of these mRNAs is necessary to ensure localized translation in response to synaptic activity (Buxbaum et al., 2015); when this regulation goes awry, autism and other neurological disorders can ensue (Kelleher and Bear, 2008).
Repression of translation is necessary during mRNA transport to dendrites, but even when localized, silencing must continue until synaptic activity occurs (Costa-Mattioli et al., 2009). This mRNA masking takes multiple forms and involves repression at initiation (Krichevsky and Kosik, 2001) and elongation (Sutton et al., 2007, Richter, 2015). Although the mechanism(s) and/or factors by which these and other processes control mRNA expression in dendrites is often unclear, it is evident that they frequently involve miRNAs (Ashraf et al., 2006, Bicker et al., 2013, Schratt, 2009) or RNA binding proteins (Darnell, 2013, Eom et al., 2013, Udagawa et al., 2015). Many of these trans-acting factors utilize deadenylation as an initiation step to silence mRNAs (Ashraf et al., 2006, Giorgi et al., 2007, Richter, 2007). Mechanistically, deadenylases repress translation by shortening poly(A) tails and thereby abrogate association of poly(A) binding protein (PABP), which is important for circularizing mRNA and recruiting the 40S ribosomal subunit (Richter, 2007). The deadenylase PARN (poly(A) ribonuclease) was presumed to be the enzyme responsible for initiating repression for at least a subset of dendritic mRNAs because it interacts with the RNA-binding protein CPEB1 (Richter, 2007, Udagawa et al., 2012). In oocytes, CPEB1 regulates translation by recruiting both Gld2, a non-canonical poly(A) polymerase, and PARN to specific mRNAs (Richter, 2007). Upon phosphorylation of CPEB, PARN is expelled from the ribonucleoprotein complex, which results in polyadenylation and subsequently translation of target mRNAs. In the brain, CPEB1 and Gld2 mediate translation in dendrites in response to synaptic stimulation (Wu et al., 1998, Zearfoss et al., 2008, Udagawa et al., 2012). However, because depletion of PARN had no discernable effect on synaptic plasticity, the deadenylase essential for repressing translation was unclear (Udagawa et al., 2012). We surmised that one or perhaps multiple deadenylases would likely govern poly(A) tail length of several different populations of dendritic mRNAs to impact learning and memory.
The carbon catabolite repression 4 negative on TATA-less (CNOT) is a conserved, multisubunit complex that functions as the major deadenylase in yeast to humans (Tucker et al., 2001, Temme et al., 2004, Schwede et al., 2008). The mammalian CNOT complex consists of four functional deadenylase enzymes: CNOT6, CNOT6L, CNOT7, and CNOT8 (Lau et al., 2009). Of these, CNOT7 and CNOT8 regulate poly(A) tail length on the majority of mRNAs (Schwede et al., 2008). Although CNOT7 and CNOT8 are 75% homologous, they have distinct targets, probably because they are differentially expressed and associate with different complexes (Lau et al., 2009). CNOT7 levels are enriched in neurons relative to CNOT8 (Chen et al., 2011); CNOT7 also associates with both the microRNA machinery (Fabian et al., 2009, Piao et al., 2010) and CPEB1 (Ogami et al., 2014). These data suggest that CNOT7 might influence translation in dendrites, and as a consequence modify synaptic transmission and higher cognitive function.
Here we identify CNOT7 as an important enzyme that regulates local translation, synapse efficacy, and learning and memory. Within 3 minutes following induction of synaptic plasticity in vitro by glycine-induced LTP, CNOT7 levels begin to decrease, which is necessary for the immediate increase in total dendritic poly(A) occurring at this time. Interestingly twenty minutes following stimulation, when CNOT7 levels are low, total dendritic poly(A) is decreased. We found that these different effects are due to short-term versus long-term depletion of CNOT7. Paradoxically, poly(A) tails are lengthened following both short-term (≤ 10 min) and long-term CNOT7 depletion (≥ 20 min), but long-term depletion induced by stimulation of LTP or knockdown of the enzyme also impairs dendritic localization of CNOT7 target mRNAs resulting in reduced dendritic poly(A). These observations indicate a critical role for CNOT7 in localization and deadenylation of dendritic mRNAs. The effect of long-term CNOT7 depletion is most apparent after four days, which in addition to the effects stated above, also resulted in reduced local translation as well as impaired synaptic plasticity. Depletion of CNOT7 in the hippocampus over several weeks following injection of AAV-expressed shRNA resulted in reduced poly(A) in the CA1 neuropil and impaired learning and memory in several cognitive tests. These and other data demonstrate that CNOT7 governs the localization, polyadenylation, and translation of specific dendritic mRNAs and that it has a key role in synaptic plasticity, learning, and memory.
Results
Depletion of CNOT7 decreases dendritic poly(A) and local protein synthesis
We analyzed the mRNA levels of all nine known cytoplasmic deadenylases (Angel1, Angel2, CNOT6, CNOT6L, CNOT7, CNOT8, Nocturnin, Pan2, and PARN) in the liver, cerebellum, hippocampus, and cortex from 40 day old mice. CNOT7 and CNOT8 were enriched in the hippocampus compared to the other enzymes (Figure S1A). CNOT7 RNA also exceeds the levels of all other deadenylases in the hippocampal CA1 neuropil, an area enriched for dendrites (Figure S1B) (Cajigas et al., 2012). Immuno-staining of cultured hippocampal neurons (DIV17) showed that CNOT7 is present throughout the cells including dendrites (Figure 1A), and is significantly reduced upon shRNA-mediated depletion (Figure S1C). Western blot analysis of mouse brain lysates revealed CNOT7 to be present in synaptosomes (Figure S1D), suggesting that it may have a synaptic function.
Figure 1.
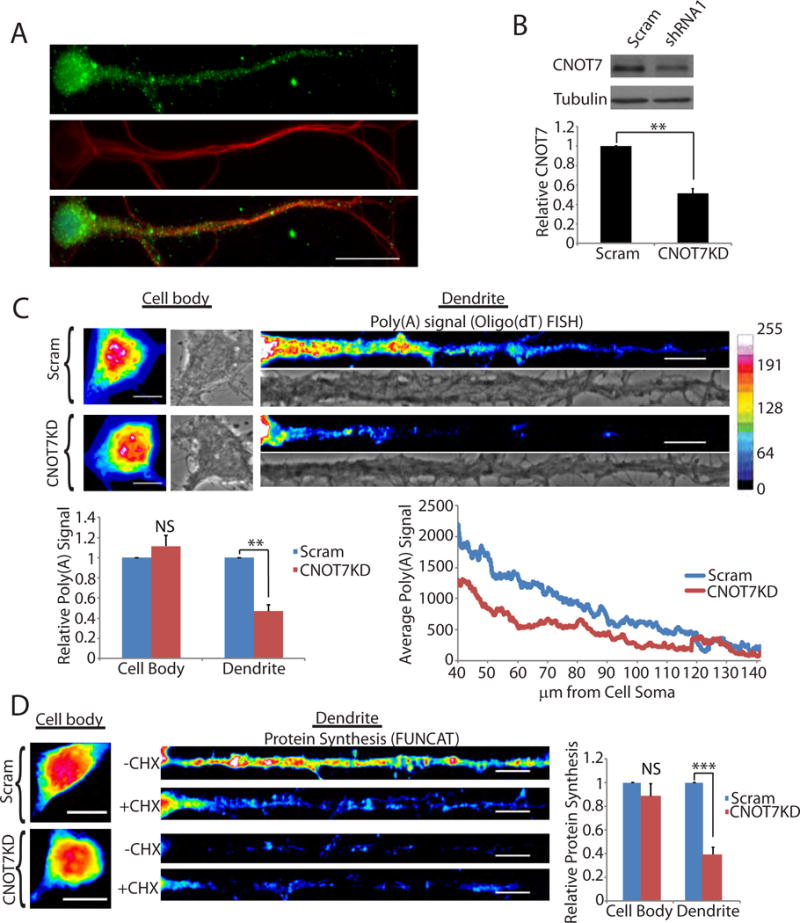
CNOT7 regulates dendritic poly(A). (A) Immunocytochemistry of cultured hippocampal neurons DIV 17 for CNOT7 (green), MAP2 (red), and DAPI (Blue). Scale bar represents 20μm (B) Representative western blot of CNOT7 (top) and tubulin (bottom). Histogram represents the average of three experiments. (C) (Top) Representative brightfield and oligo(dT) fluorescent in situ hybridization (FISH) images from the cell body (left) and dendrites (right) of scrambled (Scram) or CNOT7 knockdown (CNOT7KD) neurons. Scale bar represents 10μm. (Bottom) Bar and line graph are averages of the oligo(dT) FISH signal in ≥60 neurons/condition from three experimental replicates. (D) FUNCAT analysis of scrambled and CNOT7 knockdown neurons in the presence (+CHX) or absence (−CHX) of cycloheximide. Bar graph is the average of ≥40 neurons/condition from three experimental replicates. In these and all subsequent figures, the error bars represent SEM, *p<0.05, **p<0.01, ***p<0.001. NS, not significant. See also Figure S1.
