Abstract
Histone deacetylase inhibitors (HDACIs) are therapeutic drugs that inhibit deacetylase activity, thereby increasing acetylation of many proteins, including histones. HDACIs have antineoplastic effects in preclinical and clinical trials and are being considered for cancers with unmet therapeutic need, including neuroblastoma (NB). Uncertainty of how HDACI-induced protein acetylation leads to cell death, however, makes it difficult to determine which tumors are likely to be responsive to these agents. Here, we show that NB cells are sensitive to HDACIs, and that the mechanism by which HDACIs induce apoptosis involves Bax. In these cells, Bax associates with cytoplasmic Ku70, a protein that typically increases chemotherapy resistance. Our data show that in NB cells Ku70 binds to Bax in an acetylation-sensitive manner. Upon HDACI treatment, acetylated Ku70 releases Bax, allowing it to translocate to mitochondria and trigger cytochrome c release, leading to caspase-dependent death. This study shows that Ku70 is an important Bax-binding protein, and that this interaction can be therapeutically regulated in NB cells. Whereas the Bax-binding ability of Ku70 allows it to block apoptosis in response to certain agents, it is also a molecular target for the action of HDACIs, and in this context, a mediator of NB cell death.
Keywords: cAMP-response element-binding protein, histone acetyltransferase
Neuroblastoma (NB) is the most common extracranial solid tumor in children, and is often refractory to conventional therapies. Despite aggressive cytotoxic therapy, most children with metastatic disease are not cured (1, 2). Current treatments fail primarily because of chemotherapy resistance; thus, new drugs with novel mechanisms of action are needed. A new class of cytotoxic compounds, histone deacetylase inhibitors (HDACIs), suppresses NB tumor growth in human-mouse NB xenografts (3, 4), and several members of this therapeutic class are already being evaluated in human trials for other cancers (5). This study was performed to define the mechanism by which HDACIs kill NB cells, thereby supporting strategic clinical trial design and maximizing the efficacy of treatment.
Histone deacetylases (HDACs) repress gene transcription by modulating histone acetylation (6, 7). Some non-histone proteins, many of which are transcription factors, are also substrates of HDACs (8). The HDAC family is subdivided into three categories based on sequence similarity to the yeast proteins RPD3 (class I: HDAC1, 2, 3, and 8), HDA1 (class II: HDAC4, 5, 6, 7, 9, and 10), SIR2 (class III), and HDAC11 (class IV) (9) (10). Whereas HDACs are involved in many cellular functions, such as cell cycling and apoptosis, the best-characterized function of Class I and II HDACs is transcriptional repression. When complexed with corepressors such as nuclear hormone receptor corepressor and silencing mediator for retinoic acid and thyroid hormone receptors, HDACs are recruited to nonliganded nuclear hormone receptors to repress gene transcription (11). Class III HDACs require nicotinamide-adenine dinucleotide as a cofactor (12), and, at least in yeast, sense the metabolic state and age of the cell (12). One mammalian homolog of SIR2, SirT1, deacetylates p53, altering its function as an apoptotic protein (13), and another, SirT2, is a microtubule deacetylase (14).
Most cytotoxic compounds trigger cell death by engaging pathways that activate caspases. One such pathway, mediated by cell-surface death receptors, including Fas/CD95/Apo-1 and TRAIL, is characterized by a critical requirement for caspase-8 (15, 16). A second signaling pathway centers on a disruption of mitochondrial function, which is characterized by release of the proapoptotic mediator cytochrome c and caspase-9 activation (17). Drug resistance in NB is associated with defects that can affect either pathway, including genetic silencing of caspase-8, p53 mutations, and overexpression of Bcl-2 (18-20). Although it is apparent that HDACIs kill cells and induce apoptosis, a lack of understanding of the mechanism coupling hyperacetylation to cell death significantly limits the rational application of this therapeutic class of drugs. A mechanistic understanding sufficient to predict tumor sensitivity or resistance could accelerate optimal clinical implementation in therapeutic areas like NB.
NB treatment is complicated by the fact that these tumors are heterogeneous, being principally comprised of tumor cells that are classified as either neuronal (N)- or stromal-type cells. N-type cells express high levels of the N-myc protein, are more commonly isolated from high-risk tumor explants, form tumors in mice, and fail to express caspase-8 because of DNA methylation (21-24). Consequently, N-type cell lines were studied to model the behavior and drug responsiveness of aggressive, highly transformed NB cells that characterize high-risk tumors. In this study, we demonstrate that HDACIs are sufficient as single agents to kill NB cells; HDACIs do so by increasing the acetylation of Ku70, an autoantigen with multiple functions, including DNA repair (25). Ku70 is expressed at high levels in NB cells, and interestingly, a significant fraction of this protein is cytoplasmic where it binds and sequesters the proapoptotic protein Bax. HDACI treatment of N-type cells increases acetylation of Ku70, releasing and activating Bax, which is a critical target for proapoptotic regulation by HDACI treatment.
Materials and Methods
Transfection and Generation of Stable Expression of Ku70 Cell Lines. For transient transfection, the IMR32 cells were transfected with vector, pCMV2B Ku70 WT (Flag-tagged) (provided by S. Matsuyama, The Cleveland Clinic, Cleveland), or pCMV2B Ku70 K539R/K542R (generated by using a QuikChange site-directed mutagenesis Kit from Stratagene), along with GFP expression vector (Clontech), using the Nucleofactor kit V by Amaxa (Gaithersburg, MD) as described by the manufacturer.