Using fluorescence in situ hybridization (FISH) with labeled oligo(dT), we analyzed poly(A) in cultured hippocampal neurons four days after infection with a lentivirus expressing either a CNOT7-specific shRNA or a scrambled control (Figure 1B). Surprisingly, CNOT7 knockdown (CNOT7KD) resulted in an ~50% decrease in dendritic poly(A), not the expected increase. This result occurred mostly on dendritic mRNA because the cell body poly(A) FISH signal (Figure 1C) as well as total cellular poly(A) (Figure S1E), was not significantly affected. The decrease in dendritic poly(A) signal was replicated with a second shRNA targeting CNOT7 (Figure S1F & G). Knockdown of CNOT8, the CNOT7 paralogue, had no effect on dendritic poly(A) (Figure S1H). We ectopically expressed a catalytically-inactive form of the enzyme (D40A, mtCNOT7) as well as the wild-type enzyme (WTCNOT7) and performed oligo(dT) FISH. Neurons expressing mtCNOT7 had decreased dendritic poly(A) signal similar to the CNOT7KD neurons, while ectopic expression of WTCNOT7 produced the opposite effect (Figure S1I & J). Ectopic expression of wildtype CNOT7 was able to rescue the reduced dendritic poly(A), indicating that this dramatic effect was due to the loss of CNOT7 and not non-specific effects (Figure SK). Newly synthesized proteins, as measured by Fluorescent Non-Canonical Amino Acid Tagging (FUNCAT), decreased by ~60% in dendrites of CNOT7KD neurons but only modestly in their cell bodies (Figure 1D), similar to the observations of poly(A). Ectopic expression of WTCNOT7 resulted in increased dendritic FUNCAT signal (Figure S1L). We also treated neurons with nocodazole, a microtubule depolymerizing agent that disrupts microtubules and inhibits protein transport to dendrites (Cid-Arregui et al., 1995, Kohrmann et al., 1999, Yuen et al., 2005). This procedure distorted tubulin staining (Figure S1M), and is admittedly quite stressful to the neurons however the neurons still produced protein as shown by FUNCAT labeling and this labeling was reduced in distal dendrites of CNOT7KD neurons relative to control shRNA-expressing neurons by 32% (Figure S1M). These data indicate that CNOT7 mediates dendritic translation.
It was possible that that the decrease in dendritic poly(A) signal following CNOT7KD could be due to increased protein binding to mRNA and not reduced poly(A) (Buxbaum et al., 2014). Consequently, we treated cultured neurons with pepsin prior to FISH, which caused an ~2 fold increase in poly(A) signal in the control neurons, indicating that proteins do obscure probe hybridization to poly(A) (Figure 2A, B). CNOT7KD neurons, however, did not display an increased poly(A) signal following pepsin digestion, indicating that in dendrites, there is reduced poly(A) following CNOT7 depletion.
Figure 2.
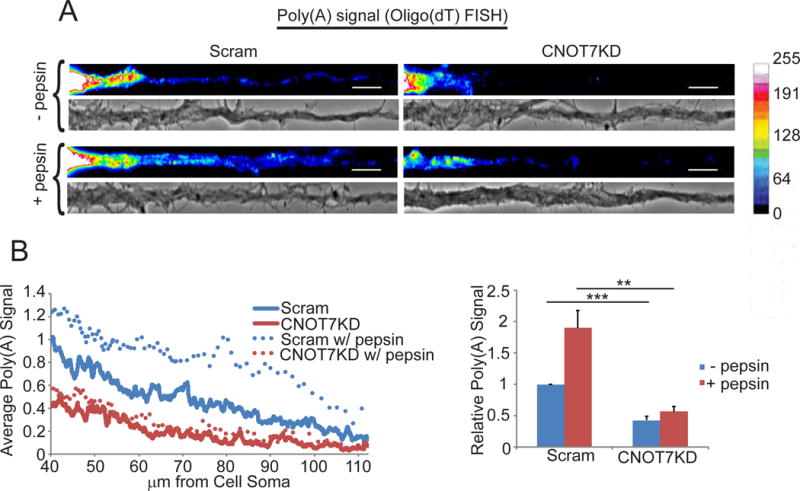
CNOT7 does not regulate masking of dendritic mRNAs. (A) Representative images and (B) quantification of oligo(dT) FISH signal in scrambled or CNOT7KD dendrites with (+) or without (−) pepsin. Scale bars represent 10μm. Bar and line graphs are the average oligo(dT) FISH signal in ≥60 dendrites/condition from 3 experimental replicates
Knockdown of CNOT7 decreases synaptic plasticity in vitro
Because poly(A) regulation in dendrites is correlated with alterations in synaptic plasticity (Udagawa et al., 2012), we tested whether CNOT7 modulates synapse function. We evoked one form of synaptic plasticity, glycine-induced LTP (also referred to as chem-LTP), in CNOT7 depleted or control neurons. Twenty minutes after glycine treatment, phosphorylation of GluR1 at S831 and S845 increased 3–4 fold in control neurons, indicating successful induction of LTP (Figure 3A & S2A) (Lee H, 2000). In CNOT7KD neurons, S831 phosphorylation modestly increased ~1.5 fold while S845 phosphorylation was virtually unchanged (Figure 3A, S2A, and S2B). Depletion of CNOT8 did not impede the increase in S845 phosphorylation following stimulation (Figure S2B), indicating that the inhibition of GluR1 phosphorylation was specific to CNOT7. Recycling of GluR1 to the membrane surface of dendrites was also impaired in CNOT7KD neurons (Figure 3B & C). This impairment was not due to decreased GluR1 because the level of this protein was unchanged between scrambled or CNOT7KD neurons (Figures 3A & S2C). These data suggest that CNOT7KD neurons have impairment in LTP induction.
Figure 3.
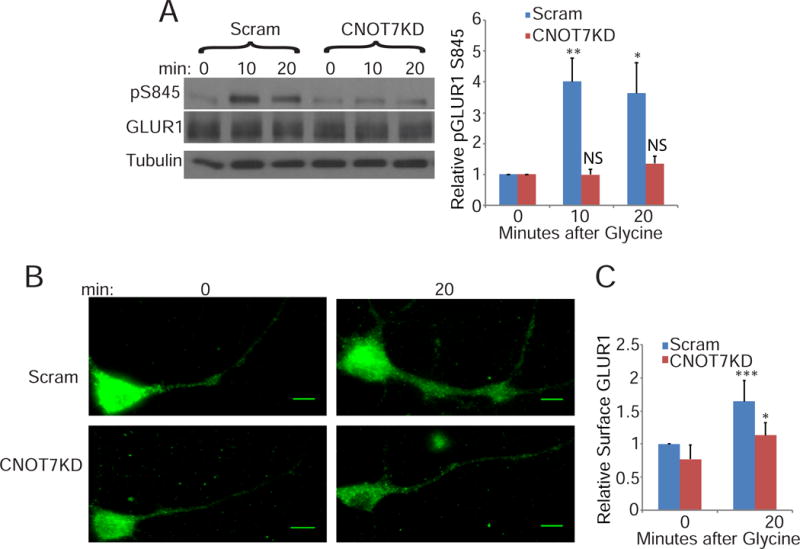
CNOT7KD inhibits long term potentiation. (A) Western blot analysis of S845 GluR1 phosphorylation in scrambled or CNOT7KD neurons at different time points following glycine stimulation. Phospho-GluR1 was normalized to total GluR1 and the bar graphs represent the average of three experiments. Time points are plotted relative to the 0 time point. (B) Representative images and (C) quantification of surface GluR1 in scrambled and CNOT7KD neurons fixed at 0 or 20 minutes following glycine stimulation. Scale bars represent 10μm. (C) Bar graph represents the average surface GluR1 in ≥50 dendrites/condition from three experimental replicates relative to the scrambled control. See also Figure S2.
CNOT7 regulates LTP induced changes in polyadenylation
We induced chem-LTP in cultured hippocampal neurons and collected protein 0, 3, 10, and 20 minutes later. Twenty minutes was chosen as the final time point because that is when both the amplitude and frequency of miniature excitatory post-synaptic currents (mEPSCs) are at their peaks (Lu et al., 2001). Western blotting revealed that CNOT7 decreased in the neurons to ~50% of pre-stimulation levels (Figure 4A). Immunocytochemistry of neurons fixed at 0 or 20 minutes post-glycine, revealed that the decrease in CNOT7 was more substantial in dendrites relative to cell bodies, 53% versus 29%, respectively (Figure 4B). The NMDA receptor antagonist MK801 prevented this in both the cell body and dendrites (Figure S3A).
Figure 4.
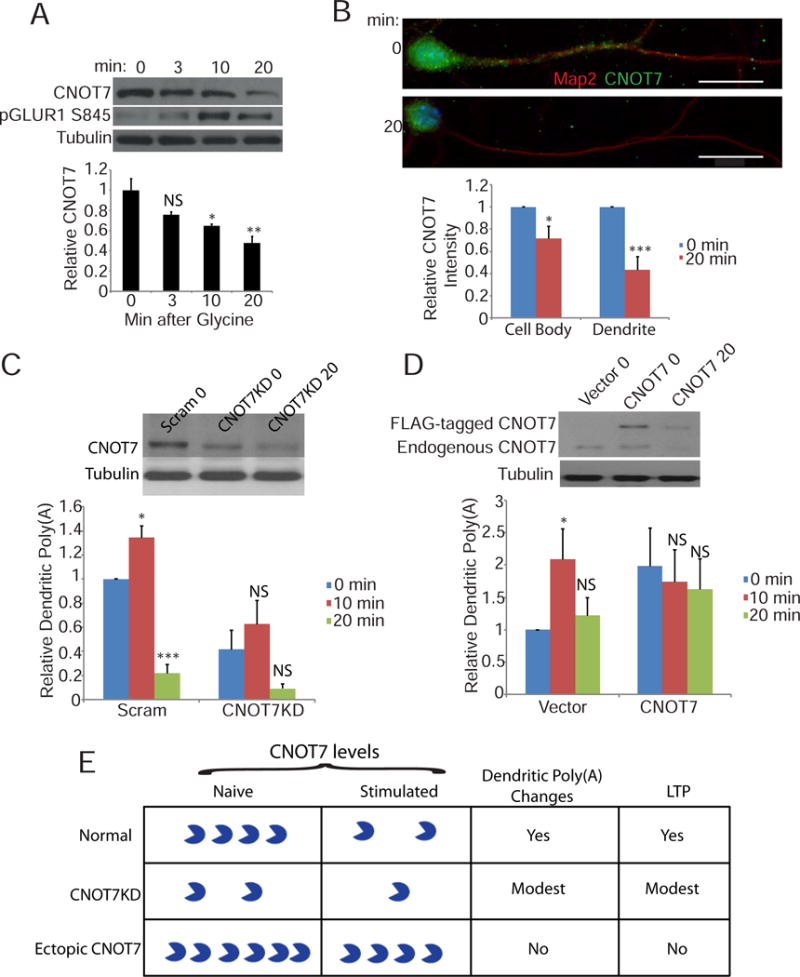
CNOT7 regulates dendritic poly(A) following synaptic plasticity. (A) Western blot analysis of CNOT7 (top), pGluR1 S845 (middle), and tubulin (bottom) at different time points following glycine stimulation. CNOT7 was normalized to tubulin and the bar graph represents the average of three experiments. Time points are plotted relative to the 0 time point. (B) Representative images of CNOT7 immunofluorescence at different time points following stimulation. Bar graphs represent the average relative CNOT7 intensity in either the soma (left) or dendrites (right) from ≥40 neurons/condition from three experimental replicates. (C) (Top) Western blot depicts CNOT7 and tubulin levels in neurons infected with scrambled shRNA (scram) 0 minutes after glycine treatment, or CNOT7 shRNA 0 or 20 min following glycine treatment. The histogram depicts average relative dendritic poly(A) signal in stimulated cells infected with either a scrambled or CNOT7 targeting shRNA from ≥24 neurons/condition from three experimental replicates. All conditions are plotted relative to the scrambled control 0 time point. (D) (Top) Western blot depicts CNOT7 and tubulin levels in neurons expressing empty vector (vector) 0 minutes following glycine stimulation, or expressing ectopic FLAG-CNOT7 at 0 or 20 min following glycine stimulation. The histogram depicts average relative dendritic poly(A) signal in stimulated cells infected with either empty vector or CNOT7-expressing lentivirus. Data are from ≥26 neurons/condition from three experimental replicates. All conditions are plotted relative to the vector control 0 time point. (E) Summary diagram showing that in normal neurons, glycine stimulation causes an ~50% decrease in CNOT7, which is correlated with a change in dendritic poly(A) and LTP induction. In CNOT7KD neurons, ~50% of normal CNOT7 is present, which decreases by an additional 50% 20 min after glycine stimulation. These CNOT7 levels are correlated with modest changes in dendritic poly(A) changes and LTP induction. Ectopic expression of FLAG-CNOT7 in neurons results in about a doubling of this protein. Twenty minutes after glycine treatment, CNOT7 levels fall to about the same level as in control (vector) neurons at time 0. Consequently, there are no changes in dendritic poly(A) and LTP is not induced. See also Figure S3.
To determine whether this decrease in CNOT7 was due to ubiquitination, we expressed hemagglutinin-tagged ubiquitin (HA-Ub) in neurons, immunoprecipitated for HA, and western blotted for CNOT7. MG132 was added to all cells to inhibit the degradation of ubiquitinated proteins. Several CNOT7-ubiquitin conjugates were identified in the HA-Ub expressing cells (denoted with numbers in the figure) that were absent from control cells not expressing HA-Ub, indicating that CNOT7 is ubiquitinated in neurons (Figure S3B). Stimulation did not increase the abundance or number of these CNOT7-ubiquitin conjugates, however, this could be due to the presence of MG132 which inhibits the proteasomal machinery, vital for LTP induction (Alvarez-Castelao and Schuman, 2015).
Oligo(dT) FISH on neurons fixed at various times following stimulation, demonstrated that dendritic poly(A) increases to ~160% of pre-stimulation levels 10 min after glycine treatment (Figure S3C). Surprisingly, dendritic poly(A) then decreases to pre-stimulation levels at the 20 minute time point, which coincides with when CNOT7 levels are at 50% of their pre-stimulation levels and when mEPSCs are at their peak (Lu et al., 2001) (Figure S3C).
The effect of glycine stimulation on dendritic poly(A) under normal conditions or upon CNOT7 depletion was examined. Figure 4C demonstrates that shRNA treatment resulted in a ~50% knockdown of CNOT7, which declined by an additional 50% upon glycine stimulation. In control (scrambled shRNA-infected) neurons, dendritic poly(A) increased at 10 minutes post-glycine but fell dramatically at 20 minutes. This same biphasic trend also occurred in CNOT7KD neurons, although the differences were not statistically different. This result is not surprising because the ~50% of control levels of CNOT7 present in these cells, is still under stimulation-induced regulation and sufficient to elicit mild changes in dendritic poly(A). Moreover, these reduced levels of CNOT7 are still adequate for glycine to promote modest LTP as assessed by phosphorylation of GluR1 and surface GluR1 immuno-staining (Figure 3).
To further investigate the importance of CNOT7 in stimulation-induced biphasic changes in dendritic poly(A), we ectopically expressed FLAG-tagged CNOT7 in neurons followed by glycine treatment (Figure 4D). CNOT7 levels were approximately double relative to those expressing only the vector (compare vector 0 and CNOT7 0). This high level of CNOT7 results in increased dendritic poly(A) (Figure 4D, Figure S1J). Glycine treatment caused destruction of exogenous and endogenous CNOT7, but the total amount of CNOT7 remaining was nearly identical to that observed in naïve (vector only expressing) cells. Maintenance of this near-control level of CNOT7 inhibited the increase and decrease in dendritic poly(A) at 10 and 20 min post-glycine treatment (Figure 4D). This near-control level of CNOT7 also impaired LTP induction as measured by pGluR1 S845 (Figure S3D). These data indicate that rapid stimulation-induced depletion of CNOT7 is necessary for stimulation-induced changes in dendritic poly(A) and induction of LTP (Figure 4E).
CNOT7 regulates polyadenylation and stability of specific neuronal mRNAs
We sought to identify specific CNOT7 target mRNAs. RNA from control or CNOT7KD neurons was incubated with poly(U) agarose; washed at 50°C to elutes mRNAs with short (< 50 nucleotides) poly(A) tails (Du and Richter, 2005) (Figure S4A); and eluted mRNAs with longer poly(A) tails (>50 nucleotides) at 75°C. Sequencing of the mRNAs with long poly(A) tails identified 97 that were differentially distributed between the scrambled and CNOT7KD samples (Figure 5A). Most of these (~65%) were enriched in the long tailed sample following CNOT7KD, suggesting that they could be direct targets of CNOT7. Gene Ontology term analysis (GO terms) indicated that many are involved in neural development and function (Figure 5B).