To generate IMR32 cell lines stably expressing WT or mutant Ku70, cells were transfected with vector, Flag-Ku70 WT, or Flag-Ku70 K539R/K542R vector with a puromycin resistance construct (provided by D. Bochar, University of Michigan, Ann Arbor) using LipofectAMINE Plus (Invitrogen) as described by the manufacturer.
Cell Viability Assays. To determine cell viability of transfected cells, we cotransfected cells with a GFP plasmid and scored GFP-positive apoptotic nuclei as described (26) after treatments. In some experiments, viability was determined by using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay as described (27).
Subcellular Fractionation. Mitochondria was isolated from IMR32 cells according to the method described in Blatt et al. (28). The purity of the fraction was determined by immunoblotting with antibodies against cytochrome oxidase subunit IV.
To separate cytosolic and nuclear fractions of IMR32 cells, we used the procedure described by Sikora et al. (29).
Western Blot Analysis and Coimmunoprecipitation. For Western analyses, the following antibodies were used: antibodies for Flag and β-tubulin were obtained from Sigma, antibodies for Ku70 and histone H1 were obtained from Santa Cruz Biochemicals, antibodies for Bax and cytochrome c were obtained from BD Pharmingen (San Diego), and cytochrome oxidase subunit IV was obtained from Molecular Probes. Western blots were developed by using the Enhanced Chemiluminescence Plus kit (Amersham Pharmacia Biotech, Piscataway, NJ).
Coimmunoprecipitation of endogenous and transfected Flag-Ku70 was performed in a 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate buffer according to the procedure described by Sawada et al. (30).
Measurement of Diploid DNA Content by Flow Cytometry. Cells were collected by using trypsinization. Measurement of DNA content was conducted after incubating cells in labeling solution (50 μg/ml propidium iodide in PBS containing 0.2% Triton X-100 and 10 μg/ml RNase A) for 30 min by using a FACS Calibur flow cytometer equipped with cellquest software (BD Biosciences, San Diego).
Results
HDACIs Kill N-Type NB Cells. To investigate the effects of deacetylase inhibition on NB cells, we used Trichostatin A (TSA), a hydroxyamic acid derivative originally developed as an antifungal agent, and sodium butyrate, a natural short-chain fatty acid (31). Both compounds inhibit class I/II HDACs, suppress growth, induce differentiation, and induce apoptosis in different tumor types (31, 32).
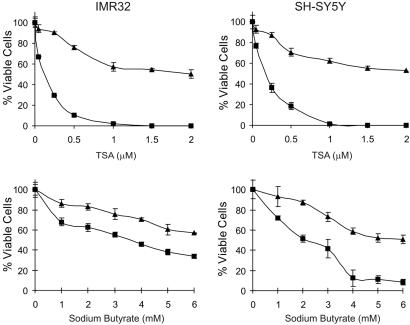
Each compound killed both SH-SY5Y and IMR32 N-type NB cells in culture (Fig. 1). By using MTT assays to measure viability, the dose- and time-dependent effects observed showed that maximal killing required 48 h of treatment. At 48 h, the EC50 for TSA against both cell types was 0.5 μM, and for sodium butyrate, it was between 2 and 3 mM. The morphology of these cells during treatment with either TSA or sodium butyrate was typical of cells undergoing apoptosis: shrunken nuclei, cytoplasmic vacuolization, and membrane blebbing (Fig. 2A); similar changes in morphology were observed when these cells were treated with cisplatin (CDDP) (10 μg/ml), a cytotoxic agent that induces apoptosis of N-type NB cells through a caspase-9-dependent process (27).
Fig. 1.
HDACI induces cell death in N-type NB cells. IMR32 and SH-SY5Y cells were treated with various concentrations of TSA or sodium butyrate as indicated for 24 (▴) or 48 (▪) h. Cell viability was determined by MTT assay. Results are expressed as the percentage of viable cells compared with vehicle-treated controls (mean ± SD, n = 3).
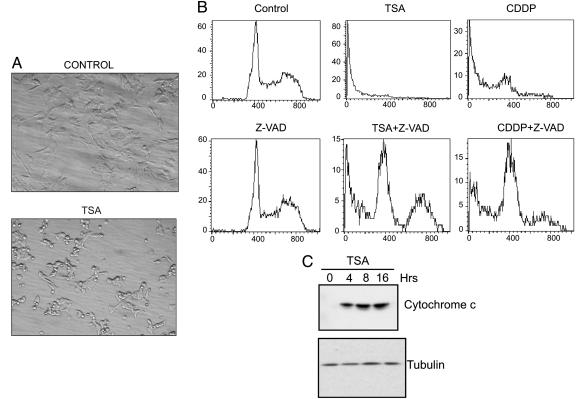
Fig. 2.
TSA induces apoptosis in IMR32 cells. (A) IMR32 cells were either treated with DMSO or 1 μM TSA for 24 h. Interference contrast microscopy was used to determine cell morphology at ×200. (B) DNA content was determined by propidium iodide staining and analyzed by flow cytometry in IMR32 cells treated with TSA (1 μM), Z-VAD (100 μM), or CDDP (10 μg/ml) as indicated for 24 h. (C) IMR32 cells were treated with 1 μM TSA for various times as indicated, and the S100 fraction was immunoblotted by using anti-cytochrome c antibodies. The same blot was reprobed with anti-β-tubulin antibodies as a loading control.