Figure 5.

CNOT7 regulates poly(A) tail length and stability of specific mRNAs. (A) Heatmap of the 97 differentially expressed mRNAs eluted from poly(U) agarose at 75° (high temp) following CNOT7KD. (B) Bar graph representing the top 5 GO terms for the 63 mRNAs enriched in the high temperature samples following CNOT7KD. (C) Venn diagram of genes differentially expressed in either the input samples (pink) or the high temperature (blue) samples. (D) (Top) Diagram depicts placement of primers (arrows) to detect mature mRNA or pre-mRNA; black boxes represent the exons and lines represent the introns. (bottom) Representative gel images and quantification of 4 different mature mRNAs (Uchl1, Cdkl2, Shisa6, or SNCA) and their corresponding pre-mRNAs in scrambled and CNOT7KD neurons. Bar graph depicts the average of three different experiments. (E) Quantification of Uchl1 mRNA in either scrambled or CNOT7KD neurons at the indicated time points following the addition of actinomycin D. The graph represents the average of two different experiments. (F) Representative northern blot analysis of Uchl1 in neurons ectopically expressing either empty vector or catalytically-inactive mutant CNOT7 (mtCNOT7). Line graph represents the Uchl1 band intensity relative to the intensity of each respective band at the 600 base mark. See also Figure S4.
Many mRNAs from neurons depleted of CNOT7 that were disproportionately eluted from poly(U) at 75° also underwent alterations in their steady state levels as assessed by RNA-seq and RT-PCR (Figure 5C & D). We examined four RNAs that increased (Uchl1 & Cdkl2) or decreased (SNCA & Shisa6) in the poly(U) 75° elution fraction in CNOT7KD neurons. RT-PCR with primers spanning an exon-exon junction was used to assess predominantly cytoplasmic RNA, and primers spanning an exon-intron junction were used to detect pre-mRNA, which serves as a proxy for transcription (Figure 5D). Because RNA levels increased in CNOT7KD cells only when analyzed with exon-exon primers, we infer that enhanced RNA stability was likely the cause of the changes in transcript levels upon CNOT7 depletion (Figure 5D). To confirm this, we examined Uchl1 (ubiquitin C-terminal hydrolase L1) RNA because CNOT7 depletion elicited a large change in the apparent poly(A) tail size as well its steady state RNA levels. Moreover, Uchl1 is an abundant mRNA that encodes a protein involved in synaptic plasticity (Gong et al., 2006, Hegde et al., 1997). To measure the decay rate of Uchl1 mRNA, control or CNOT7KD neurons were treated with actinomycin D to inhibit transcription and cells were collected 0–9 hours later. Figure 5E shows that although there was little change in the Uchl1 RNA in CNOT7KD cells, the transcript underwent a steady decline in control cells, confirming that this deadenylating enzyme mediates RNA instability. Furthermore, ectopic expression of CNOT7 in CNOT7KD cells appeared to partially rescue the increase in both Uchl1 and Cdkl2 mRNA, indicating that this increased stability was due to CNOT7 depletion (Figure S4B).
We attempted to assess Uchl1 RNA poly(A) tail size by northern blotting (Figure S4C) or with a RT-PCR based poly(A) tail-length assay (Figure S4D). The large increase in Uchl1 mRNA following CNOT7KD obscured any tail size changes. Because of the slower kinetics involved in ectopic expression of a dominant-negative catalytically-inactive form of CNOT7 (D40A), we suspected this method would allow us to detect Uchl1 after it gained a poly(A) tail but before it had time to accumulate due to increased stability (Figure 5E). Because Uchl1 is a large transcript (1156 bases), we annealed RNA from control and CNOT7 D40A-expressing neurons with an antisense oligonucleotide positioned 606 nucleotides from the 3′ end, which was followed by RNAse H cleavage and northern analysis. Figures 5F, S4E, and S4F show that the Uchl1 median tail size lengthened from ~49nt to ~118nt following the D40A mutant expression. We also demonstrated that FLAG-mtCNOT7 interacts directly with Uchl1 mRNA by formaldehyde crosslinking, FLAG immunopecipitation, and RT-PCR for Uclhl1 and GAPDH mRNAs (Figure S4G). Wild-type CNOT7 was not used, because it would increase deadenylation and likely degradation of Uchl1 RNA. These results indicate that CNOT7 directly regulates the poly(A) tail length and overall stability of Uchl1 and likely many other RNAs identified in Figure 5.
Differential localization and polyadenylation of CNOT7 targets following long-term CNOT7 depletion
Our data seem paradoxical: CNOT7 knockdown or ectopic expression of a catalytically-inactive protein causes a reduction in dendritic poly(A) when analyzed by oligo(dT) FISH (Figures 1 and 2), yet also results in increased poly(A) tail size and/or stability of specific RNAs (Figure 5). To resolve this issue, we repeated the FISH experiments but examined specific mRNAs whose poly(A) and/or stability is regulated by CNOT7 (Figure 5). Our reasoning was that a possible differential localization of specific RNAs would not be discernable by oligo(dT) FISH, yet could at least partially explain the loss of dendritic poly(A) upon CNOT7 knockdown. For these experiments we utilized the ViewRNA ISH kit, which utilizes ~20 oligonucleotide pairs/target and only when the oligo pairs bind side by side is there fluorescent signal. This provides high specificity to this technique. FISH for Uchl1 RNA in control and CNOT7KD neurons shows that CNOT7 depletion resulted in decreased Uchl1 puncta in dendrites but increased signal in the cell body (Figure 6 A, B, and C). The RNA encoding Cdkl2 (cyclin dependent kinase like 2) also shifted from a dendritic to a cell body localization following CNOT7 knockdown. Thus, RNAs that gain poly(A) and/or are stabilized by CNOT7 knockdown also accumulate in the cell body over the four days of knockdown, at the expense of decreased localization to dendrites. On the other hand, SNCA (synuclein α) and Shisa6, two RNAs that had reduced poly(A) and/or steady state levels following CNOT7 knockdown, displayed decreased FISH signals in both dendrites and cell bodies (Figure 6A–C). We are confident these signals are specific, as they matched perfectly the changes we observed via qPCR (Figure 5C). CaMKIIα, (calmodulin-dependent kinase IIα) and PPIB (peptidylprolyl isomerase B) RNAs, whose poly(A) tail length and stability were not altered by CNOT7, exhibited no change in their localization upon CNOT7 depletion(Figure S5A). Control cells treated with all FISH reagents except for the targeting probe resulted in no detectable fluorescence (Figure S5B). Importantly, ectopic expression of CNOT7 caused an increase in dendritic localization of Uchl1 RNA (Figure S5C), opposite from what was observed in CNOT7 knockdown neurons. These data indicate that CNOT7 regulates dendritic localization of specific target RNAs.
Figure 6.
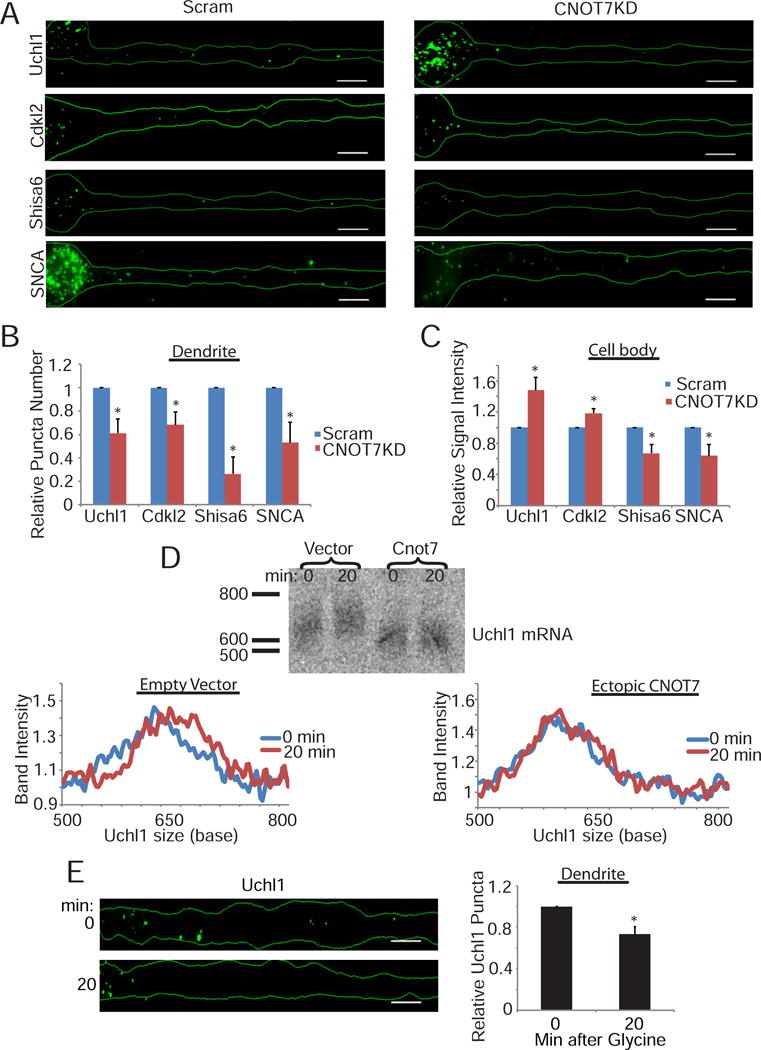
CNOT7 regulates its target’s localization and poly(A) tail length. (A) Representative images of FISH for specific targets (Uchl1, Cdkl2, Shisa6, or SNCA) in scrambled or CNOT7KD neurons. Scale bars represent 10μm. Bar graphs represent the average quantification of either the relative number of dendritic puncta (B) or relative cell body signal intensity (C) for the specific targets in either scrambled (blue) or CNOT7KD (red) neurons. Bar graphs represent the average signal from ≥40 neurons/condition plotted relative to the scrambled control. (D) (top) Northern blot of Uchl1 zero or twenty minutes following stimulation in either vector or CNOT7 expressing neurons. (bottom) Line graph represents the Uchl1 band intensity relative to the intensity of each respective band at the 500 base mark. (E) (left) Representative images for Uchl1 mRNA FISH in dendrites of neurons fixed either 0 minutes or 20 minutes following stimulation. (right) Bar graphs represent the average signal from ≥30 neurons/condition plotted relative to the 0 minute control. See also Figure S5.
RNA from neurons (DIV17–19) collected 0–20 minutes following chem-LTP was used for northern analysis of Uchl1. Chem-LTP induced a gradual, NMDA-dependent increase in Uchl1 RNA poly(A) over the 20 minutes following stimulation (Figure S5D & E). To determine whether this polyadenylation is mediated by the rapid depletion of CNOT7 as suggested by the data in Figure 4, wild type CNOT7 was ectopically expressed in cultured neurons followed by chem-LTP. Ectopic CNOT7 prevented the glycine-induced increase in Uchl1 poly(A) (Figure 6D). We conclude that rapid destruction of CNOT7 is essential for stimulation-induced polyadenylation of target mRNAs. We next performed FISH for Uchl1 RNA in dendrites fixed at 0 or 20 minutes post-stimulation. Uchl1 RNA exhibited decreased dendritic localization in response to stimulation (Figure 6E). These data indicate that depletion of CNOT7 following stimulation could activate translation through lengthening of dendritic mRNA poly(A) tails and, over time (i.e. long-term depletion), inhibit translation by impairing localization of new mRNAs to dendrites. Because both long-term stimulation-induced depletion and CNOT7 knockdown have similar effects, we consider long-term depletion of CNOT7 to be 20 minutes to several days.
Depletion of CNOT7 in the hippocampus decreases poly(A) in the neuropil and impairs cognitive function
We injected AAV-expressing CNOT7 or scrambled shRNAs into the hippocampus of wild type mice (Figure S6A). The CNOT7KD mice exhibited significantly reduced CNOT7 in the CA1 region (Figure S6B). Two CNOT7 target RNAs, Uchl1 and Cdkl2, were increased in CA1 of these mice (Figure S6B), indicating that in vivo, CNOT7 activity is similar to that observed in cultured neurons. Oligo(dT) FISH signal was detected in hippocampal CA1 dendritic projections emanating from the cell body of scrambled shRNA-injected mice (Figure 7A, arrows). However, there was a strong reduction in FISH signal in these projections in CA1 from the CNOT7 shRNA-injected mice (Figure 7A). The average oligo(dT) signal in the neuropil relative to the corresponding somatic region was reduced by ~40%, indicating impaired dendritically localized poly(A) in vivo (Figure S6C).
Figure 7.
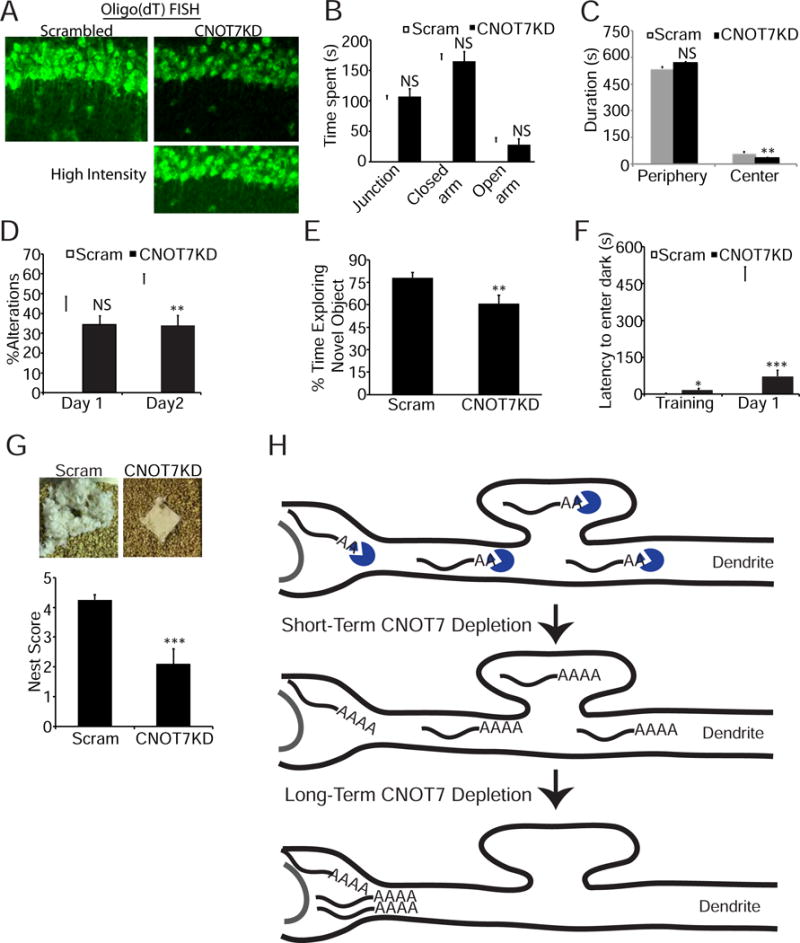
CNOT7KD in the hippocampus decreases poly(A) localization and impairs cognitive function. (A) Magnified images of oligo(dT) FISH signal in the CA1 neuropil of set 1 mice (Figure S6) injected with either scrambled or CNOT7 targeting shRNA. White arrows show examples of neurite-localized signal. Bottom right image is the CNOT7KD image with brightness level increased. (B) Bar graphs represent the average time (seconds) spent in the junction, closed, or open arms of an elevated plus maze (14 mice per condition). (C) Bar graph represents the average time in seconds spent in either the periphery or the center of an open field (14 mice/condition). (D) Bar graph represents the percent alterations performed in a T-maze (8–10 mice/condition). (E) Bar graph represents the percent time animals spent exploring a novel object (12–14 mice/condition). (F) Bar graph represents the latency to enter the dark compartment either on a training day or 24 hours after a foot shock in the dark (10–12 mice/condition). (G) Representative images (top) of nestlets from either scrambled or CNOT7KD mice. Bar graph (bottom) represents the average nest scores (10 mice/condition). (H) Model of major CNOT7 activities in neurons. The top diagram depicts mRNAs throughout a dendrite and cell body whose poly(A) tails are shortened by CNOT7 (blue). The middle diagram depicts a dendrite shortly after CNOT7 depletion, which occurs during glycine-induced LTP. Dendritic mRNAs have lengthened poly(A) tails. The bottom diagram depicts a dendrite after long-term CNOT7 depletion, such as by shRNA-mediated knockdown or ≥ 20min following stimulation. CNOT7 target RNAs retain long poly(A) tails but transport to dendrites is impeded; RNAs extant in dendrites are likely degraded. See also Figure S6.
We next tested the AAV-injected mice for anxiety by the elevated plus maze (EPM) and the open field test (OFT). For the EPM, mice choose to spend time in the closed or open arms of an elevated platform. More time spent in the closed arms indicates increased anxiety. CNOT7KD mice spent equal time in the closed arms as the scrambled controls (Figure 7B). The OFT, which measures the time mice spend in the center of an open field, showed that CNOT7KD mice spent 50% less time (31s vs 62s) in the center compared to control mice (Figure 7C). These results are not due to decreased locomotor activity as distance traveled in the open field test and the total number of entries into the arms of the elevated plus maze were comparable between groups (Figure S6D & E).