To confirm that HDACI-induced NB cell death is apoptotic and is indeed caspase-dependent, we treated IMR32 cells with TSA (1 μM) in the presence or absence of Z-VAD-FMK (Z-VAD) (100 μM), a pan-caspase inhibitor. After 24 h, apoptotic DNA fragmentation was assessed by using flow cytometry to measure the DNA content of semipermeabilized cells after propidium iodide staining. As shown in Fig. 2B, a large fraction (78%) of TSA-treated cells had hypodiploid DNA content, similar to the response observed with CDDP. When Z-VAD was combined with either TSA or CDDP, the apoptotic response was significantly reduced. Together with the morphologic evidence, these results support the conclusion that HDACI treatment kills NB cells through an apoptotic mechanism.
HDACI-Induced Apoptosis Is Mediated by Bax. Chemical-induced apoptosis frequently involves a mitochondria-centered apoptotic response, although death-receptor-coupled caspase-8 activation has also been observed in certain cells. Because N-type NB cells do not express caspase-8, we focused our experiments on the mitochondria-dependent pathway. First, we measured the release of cytochrome c from the mitochondria to the cytoplasm, a hallmark of initiation of this death pathway (33, 34). Whereas cytochrome c is absent in the cytoplasm of untreated cells, it appears 4 h after TSA treatment, and the increase persists for up to 16 h (Fig. 2C). These results suggest that HDACI treatment induces apoptosis in IMR32 cells by activating the mitochondria/caspase-9 pathway.
Cytochrome c is released by mechanisms involving opening of the permeability transition pore, collapse of the electrochemical gradient across the inner mitochondrial membrane, or insertion of proapoptotic molecules (e.g., BH3 domain-containing proteins) into mitochondria that form channels or trigger permeability pore opening. We focused on the possibility that a BH3 domain-containing protein, Bax, was involved. Recent work has shown that acetylation controls Bax activation, which is regulated by the cAMP-response element-binding protein (CREB) binding protein (CBP), a histone acetyltransferase. CBP activates Bax indirectly by acetylating Ku70, a multifunctional protein that sequesters Bax only when it is not acetylated (35).
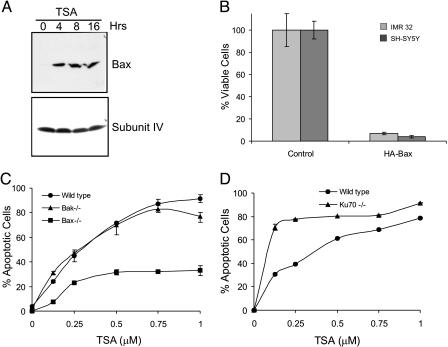
To test whether Bax is involved in inducing apoptosis of N-type NB cells in response to HDACI, we first measured the level of Bax in the mitochondria of TSA-treated IMR32 cells. The level of Bax increases at 4 h after TSA treatment and persists up to 16 h (Fig. 3A), a time course similar to the appearance of cytochrome c after TSA treatment in the cytoplasm (Fig. 2C). We next asked whether increase of Bax alone in NB cells is sufficient to induce apoptosis. Indeed, transient transfection of a Bax expression plasmid killed both SH-SY5Y and IMR32 cells (Fig. 3B; see also Fig. 7, which is published as supporting information on the PNAS web site). Finally, because pilot experiments revealed that mouse embryonic fibroblasts (MEFs) are sensitive to TSA over a similar range of TSA concentrations as N-type cells, we used Bax-/- MEFs to determine whether Bax is necessary for HDACI-induced death. As shown in Fig. 3C, Bax-/- cells were resistant to TSA-induced apoptosis compared with WT or Bak-/- MEFs. Collectively, this series of experiments demonstrates that (i) Bax is activated by HDACIs in NB cells; (ii) Bax is sufficient to kill NB cells; and (iii) Bax is required, at least in MEFs, for HDACI-induced killing.
Fig. 3.
Bax mediates TSA-induced apoptosis. (A) Mitochondrial fractions of IMR32 cells isolated after treatment with 1 μM TSA for various times as indicated were immunoblotted by using anti-Bax antibodies. The same blot was probed for cytochrome oxidase subunit IV as a loading control. (B) IMR32 and SH-SY5Y were transfected with either empty vector or HA-tagged Bax. Cell viability was determined by MTT assay 48 h after transfection. Results are expressed as the percentage of control (mean ± SD, n = 3). (C) WT, Bax-/-, or Bak-/- MEF cells were treated with various concentrations of TSA as indicated for 24 h. Percent of apoptotic cells was derived quantitatively by measuring the percentage of subG1 population using flow cytometry. Results are expressed as mean ± SD (n = 3). (D) WT and Ku70-/- MEF cells had the same TSA treatment as described in C. Cell viability was determined by MTT assay.
Ku70 Is Expressed and Bound with Bax in NB Cells. Ku70 is known as a nuclear protein but is also identified in the cytoplasm of cells, such as HEK293T cells, where it is associated with Bax and prevents Bax activation (30). Interestingly, MEF cells deficient in Ku70 (Ku70-/-) enhance the sensitivity of the MEF cells to TSA treatment (Fig. 3D), suggesting that the interaction between Bax and Ku70 may be a determining factor in the sensitivity to TSA.
To assess whether Ku70 mediates HDACI-induced Bax activation in NB cells, we determined Ku70 expression in N-type NB cells. IMR32 cells were fractionated by differential centrifugation. Immunoblotting showed that Ku70 is indeed present in the cytoplasm and nuclei of these cells, and, as expected, Bax was found only in the cytoplasm (Fig. 4A).
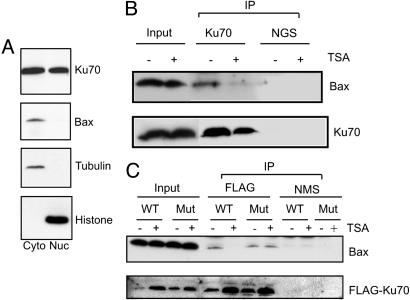
Fig. 4.
Bax is released from Ku70 in response to TSA. (A) Cytoplasmic and nuclear fractions of IMR32 cells were isolated. Equal amounts of proteins were separated on a 4-20% SDS/PAGE gradient gel and then immunoblotted by using anti-Ku70, anti-Bax, anti-β-tubulin, or anti-histone H1 antibodies as indicated. (B) Endogenous Ku70 before or after 4 h TSA treatment was immunoprecipitated by using goat anti-Ku70 antibodies or normal goat serum (NGS) as a control. The immunoprecipitates were separated by SDS/PAGE and probed with anti-Bax or anti-Ku70 antibodies. Input represents 20% of the samples used in immunoprecipitation. (C) IMR32 cells were transfected with either Flag-Ku70 WT or Flag-Ku70 K539R/K542R mutant expression vectors. Twenty-four hours after transfection, cells were treated with 1 μM TSA for an additional 4 h. The transfected Flag-Ku70s were immunoprecipitated by using anti-Flag antibodies or normal mouse serum (NMS) as a control, separated by SDS/PAGE, and probed with anti-Bax or anti-Flag antibodies. Input represents 20% of the samples used in immunoprecipitation.
To determine whether Ku70 interacts with Bax in N-type NB cells, we precipitated endogenous Ku70 by using a Ku70-specific antibody raised in goat and used normal goat serum as the control. The results shown in Fig. 4B indicate that Ku70 associates with endogenous Bax, but this interaction is disrupted by 4 h of TSA treatment. This result is consistent with a model in which TSA treatment increases the acetylation of Ku70, and the acetylation of Ku70 disrupts its interaction with Bax.
To further test this model, we constructed a Ku70 mutant in which Lys-539 and Lys-542 were replaced with arginines. These two lysines were shown to be critical acetylation-sensitive sites for Ku70-Bax binding (35). Arginine substitution maintains the positive charge at these positions but prevents acetylation. We tested the ability of this mutant to interact with Bax and whether its binding to Bax was sensitive to TSA treatment. Flag-Ku70 WT or Flag-Ku70 double-lysine mutant was transfected into IMR32 cells, and protein complexes were precipitated by using anti-Flag antibodies. As shown in Fig. 4C, the double-lysine mutant and the Flag-Ku70 WT associate with Bax in control-treated cells. After 4 h of TSA treatment, Bax is no longer coprecipitated with Ku70 WT, but it is coprecipitated with the double-lysine mutant. These results show that these two acetylation-sensitive residues (K539 and K542) underlie the TSA sensitivity of the interaction between Ku70 and Bax, supporting the hypothesis that TSA-induced Bax activation is mediated by Ku70 acetylation.
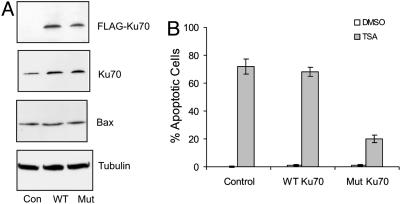
Lys-539 and Lys-542 of Ku70 Are Critical for TSA-Induced Apoptosis. Because the interaction of Bax with the Ku70 double-lysine mutant is insensitive to TSA treatment, we hypothesized that if acetylation of Ku70 is required for HDACIs to kill NB cells, then the double-lysine mutant would act in a dominant-negative fashion to block TSA-induced cell death. To test this hypothesis, IMR32 cells were cotransfected with vector alone, Flag-Ku70 WT, or Flag-Ku70 double-lysine mutant expression vectors in 10-fold excess with a GFP expression vector that marked transfected cells. Twenty-four hours after transfection, identical samples were either analyzed for protein expression or treated to determine TSA responsiveness. Protein expression was determined by first sorting cells by flow cytometry to isolate the GFP-positive (transfected) cells. Expression of transfected Flag-Ku70 proteins, total Ku70, and Bax was analyzed by immunoblotting. As shown in Fig. 5A, the level of expression of Flag-Ku70 WT and Flag-Ku70 double-lysine mutant was similar, and appeared to be ≈2-fold higher than the level of endogenous Ku70. Bax expression was equivalent in all transfected cells.
Fig. 5.
Lys-539 and Lys-542 of Ku70 are critical for Ku70-dependent, TSA-induced apoptosis. IMR32 cells were transfected with a GFP expression vector plus control vector, Flag-Ku70 WT, or Flag-Ku70 double-lysine mutant expression vector. Twenty-four hours after transfection, cells were treated with vehicle (DMSO) or 1 μM TSA for 24 h. (A) The GFP-positive cells were sorted by flow cytometry. Equal amounts of extracts were separated by 4-20% gradient SDS/PAGE and then the blot was probed with anti-Flag, anti-Ku70, anti-Bax, or anti-β-tubulin antibodies as shown. (B) Apoptotic cells were determined by staining the cells with Hoechst dye and counting cells showing DNA condensation per 100 GFP-positive cells. The results are from three different experiments and expressed as mean ± SD (n = 3).