We used several assays to measure working (short term) and long-term memory. Working memory was assessed using the T-maze, where the spontaneous alternations of mice are measured in a T-shaped apparatus. On the habituation day one, the percent alternations were comparable between both sets of mice. On day 2, CNOT7KD mice had significantly decreased alterations compared to controls (33.6% vs 54.7%), indicating impaired working memory (Figure 7D). Long-term memory was assessed using two assays: novel object recognition and passive avoidance. In novel object recognition, mice were placed in an arena with a novel object and a familiar object. Mice that remember the familiar object spend more time with the novel object. CNOT7KD mice spent 61% of their time exploring the novel object compared to 78% for the control animals (Figure 7E). In the passive avoidance assay, mice were placed in a light chamber connected to a dark chamber by a door. The mice prefer the dark and immediately enter the dark chamber, after which the door closes and the mice were given a mild foot shock. Twenty-four hours later the mice were placed back in the light chamber and the latency to enter the dark was measured, with a longer latency indicating memory of the foot shock. Control animals had an average latency of 459 seconds while CNOT7KD animal latency was only 72 seconds (Figure 7F). Nest building was also impaired in CNOT7KD animals (Figure 7G). Increased anxiety, impaired learning and memory, and impaired nest building are shared features of various autistic models, which could suggest a role for CNOT7 in this disorder.
Discussion
This study identifies CNOT7 as a coordinator of mRNA transport and translation in dendrites and does so by modulating RNA poly(A) tail length and stability. Figure 7H shows a model that depicts some of the most salient activities of this enzyme in neurons. Although it is expressed throughout the cells, CNOT7 has a particularly important function in dendrites. In response to synaptic stimulation, it is gradually destroyed. This leads to polyadenylation of specific dendritic mRNAs and is likely followed by a burst of translation in dendrites (Udagawa et al., 2012). Twenty minutes after stimulation, the localization of these specific polyadenylated mRNAs are decreased in dendrites resulting in reduced overall dendritic poly(A), which is caused by the destruction of CNOT7. This event is recapitulated when CNOT7 is knocked down for 4 days, indicating stimulation-induced long-term (20 minutes) depletion of CNOT7 and shRNA-mediated knockdown in vivo for several days result in similar outcomes. When CNOT7 is depleted for 4 days, the enzyme’s target RNAs become more stable and are mostly confined to the cell body. The dendritic RNAs, at least in part, are then destroyed, resulting in decreased protein synthesis. Two additional events occur upon prolonged CNOT7 knockdown in vivo: reduction of poly(A) in CA1 dendrites and a decline in the performance in several cognitive tasks, indicating impaired learning and memory.
We were unable to confirm a physical link between CPEB1 and CNOT7 as shown by Ogami et al (2014), and consequently surmise that these proteins act independently, probably on unique sets of mRNAs to control poly(A) tail length and translation in dendrites. CPEB1 requires a 3′ UTR cytoplasmic polyadenylation element (CPE) to affect polyadenylation. The RNAs whose poly(A) tails are controlled by CNOT7, at least as identified by differential thermal elution from poly(U) beads, are not enriched for this element relative to total RNA. How CNOT7 might be tethered to specific RNAs such as Uchl1 is unknown but it may involve unique RNA binding proteins. These RNA binding proteins are likely unique to CNOT7 because depletion of CNOT8, the CNOT7 paralogue, has no obvious effect on dendritic poly(A) or chem-LTP. In non-neuronal cells, CNOT8 apparently compensates for the loss of CNOT7 (Aslam et al., 2009, Doidge et al., 2012), which we do not observe. Of course, CNOT8 or other deadenylating enzymes such as PARN or PAN2 (Udagawa et al., 2015) could modulate poly (A) tail length in dendrites, but a consequential change in synaptic efficacy may not necessarily occur.
CNOT7 regulates a dynamic translational landscape in neurons and thus has multiple roles in RNA expression and consequent changes in synaptic function. For example, soon after glycine activation of LTP (≤10 min), CNOT7 levels are moderately reduced, resulting in polyadenylation of target RNAs in dendrites. At longer times (≥20 min) after treatment with glycine, substantial CNOT7 destruction takes place, which evokes further polyadenylation and a reduction in transcript level in dendrites. This effect of long-term CNOT7 depletion is more dramatically evident when CNOT7 is depleted for 4 days by treatment of neurons with a shRNA. Such time-dependent effects of CNOT7 at least partially explain the bimodal changes in dendritic poly(A) and translation that occur after synaptic stimulation.
The key event that mediates this mRNA regulation is CNOT7 destruction. Some evidence suggests that a reduction of CNOT7 takes place with other types of stimulation in cultured neurons (Schanzenbacher et al., 2016) and even during learning and memory in living animals (Cho et al., 2015). Although it is clear from our data that CNOT7 is rapidly destroyed upon LTP induction, reduced CNOT7 synthesis may also occur at this time. CNOT7 destruction is most likely mediated by the ubiquitin-proteasome system, which is known to regulate synaptic function and the dendritic proteome (Lee et al., 2012, Huang et al., 2015, Hegde, 2016, Alvarez-Castelao and Schuman, 2015).
Depletion of CNOT7 from the hippocampus results in deficient short and long term memory, which may be causally linked to the decreased poly(A) in the CA1 neuropil we observed following knockdown. It was recently demonstrated that contextual fear learning in mice correlated with a significant decrease in the translational efficiency of several mRNAs including that for CNOT7 (Cho et al., 2015). This decrease occurred within five minutes and was moderately maintained for up to four hours. However, the entire hippocampus was analyzed and not just the neurite-rich neuropil, which our data suggest would be the region where CNOT7 levels would decrease most dramatically. Taken together these data indicate that the changes we observed in cultured neurons may represent similar events that occur during learning in the living animal, and that dynamic control of CNOT7 destruction may regulate dendritic translation and higher cognitive function.
Experimental Procedures
Mouse Maintenance
Mouse protocols were reviewed and approved by the institutional animal care and use committee (IACUC) and all colonies were maintained following animal research guidelines. Only C57Bl/6 wild-type mice were used in this study with the ages indicated for each experiment in the method details.
Hippocampal Neuron Culture and Drug Treatments
Hippocampal neurons were cultured and maintained exactly as described in (Huang and Richter, 2007). For chem-LTP, 17–19 days in vitro (DIV) hippocampal neurons were stimulated with 200μM glycine as described in Lu et al 2001. Actinomycin D (Sigma) was added to DIV 17–19 hippocampal neurons at a concentration of 2.5μg/mL.
Immunocytochemistry
Hippocampal neurons were grown on coverslips, permeabilized with 0.1% Triton-X-100, fixed with 4% paraformaldehyde (PFA)/4% sucrose, blocked with 10% bovine serum albumin, and immunostained for CNOT7 1:100, Map2 1:500, tubulin 1:1000 or GluR1 1:10. Surface staining of GluR1 was performed as described above except without permeabilization. Images were acquired with a Zeiss Axiovert 200M microscope and a Hammatsu ORCA-ER camera using a 100X oil objective. The Z stack maximum projection images as well as the straightened dendrites were obtained using Image J software. To quantify fluorescence intensity, a 20 pixel wide line that was either 100 pixels long (cell body) or 1800 pixels long (dendrite) was drawn over the desired cell region and the fluorescence intensity under this line quantified.
Fluorescent In-Situ Hybridization (FISH)
Oligo(dT) FISH was performed as described in Swanger et al 2011 (Swanger et al., 2011) with a 50-mer oligo(dT) probe labeled using the Cy5 Mono-Reactive Dye Pack (GE). Pepsin digestion was performed post- fixation with 0.05mg/mL pepsin for 45sec on ice. For FISH of specific targets, the ViewRNA ISH (Affymetrix) kit was used according to manufacturer’s instructions except the protease step was omitted.
Fluorescent Non-Canonical Amino Acid Tagging (FUNCAT)
FUNCAT was performed using the Click-iT kit (ThermoFisher). For control cells, 100μg/mL cycloheximide was added 15 minutes prior to the 1 hour AHA incubation. Cells were then fixed with 4% PFA/4%sucrose and the FUNCAT reaction was performed (Dietrich et al., 2010).
Poly(U) Chromatography and Thermal Elution
Extracted RNA was denatured in CSB Buffer (25% formamide, 700mM NaCl, 50mM Tris-Cl pH 7.5, and 1mM EDTA). Forty micrograms of RNA was incubated with 0.025g of poly(U) agarose (Sigma) for 1–2hrs. The agarose was then washed with room temperature LSB buffer (25% formamide, 0.1M NaCl, 50mM Tris-Cl pH 7.5, 10mM EDTA). To elute mRNAs with short poly(A) tails (Du and Richter, 2005, Udagawa et al., 2012), the agarose was washed again with 50°C LSB buffer (this step was omitted in the total RNA samples). Long-tailed mRNAs were then eluted with 75°C LSB buffer.