The apoptotic response of transfected cells to TSA was determined by fluorescent microscopy 24 h after treatment by scoring the nuclear morphology of Hoechst-stained GFP-positive cells as described (30). Seventy percent of cells transfected with vector alone or Flag-Ku70 WT were apoptotic, but only 20% of cells transfected with the Ku70 double-lysine mutant were apoptotic (Fig. 5B). Similar results were obtained when GFP-expressing cells were first isolated by cell sorting, treated with TSA, and then their viability was measured by MTT assay (data not shown). From these results, showing that a modest level of expression of the double-lysine mutant confers resistance to TSA, we conclude that the acetylation-sensitive residues K539 and K542 are critical to HDACI-induced NB cell killing.
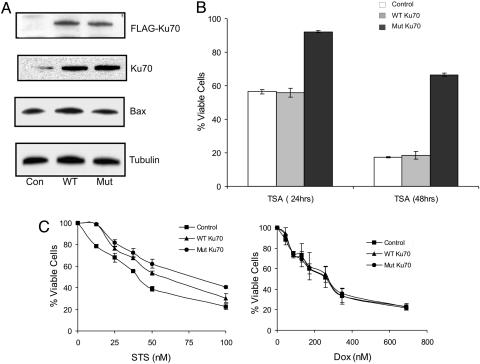
To test this model further, we prepared IMR32 cells that are stably transfected with Flag-Ku70 WT or Flag-Ku70 double-lysine mutant expression vector. As expected, stable cell lines were isolated that achieved a greater level of transfected gene expression relative to endogenous gene expression (Fig. 6A). Although Flag-Ku70 WT and Flag-Ku70 double-lysine mutant were expressed at nearly equal levels, both transfected genes were expressed at levels several fold higher than endogenous Ku70. Similar to the results above, there were no differences in the level of Bax expression in control and Ku70-overexpressing cells. These stably transfected cells were treated with either vehicle or TSA (1 μM) for 24 or 48 h, after which viability was determined by MTT assay (Fig. 6B). At 24 h, the viability of control and Ku70 WT-expressing cells was only 50%, whereas 95% of cells expressing the Ku70 double-lysine mutation were alive. After 48 h, the viability of control cells and Ku70 WT-expressing cells was only 15% compared with 65% in Flag-Ku70 double-lysine mutant-transfected cells.
Fig. 6.
Overexpression of Ku70 WT and the double-lysine mutant has differential effects in blocking TSA, STS, or Dox-induced apoptosis. (A) Expression of Flag-Ku70 WT or the double-lysine mutant in the stable IMR32 cell lines was determined by immunoblotting with anti-Flag, anti-Ku70, anti-Bax, or anti-β-tubulin antibodies as indicated. (B) After 24 or 48 h of 1 μM TSA treatment, the viability of IMR32 cells stably expressing Flag-Ku70 WT or the double-lysine mutant was determined by MTT assay. Results are presented as mean ± SD (n = 3). (C) After 24 h of STS or Dox treatment, the viability of IMR32 cells stably expressing Flag-Ku70 WT or the double-lysine mutant was determined by MTT assay. Results are expressed as the percentage of respective controls (mean ± SD, n = 3).
We examined the sensitivity of these stably transfected cells to other agents. Staurosporine (STS), a protein kinase inhibitor, is a strong inducer of apoptosis in many different cell types where it activates Bax and a mitochondrial caspase-dependent pathway (36). In contrast, doxorubicin (Dox) activates p53, leading to NF-κB dependent apoptosis in N-type NB cells and has no known dependence on Bax. High-level expression of Ku70 WT, and to an even greater extent, the double-lysine mutant blocked killing by STS (Fig. 6C Left). In contrast, killing by Dox was unaffected by expression of either WT or the double-lysine mutant of Ku70 (Fig. 6C Right). Collectively, these results confirm the importance of Ku70 in mediating TSA-induced cell death. Cells overexpressing Ku70 WT were equally sensitive to TSA as vector control cells, showing that increased Ku70, in the context of stable Bax levels, does not alter sensitivity to HDACIs. In contrast, killing by STS is reduced by increased Ku70 expression. From these results, we conclude that K539 and K542, two acetylation sites on Ku70, are indeed required for TSA-induced death. Moreover, because HDACIs directly target Ku70-Bax binding through acetylation of critical residues, their cytotoxic effectiveness is not limited by high-level Ku70 expression, which can modulate killing by other agents such as STS. Presumably when Bax is activated by STS, the presence of increasing amounts of nonacetylated Ku70 act competitively to bind and sequester Bax reducing its mitochondrial translocation.
Discussion
Previous studies have shown that HDACIs kill NB cells in vitro and shrink NB tumors in mice (3, 4, 37); however, the mechanism leading to these effects remained unknown. In this study, we provide a mechanistic link between HDACIs and their antitumor effects. Our results demonstrate a key role for Ku70. Specifically, inhibition of HDAC activity leads to increased acetylation of Ku70, which disrupts its binding to Bax. In turn, Bax is released from Ku70, translocates to mitochondria, and triggers the release of cytochrome c and caspase-dependent apoptosis.
Ku70 is a protein with multiple functions. Originally discovered as an autoantigen (38, 39), it is essential for the repair of nonhomologous DNA double-strand breaks. In this role, Ku70 complexes with Ku80 to form the DNA-binding component of the DNA-dependent protein kinase (40, 41). More recent work has shown that Ku70 is also an acetylation-sensitive binding partner for Bax (35). By screening a human cDNA library to identify inhibitors of Bax, Sawada et al. (42) identified the Bax-binding and Bax-inhibiting properties of Ku70. They showed that Ku70 binds to Bax through its C terminus (42). Five lysine residues within this domain (539, 542, 544, 553, and 556) are acetylated by CBP and p300/CBP-associated factor. Moreover, inhibition of HDAC activity by using TSA (to inhibit class I and II HDACs) and nicotinamide (to inhibit class III HDACs), results in increased acetylation of these residues (35), suggesting that, on balance, the acetylation status of these same residues is also determined by HDACs (26).
In terms of Bax binding, the five acetylated C-terminal lysines are not equivalent. Mutating K539 and K542 to glutamine (a substitution that structurally mimics acetylated lysine) renders Ku70 ineffective in blocking Bax-induced cell death (35), whereas mutation of the other lysines has no significant effect. Based on these findings, we expressed a mutant Ku70 in which K539 and K542 were substituted with arginine. In contrast to glutamine, this change preserves the positive charge of lysine and prevents acetylation by CBP, thus effectively mimicking the nonacetylated state of the WT protein (43). Predictably, this mutation does not prevent Ku70 from interacting with Bax (Fig. 4C); unlike the wild type, its interaction with Bax is not sensitive to HDACIs. These results argue that acetylation of K539 and K542 determines the extent of Bax binding and indicate that these two residues are critically important for the proapoptotic action of HDACIs in NB cells. Additional structural information is necessary to understand how acetylation at these sites causes Bax release.
Besides mitochondrial translocation, Bax-induced apoptosis requires that it undergoes a conformational change, exposing its N-terminal epitope and allowing insertion and oligomerization in mitochondrial membranes. Cohen et al. (35) showed with HEK293T and HeLa cells that combined class I/II and class III HDAC inhibition resulted in Bax-Ku70 dissociation but did not induce apoptosis unless cells were additionally primed by overexpressing Bax (35). Because our results show that inhibition of class I/II HDAC activity by TSA in NB cells is sufficient to trigger Bax-induced apoptosis (Fig. 1), and that overexpression of Bax is also sufficient to kill these cells (Fig. 3B), we hypothesize that conformational change of Bax is regulated by additional factors that either activate or suppress Bax in the cytosol. These factors are sensitive to HDACI treatment and overexpression of Bax, which may result in an increased level of Bax available, titrating putative Bax suppressors in cells. It is possible that HEK293T and HeLa cells may contain one or more Bax inhibitors in addition to Ku70. Thus, overexpression would be necessary to overcome these inhibitors. Additional studies are needed to further develop these hypotheses.
Ku70 expression in tumors is associated with unfavorable responses to conventional chemotherapy and radiation treatment. For example, Ku70 expression in cervical carcinomas inversely correlates with radiation sensitivity and patient survival (44). Experimentally, ectopic overexpression of Ku70 leads to resistance to agents, including curcumin, gamma radiation (45), and STS (see Fig. 6C). Conversely, strategies that lower Ku70 enhance chemo- and radiation sensitivity of cells and tissues (46-48). The ability of Ku70 to bind and sequester Bax offers an appealing explanation for its antiapoptotic effects. Because acetylation specifically interferes with Bax-Ku70 binding, HDACI treatment modifies this protein's function in a highly specific way such that it becomes a source from which Bax is released and activated. This model consequently predicts that high levels of Ku70 expression, which limit cellular sensitivity to radiation and some types of chemotherapy, will actually facilitate sensitivity to HDACIs. Experiments are under-way to determine the extent that Ku70, Bax levels, and the expression of other Bax-binding proteins coordinate to regulate HDACI responsiveness in NB tumors.
The mechanistic understanding developed here is relevant to the design of clinical trials investigating the efficacy of HDACIs against cancers, and NB in particular. The mechanism predicts NB tumors with abnormal p53 or caspase-8 expression, important factors in chemoresistance, may remain sensitive to HDACI treatment. The results support testing HDACIs in patients with NB, including those with tumors that are resistant to other agents, and argue that clinical investigations should evaluate Ku70 and Bax as biomarkers to predict response.
Supplementary Material
Acknowledgments
We thank Drs. S. Matsuyama and D. Bochar for expression vectors; Drs. M. Armstrong, J. Jarzembowski, and J. Broadbent for their constructive comments on the manuscript; and A. Boitano for technical support. This work was supported in part by National Institutes of Health Grant CA697276-04, the Janette Ferrantino Hematology Research Fund (to V.P.C.), and the Ravitz Foundation (to A.W.O. and V.P.C.). This work used the Sequencing Core of the Michigan Diabetes Research and Training Center, which was funded by National Institute of Diabetes and Digestive and Kidney Diseases Grant NIH5P60 DK20572.