Behavioral assays
Adult male wild type mice from the C56BL/6 background were used for all mouse studies. For AAV injections, 10–12 week old mice were injected bilaterally with 2 × 1011 viral particles in the hippocampal CA1 region and behavioral experiments were performed three weeks later. The elevated plus maze, open field test, novel object recognition, T-maze, and Passive avoidance were performed as described in (Mansur et al., 2016). For the nest building assay, ~2.5g nestlets were placed in the cages of single housed animals one hour prior to the dark cycle. Sixteen hours later the nestlets were collected to determine a nest building score (Deacon, 2006). Nests were scored blinded.
Quantification and Statistical Analysis
Statistical details of each experiment are provided in the figure legends for that experiment. Student’s t-test was used to determine the significance for all figures except Figure 3A, 4A, 4C, 4D, and S3B where the ANOVA test was used. Significance was defined as a p-value < 0.05. On all figures, error bars represent the standard error of the mean and *p≤0.05, **p≤0.01, ***p≤0.001, NS = Non-signficant. Please refer to our supplemental information for sequence-based reagents and resource table.
Supplementary Material
Highlights.
CNOT7 regulates dendritic mRNA transport and local translation
CNOT7 regulates synaptic plasticity in cultured neurons
Dendritic CNOT7 is destroyed during synaptic stimulation
Depletion of CNOT7 from the hippocampus impairs cognitive function
Acknowledgments
We thank J Shin, KY Paek, SA Swanger, B Liu, A Hien, E Stackpole, E Donnard, SD Redick, SJ Doxsey, and AR Buxbaum for technical help and advice, and Y Edwards and S Gujja in the bioinformatics core, Program in Molecular Medicine for assistance with the bioinformatics, GJ Bassell for providing the AAVs, AR Tapper for the use of the stereotactic injection and elevated plus maze equipment, and RP Alencar for graphic design expertise. RM was supported by NIH predoctoral fellowship (F31NS092415). FM was supported by a Science Without Borders Fellowship, CNPq, Brazil. This work was supported by NIH grant R01NS079415 to JDR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
RLM and JDR designed the experiments. RLM performed all the experiments except for those involving RNA-IP, stereotactic injections and animal behaviors, which were performed by FM. RLM and JDR wrote the manuscript.
Accession Numbers
The RNA sequencing data can be accessed with accession number GEO: GSE88777.
References
- ALVAREZ-CASTELAO B, SCHUMAN EM. The Regulation of Synaptic Protein Turnover. J Biol Chem. 2015;290:28623–30. doi: 10.1074/jbc.R115.657130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ASHRAF SI, MCLOON AL, SCLARSIC SM, KUNES S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124:191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]
- ASLAM A, MITTAL S, KOCH F, ANDRAU JC, WINKLER GS. The Ccr4-NOT deadenylase subunits CNOT7 and CNOT8 have overlapping roles and modulate cell proliferation. Mol Biol Cell. 2009;20:3840–50. doi: 10.1091/mbc.E09-02-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BICKER S, KHUDAYBERDIEV S, WEISS K, ZOCHER K, BAUMEISTER S, SCHRATT G. The DEAH-box helicase DHX36 mediates dendritic localization of the neuronal precursor-microRNA-134. Genes Dev. 2013;27:991–6. doi: 10.1101/gad.211243.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRADSHAW K, EMPTAGE N, BLISS T. A role for dendritic protein synthesis in hippocampal late LTP. European Journal of Neuroscience. 2003:3150–52. doi: 10.1111/j.1460-9568.2003.03054.x. [DOI] [PubMed] [Google Scholar]
- BUXBAUM AR, HAIMOVICH G, SINGER RH. In the right place at the right time: visualizing and understanding mRNA localization. Nat Rev Mol Cell Biol. 2015;16:95–109. doi: 10.1038/nrm3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUXBAUM AR, WU B, SINGER RH. Single beta-actin mRNA detection in neurons reveals a mechanism for regulating its translatability. Science. 2014;343:419–22. doi: 10.1126/science.1242939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAJIGAS IJ, TUSHEV G, WILL TJ, TOM DIECK S, FUERST N, SCHUMAN EM. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron. 2012;74:453–66. doi: 10.1016/j.neuron.2012.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEN C, ITO K, TAKAHASHI A, WANG G, SUZUKI T, NAKAZAWA T, YAMAMOTO T, YOKOYAMA K. Distinct expression patterns of the subunits of the CCR4-NOT deadenylase complex during neural development. Biochem Biophys Res Commun. 2011;411:360–4. doi: 10.1016/j.bbrc.2011.06.148. [DOI] [PubMed] [Google Scholar]
- CHO J, YU NK, CHOI JH, SIM SE, KANG SJ, KWAK C, LEE SW, KIM JI, CHOI DI, KIM VN, KAANG BK. Multiple repressive mechanisms in the hippocampus during memory formation. Science. 2015;350:82–7. doi: 10.1126/science.aac7368. [DOI] [PubMed] [Google Scholar]
- CID-ARREGUI A, PARTON RG, SIMONS K, DOTTI CG. Nocodazole-dependent transport, and brefeldin A–sensitive processing and sorting, of newly synthesized membrane proteins in cultured neurons. J Neurosci. 1995;15:4259–69. doi: 10.1523/JNEUROSCI.15-06-04259.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COSTA-MATTIOLI M, SOSSIN WS, KLANN E, SONENBERG N. Translational control of long-lasting synaptic plasticity and memory. Neuron. 2009;61:10–26. doi: 10.1016/j.neuron.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DARNELL RB. RNA protein interaction in neurons. Annu Rev Neurosci. 2013;36:243–70. doi: 10.1146/annurev-neuro-062912-114322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DEACON RM. Assessing nest building in mice. Nat Protoc. 2006;1:1117–9. doi: 10.1038/nprot.2006.170. [DOI] [PubMed] [Google Scholar]
- DIETRICH DC, HODAS JJL, GOUZER G, SHADRIN IY, NGO JT, TRILLER A, TIRRELL DA, SCHUMAN EM. In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nature Neuroscience. 2010:897–905. doi: 10.1038/nn.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOIDGE R, MITTAL S, ASLAM A, WINKLER GS. The anti-proliferative activity of BTG/TOB proteins is mediated via the Caf1a (CNOT7) and Caf1b (CNOT8) deadenylase subunits of the Ccr4-not complex. PLoS One. 2012;7:e51331. doi: 10.1371/journal.pone.0051331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DU L, RICHTER JD. Activity-Dependent polyadenylation in neurons. RNA. 2005:1340–1347. doi: 10.1261/rna.2870505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EOM T, ZHANG C, WANG H, LAY K, FAK J, NOEBELS JL, DARNELL RB. NOVA-dependent regulation of cryptic NMD exons controls synaptic protein levels after seizure. Elife. 2013;2:e00178. doi: 10.7554/eLife.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FABIAN MR, MATHONNET G, SUNDERMEIER T, MATHYS H, ZIPPRICH JT, SVITKIN YV, RIVAS F, JINEK M, WOHLSCHLEGEL J, DOUDNA JA, CHEN CY, SHYU AB, YATES JR, 3RD, HANNON GJ, FILIPOWICZ W, DUCHAINE TF, SONENBERG N. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell. 2009;35:868–80. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GIORGI C, YEO GW, STONE ME, KATZ DB, BURGE C, TURRIGIANO G, MOORE MJ. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007;130:179–91. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- GONG B, CAO Z, ZHENG P, VITOLO OV, LIU S, STANISZEWSKI A, MOOLMAN D, ZHANG H, SHELANSKI M, ARANCIO O. Ubiquitin hydrolase Uch-L1 rescues beta-amyloid-induced decreases in synaptic function and contextual memory. Cell. 2006;126:775–88. doi: 10.1016/j.cell.2006.06.046. [DOI] [PubMed] [Google Scholar]
- HEGDE AN. Proteolysis, synaptic plasticity and memory. Neurobiol Learn Mem. 2016 doi: 10.1016/j.nlm.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HEGDE AN, INOKUCHI K, PEI W, CASADIO A, GHIRARDI M, CHAIN DG, MARTIN KC, KANDEL ER, SCHWARTZ JH. Ubiquitin C-terminal hydrolase is an immediate-early gene essential for long-term facilitation in Aplysia. Cell. 1997;89:115–26. doi: 10.1016/s0092-8674(00)80188-9. [DOI] [PubMed] [Google Scholar]
- HUANG J, IKEUCHI Y, MALUMBRES M, BONNI A. A Cdh1-APC/FMRP Ubiquitin Signaling Link Drives mGluR-Dependent Synaptic Plasticity in the Mammalian Brain. Neuron. 2015;86:726–39. doi: 10.1016/j.neuron.2015.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUANG YS, RICHTER JD. Analysis of mRNA Translation in Cultured Hippocanmpal Neurons. Methods in Enzymology. 2007:143–162. doi: 10.1016/S0076-6879(07)31008-2. [DOI] [PubMed] [Google Scholar]
- HUBER KM, BEAR MF. Role for rapid dendritic protein synthesis in hippocampal mGLUR-dependent long-term depression. Science. 2000;273:1254–1257. doi: 10.1126/science.288.5469.1254. [DOI] [PubMed] [Google Scholar]
- KANDEL ER. The molecular biology of memory storage: a dialogue between genes and synapses. Science. 2001:1030–8. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- KANG H,SE. A requirement for local protein synthesis in neutrophin-induced hippocampal synaptic plasticity. Science. 1996:1402–1406. doi: 10.1126/science.273.5280.1402. [DOI] [PubMed] [Google Scholar]
- KELLEHER RJ, 3RD, BEAR MF. The autistic neuron: troubled translation? Cell. 2008;135:401–6. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- KOHRMANN M, LUO M, KAETHER C, DESGROSEILLERS L, DOTTI CG, KIEBLER MA. Microtubule-dependent recruitment of Staufen-green fluorescent protein into large RNA-containing granules and subsequent dendritic transport in living hippocampal neurons. Mol Biol Cell. 1999;10:2945–53. doi: 10.1091/mbc.10.9.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KRICHEVSKY AM, KOSIK KS. Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron. 2001;32:683–96. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- LAU NC, KOLKMAN A, VAN SCHAIK FM, MULDER KW, PIJNAPPEL WW, HECK AJ, TIMMERS HT. Human Ccr4-Not complexes contain variable deadenylase subunits. Biochem J. 2009;422:443–53. doi: 10.1042/BJ20090500. [DOI] [PubMed] [Google Scholar]
- LEE H,BM, KAMEYAMA K, BEAR MF, HUGANIR RL. Regulation of distinct AMPA receptor phosphorylation sites during bidirectional synaptic plasticity. Nature. 2000:955–959. doi: 10.1038/35016089. [DOI] [PubMed] [Google Scholar]
- LEE SH, KWAK C, SHIM J, KIM JE, CHOI SL, KIM HF, JANG DJ, LEE JA, LEE K, LEE CH, LEE YD, MINIACI MC, BAILEY CH, KANDEL ER, KAANG BK. A cellular model of memory reconsolidation involves reactivation-induced destabilization and restabilization at the sensorimotor synapse in Aplysia. Proc Natl Acad Sci U S A. 2012;109:14200–5. doi: 10.1073/pnas.1211997109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LU W, MAN H, JU W, TRIMBLE WS, MACDONALD JF, WANG YT. Activation of synaptic NMDA receptors induces membrane insertion of new AMPA receptors and LTP in cultured hippocampal neurons. Neuron. 2001;29:243–54. doi: 10.1016/s0896-6273(01)00194-5. [DOI] [PubMed] [Google Scholar]
- MANSUR F, IV, SHINA M, GU W, SCHAEVITZ L, STACKPOLE E, GUJJA S, EDWARDS YJ, RICHTER JD. Gld2-catalyzed 3′ monoadenylation of miRNAs in the hippocampus has no detectable effect on their stability or on animal behavior. RNA. 2016;22:1492–9. doi: 10.1261/rna.056937.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARTIN KC, KANDEL ER. Cell adhesion molecules, CREB, and the formation of new synaptic connections. Neuron. 1996;17:567–70. doi: 10.1016/s0896-6273(00)80188-9. [DOI] [PubMed] [Google Scholar]
- MILLER S, YASUDA M, COATS JK, JONES Y, MARTONE ME, MAYFORD M. Disruption of Dendritic Translation of CaMKIIα Impairs Stabilization of Synaptic Plasticity and Memory Consolidation. Neuron. 2002:507–519. doi: 10.1016/s0896-6273(02)00978-9. [DOI] [PubMed] [Google Scholar]
- OGAMI K, HOSODA N, FUNAKOSHI Y, HOSHINO S. Antiproliferative protein Tob directly regulates c-myc proto-oncogene expression through cytoplasmic polyadenylation element-binding protein CPEB. Oncogene. 2014;33:55–64. doi: 10.1038/onc.2012.548. [DOI] [PubMed] [Google Scholar]
- PIAO X, ZHANG X, WU L, BELASCO JG. CCR4-NOT deadenylates mRNA associated with RNA-induced silencing complexes in human cells. Mol Cell Biol. 2010;30:1486–94. doi: 10.1128/MCB.01481-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RICHTER JD. CPEB: a life in translation. Trends Biochem Sci. 2007;32:279–85. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- RICHTER JDCJ. Pausing on Polyribosomes: Make Way for Elongation in Translational Control. Cell. 2015;163:8. doi: 10.1016/j.cell.2015.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHANZENBACHER CT, SAMBANDAN S, LANGER JD, SCHUMAN EM. Nascent Proteome Remodeling following Homeostatic Scaling at Hippocampal Synapses. Neuron. 2016;92:358–371. doi: 10.1016/j.neuron.2016.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHRATT G. microRNAs at the synapse. Nat Rev Neurosci. 2009;10:842–9. doi: 10.1038/nrn2763. [DOI] [PubMed] [Google Scholar]
- SCHWEDE A, ELLIS L, LUTHER J, CARRINGTON M, STOECKLIN G, CLAYTON C. A role for Caf1 in mRNA deadenylation and decay in trypanosomes and human cells. Nucleic Acids Res. 2008;36:3374–88. doi: 10.1093/nar/gkn108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUTTON MA, SCHUMAN EM. Dendritic Protein Synthesis, Synaptic Plasticity, and Memory. Cell. 2006:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- SUTTON MA, TAYLOR AM, ITO HT, PHAM A, SCHUMAN EM. Postsynaptic decoding of neural activity: eEF2 as a biochemical sensor coupling miniature synaptic transmission to local protein synthesis. Neuron. 2007;55:648–61. doi: 10.1016/j.neuron.2007.07.030. [DOI] [PubMed] [Google Scholar]
- SWANGER SA, BASSELL GJ, GROSS C. High-Resolution Fluorescence In Situ Hybridization to Detect mRNAs In Neuronal Compartments In Vitro and In Vivo. Methods Molecular Biology. 2011:103–23. doi: 10.1007/978-1-61779-005-8_7. [DOI] [PubMed] [Google Scholar]
- TEMME C, ZAESSINGER S, MEYER S, SIMONELIG M, WAHLE E. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J. 2004;23:2862–71. doi: 10.1038/sj.emboj.7600273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TUCKER M, VALENCIA-SANCHEZ MA, STAPLES RR, CHEN J, DENIS CL, PARKER R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–86. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- UDAGAWA T, FUJIOKA Y, TANAKA M, HONDA D, YOKOI S, RIKU Y, IBI D, NAGAI T, YAMADA K, WATANABE H, KATSUNO M, INADA T, OHNO K, SOKABE M, OKADO H, ISHIGAKI S, SOBUE G. FUS regulates AMPA receptor function and FTLD/ALS-associated behaviour via GluA1 mRNA stabilization. Nat Commun. 2015;6:7098. doi: 10.1038/ncomms8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UDAGAWA T, SWANGER SA, TAKEUCHI K, KIM JH, NALAVADI V, SHIN J, LORENZ LJ, ZUKIN RS, BASSELL GJ, RICHTER JD. Bidirectional control of mRNA translation and synaptic plasticity by the cytoplasmic polyadenylation complex. Mol Cell. 2012;47:253–66. doi: 10.1016/j.molcel.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WU L, WELLS D, TAY J, MENDIS D, ABBOTT MA, BARNITT A, QUINLAN E, HEYNEN A, FALLON JR, RICHTER JD. CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of alpha-CaMKII mRNA at synapses. Neuron. 1998;21:1129–39. doi: 10.1016/s0896-6273(00)80630-3. [DOI] [PubMed] [Google Scholar]
- YUEN EY, JIANG Q, FENG J, YAN Z. Microtubule regulation of N-methyl-D-aspartate receptor channels in neurons. J Biol Chem. 2005;280:29420–7. doi: 10.1074/jbc.M504499200. [DOI] [PubMed] [Google Scholar]
- ZEARFOSS NR, ALARCON JM, TRIFILIEFF P, KANDEL E, RICHTER JD. A molecular circuit composed of CPEB-1 and c-Jun controls growth hormone-mediated synaptic plasticity in the mouse hippocampus. J Neurosci. 2008;28:8502–9. doi: 10.1523/JNEUROSCI.1756-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