Author contributions: C.S., A.W.O., V.P.C., and R.P.S.K. designed research; C.S. and X.B. performed research; C.S. and A.W.O. contributed new reagents/analytic tools; C.S., A.W.O., V.P.C., and R.P.S.K. analyzed data; and C.S., A.W.O., V.P.C., and R.P.S.K. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HDAC, histone deacetylase; HDACI, HDAC inhibitor; NB, neuroblastoma; N, neuronal; TSA, Trichostatin A; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide; CDDP, cisplatin; MEF, mouse embryonic fibroblast; STS, staurosporine; Dox, doxorubicin; CREB, cAMP-response element-binding protein; CBP, CREB-binding protein.
References
- 1.Frappaz, D., Michon, J., Coze, C., Berger, C., Plouvier, E., Lasset, C., Bernard, J. L., Stephan, J. L., Bouffet, E., Buclon, M., et al. (2000) J. Clin. Oncol. 18, 468-476. [DOI] [PubMed] [Google Scholar]
- 2.Schmidt, M. L., Lukens, J. N., Seeger, R. C., Brodeur, G. M., Shimada, H., Gerbing, R. B., Stram, D. O., Perez, C., Haase, G. M. & Matthay, K. K. (2000) J. Clin. Oncol. 18, 1260-1268. [DOI] [PubMed] [Google Scholar]
- 3.Coffey, D. C., Kutko, M. C., Glick, R. D., Butler, L. M., Heller, G., Rifkind, R. A., Marks, P. A., Richon, V. M. & La Quaglia, M. P. (2001) Cancer Res. 61, 3591-3594. [PubMed] [Google Scholar]
- 4.Jaboin, J., Wild, J., Hamidi, H., Khanna, C., Kim, C. J., Robey, R., Bates, S. E. & Thiele, C. J. (2002) Cancer Res. 62, 6108-6115. [PubMed] [Google Scholar]
- 5.Marks, P., Rifkind, R. A., Richon, V. M., Breslow, R., Miller, T. & Kelly, W. K. (2001) Nat. Rev. Cancer 1, 194-202. [DOI] [PubMed] [Google Scholar]
- 6.Lee, D. Y., Hayes, J. J., Pruss, D. & Wolffe, A. P. (1993) Cell 72, 73-84. [DOI] [PubMed] [Google Scholar]
- 7.Wolffe, A. P. & Guschin, D. (2000) J. Struct. Biol. 129, 102-122. [DOI] [PubMed] [Google Scholar]
- 8.Bannister, A. J. & Miska, E. A. (2000) Cell Mol. Life Sci. 57, 1184-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thiagalingam, S., Cheng, K. H., Lee, H. J., Mineva, N., Thiagalingam, A. & Ponte, J. F. (2003) Ann. N.Y. Acad. Sci. 983, 84-100. [DOI] [PubMed] [Google Scholar]
- 10.Gregoretti, I. V., Lee, Y. M. & Goodson, H. V. (2004) J. Mol. Biol. 338, 17-31. [DOI] [PubMed] [Google Scholar]
- 11.Ng, H. H. & Bird, A. (2000) Trends Biochem. Sci. 25, 121-126. [DOI] [PubMed] [Google Scholar]
- 12.Grozinger, C. M. & Schreiber, S. L. (2002) Chem. Biol. 9, 3-16. [DOI] [PubMed] [Google Scholar]
- 13.Vaziri, H., Dessain, S. K., Ng Eaton, E., Imai, S. I., Frye, R. A., Pandita, T. K., Guarente, L. & Weinberg, R. A. (2001) Cell 107, 149-159. [DOI] [PubMed] [Google Scholar]
- 14.North, B. J., Marshall, B. L., Borra, M. T., Denu, J. M. & Verdin, E. (2003) Mol. Cell 11, 437-444. [DOI] [PubMed] [Google Scholar]
- 15.Ashkenazi, A. & Dixit, V. M. (1998) Science 281, 1305-1308. [DOI] [PubMed] [Google Scholar]
- 16.Chinnaiyan, A. M., O'Rourke, K., Tewari, M. & Dixit, V. M. (1995) Cell 81, 505-512. [DOI] [PubMed] [Google Scholar]
- 17.Green, D. R. & Reed, J. C. (1998) Science 281, 1309-1312. [DOI] [PubMed] [Google Scholar]
- 18.Iolascon, A., Borriello, A., Giordani, L., Cucciolla, V., Moretti, A., Monno, F., Criniti, V., Marzullo, A., Criscuolo, M. & Ragione, F. D. (2003) Cancer Genet. Cytogenet. 146, 41-47. [DOI] [PubMed] [Google Scholar]
- 19.Tweddle, D. A., Pearson, A. D., Haber, M., Norris, M. D., Xue, C., Flemming, C. & Lunec, J. (2003) Cancer Lett. (Shannon, Irel.) 197, 93-98. [DOI] [PubMed] [Google Scholar]
- 20.Fulda, S. & Debatin, K. M. (2003) Cancer Lett. (Shannon, Irel.) 197, 131-135. [DOI] [PubMed] [Google Scholar]
- 21.Ciccarone, V., Spengler, B. A., Meyers, M. B., Biedler, J. L. & Ross, R. A. (1989) Cancer Res. 49, 219-225. [PubMed] [Google Scholar]
- 22.Ambros, I. M. & Ambros, P.F. (2000) in Neuroblastoma, eds. Bordeur, G. M., Sawada, T., Tsuchida, Y. & Voute, P.A. (Elsevier, Amsterdam), pp. 233-235.
- 23.Ross, R. A., John, T. H., Reis, D. J., Spengler, B. A. & Biedler, J. R. (1980) in Advances in Neuroblastoma Research, ed. Evans, A. E. (Raven, New York), pp. 151-160.
- 24.Teitz, T., Wei, T., Valentine, M. B., Vanin, E. F., Grenet, J., Valentine, V. A., Behm, F. G., Look, A. T., Lahti, J. M. & Kidd, V. J. (2000) Nat. Med. 6, 529-535. [DOI] [PubMed] [Google Scholar]
- 25.Jackson, S. P. (2002) Carcinogenesis 23, 687-696. [DOI] [PubMed] [Google Scholar]
- 26.Cohen, H. Y., Miller, C., Bitterman, K. J., Wall, N. R., Hekking, B., Kessler, B., Howitz, K. T., Gorospe, M., de Cabo, R. & Sinclair, D. A. (2004) Science 305, 390-392. [DOI] [PubMed] [Google Scholar]
- 27.Bian, X., McAllister-Lucas, L. M., Shao, F., Schumacher, K. R., Feng, Z., Porter, A. G., Castle, V. P. & Opipari, A. W., Jr. (2001) J. Biol. Chem. 276, 48921-48929. [DOI] [PubMed] [Google Scholar]
- 28.Blatt, N. B., Bednarski, J. J., Warner, R. E., Leonetti, F., Johnson, K. M., Boitano, A., Yung, R., Richardson, B. C., Johnson, K. J., Ellman, J. A., et al. (2002) J. Clin. Invest. 110, 1123-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sikora, E., Kaminska, B., Radziszewska, E. & Kaczmarek, L. (1992) FEBS Lett. 312, 179-182. [DOI] [PubMed] [Google Scholar]
- 30.Sawada, M., Sun, W., Hayes, P., Leskov, K., Boothman, D. A. & Matsuyama, S. (2003) Nat. Cell Biol. 5, 320-329. [DOI] [PubMed] [Google Scholar]
- 31.Marks, P. A., Richon, V. M. & Rifkind, R. A. (2000) J. Natl. Cancer Inst. 92, 1210-1216. [DOI] [PubMed] [Google Scholar]
- 32.Johnstone, R. W. (2002) Nat. Rev. Drug Discov. 1, 287-299. [DOI] [PubMed] [Google Scholar]
- 33.Adams, J. M. & Cory, S. (2002) Cur. Opin. Cell Biol. 14, 715-720. [DOI] [PubMed] [Google Scholar]
- 34.Adrain, C. & Martin, S. J. (2001) Trends Biochem. Sci. 26, 390-397. [DOI] [PubMed] [Google Scholar]
- 35.Cohen, H. Y., Lavu, S., Bitterman, K. J., Hekking, B., Imahiyerobo, T. A., Miller, C., Frye, R., Ploegh, H., Kessler, B. M. & Sinclair, D. A. (2004) Mol. Cell 13, 627-638. [DOI] [PubMed] [Google Scholar]
- 36.Tafani, M., Minchenko, D. A., Serroni, A. & Farber, J. L. (2001) Cancer Res. 61, 2459-2466. [PubMed] [Google Scholar]
- 37.Coffey, D. C., Kutko, M. C., Glick, R. D., Swendeman, S. L., Butler, L., Rifkind, R., Marks, P. A., Richon, V. M. & LaQuaglia, M. P. (2000) Med. Pediatr. Oncol. 35, 577-581. [DOI] [PubMed] [Google Scholar]
- 38.Mimori, T., Hardin, J. A. & Steitz, J. A. (1986) J. Biol. Chem. 261, 2274-2278. [PubMed] [Google Scholar]
- 39.Rathmell, W. K. & Chu, G. (1994) Proc. Natl. Acad. Sci. USA 91, 7623-7627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dvir, A., Peterson, S. R., Knuth, M. W., Lu, H. & Dynan, W. S. (1992) Proc. Natl. Acad. Sci. USA 89, 11920-11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gottlieb, T. M. & Jackson, S. P. (1993) Cell 72, 131-142. [DOI] [PubMed] [Google Scholar]
- 42.Sawada, M., Hayes, P. & Matsuyama, S. (2003) Nat. Cell Biol. 5, 352-357. [DOI] [PubMed] [Google Scholar]
- 43.Li, M., Luo, J., Brooks, C. L. & Gu, W. (2002) J. Biol. Chem. 277, 50607-50611. [DOI] [PubMed] [Google Scholar]
- 44.Wilson, C. R., Davidson, S. E., Margison, G. P., Jackson, S. P., Hendry, J. H. & West, C. M. (2000) Br. J. Cancer 83, 1702-1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rashmi, R., Kumar, S. & Karunagaran, D. (2004) Carcinogenesis 25, 1867-1877. [DOI] [PubMed] [Google Scholar]
- 46.Omori, S., Takiguchi, Y., Suda, A., Sugimoto, T., Miyazawa, H., Tanabe, N., Tatsumi, K., Kimura, H., Pardington, P. E., Chen, F., et al. (2002) DNA Repair (Amst) 1, 299-310. [DOI] [PubMed] [Google Scholar]
- 47.Gu, Y., Jin, S., Gao, Y., Weaver, D. T. & Alt, F. W. (1997) Proc. Natl. Acad. Sci. USA 94, 8076-8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li, G. C., He, F., Shao, X., Urano, M., Shen, L., Kim, D., Borrelli, M., Leibel, S. A., Gutin, P. H. & Ling, C. C. (2003) Cancer Res. 63, 3268-3274. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.