Abstract
Legionella pneumophila invades and replicates intracellularly in human and protozoan hosts. The bacteria use the Icm/Dot type IVB secretion system to translocate effectors that inhibit phagosome maturation and modulate host vesicle trafficking pathways. To understand how L. pneumophila modulates organelle trafficking in host cells, we carried out pathogen effector protein screening in yeast, identifying L. pneumophila genes that produced membrane trafficking [vacuole protein sorting (VPS)] defects in yeast. We identified four L. pneumophila DNA fragments that perturb sorting of vacuolar proteins. Three encode ORFs of unknown function that are translocated via the Icm/Dot transporter from Legionella into macrophages. VPS inhibitor protein (Vip) A is a coiled-coil protein, VipD is a patatin domain-containing protein, and VipF contains an acetyltransferase domain. Processing studies in yeast indicate that VipA, VipD, and VipF inhibit lysosomal protein trafficking by different mechanisms; overexpressing VipA has an effect on carboxypeptidase Y trafficking, whereas VipD interferes with multivesicular body formation at the late endosome and endoplasmic reticulum-to-Golgi body transport. Such differences highlight the multiple strategies L. pneumophila effectors use to subvert host trafficking processes. Using yeast as an effector gene discovery tool allows for a powerful, genetic approach to both the identification of virulence factors and the study of their function.
Keywords: bacterial pathogenesis, host-pathogen interaction, virulence
Legionella pneumophila is a Gram-negative facultative intracellular pathogen of protozoa and human alveolar macrophages. Human infection results in a severe pneumonia known as Legionnaires' disease. L. pneumophila alters host cell trafficking pathways in both types of host cells. The bacteria reside in a vacuole that does not fuse with lysosomes or acidify but recruits endoplasmic reticulum (ER)-derived vesicles (1-4). These properties require a type IVB secretion system (TFBSS) known as the Icm/Dot system (5, 6). The essential nature of the TFBSS predicts that Icm/Dot translocated effectors modulate host organelle trafficking by interacting with host components to inhibit phagolysosome formation and promote establishment of a permissive, replicative vacuole. Several Icm/Dot substrates have been identified: LidA, RalF, LepA, LepB, SdeB, SdeC, SdhB, and SidA-SidH (7-10). Among these, only RalF has been shown to directly perturb vesicle trafficking by inhibiting acquisition of the ARF1 GTPase to the Legionella vacuole. The LepA and LepB proteins are involved in the release of Legionella from protozoa by an unknown mechanism. Mutants in all of the above effectors have no discernible defects in inhibition of phagolysosome maturation or intracellular growth, which suggests the presence of additional effectors that interact with host trafficking factor(s) and contribute to modulation of host organelle traffic.
To identify effectors that alter eukaryotic trafficking pathways, we took advantage of a well characterized system in budding yeast used to study organelle trafficking pathways. In Saccharomyces cerevisiae, vacuole protein sorting (VPS) pathway components (VPS proteins) control several distinct vesicle trafficking pathways (11-13). Moreover, numerous mammalian VPS orthologs play similar roles in endosomal trafficking pathways. Examples include the following: hVPS34, a human phosphatidylinositol 3-kinase involved in endosomal transport events including phagosome maturation (14, 15); Tsg101, a human homolog of Vps23p (16); and SKD1 and mVps18p, mouse homologs of Vps4p and Vps18p, respectively (17, 18).
We screened a L. pneumophila genomic library for genes that induce a Vps- phenotype in yeast as candidate effectors capable of altering endosomal traffic. Using pathogen effector protein screening in yeast (PEPSY), we identified three genes [VPS inhibitor protein (vip) A, vipD, and vipF] that encode proteins that specifically interfere with trafficking in yeast and are translocated from L. pneumophila into host macrophages via the Icm/Dot TFBSS.
Materials and Methods
Bacterial and Yeast Strains. Bacterial strains, media, and antibiotic concentrations were used as previously described (9). The diploid NSY01 was generated from the haploid BHY10 (19) by homothallic switching endonuclease (HO endonuclease) induction (20). BHY10 was transformed with YCp50::HOc12 [a gift of Aaron Mitchell (Columbia University, New York)], and the diploid was isolated (20).
Plasmid Construction. Genomic DNA was isolated from stationary phase L. pneumophila Philadelphia 1 isolate grown overnight in N-(2-acetamido)-2-aminoethanesulfonic acid (ACES)-buffered yeast extract (AYE) broth by using the DNeasy Kit (Qiagen, Valencia, CA). Thirty micrograms of genomic DNA was partially digested with Sau3AI for 15 min at 37°C. Digested DNA was run on an agarose gel, the region corresponding to ≈0.8-5 kb was excised, and DNA was purified. pWS93 [a gift of Marian Carlson (Columbia University)] was linearized by BamHI/BglII digestion to remove a 3xHA tag located between those two sites. Sau3AI-digested L. pneumophila DNA was ligated to linearized pWS93 and transformed into DH5α. Carbenicillin-resistant colonies (≈35,000) were scraped, and plasmid DNA was prepared. Plasmid DNA was transformed into NSY01, and transformants were plated on synthetic complete medium without uracil (SC-Ura) plates with 2% fructose as the only carbon source. pNS00 was generated by self-ligating pWS93 linearized by BamHI/BglII digestion. VPS4E233Q was amplified from pMB66 (21) and cloned into the SalI site of pNS00. To make cyclase fusions, vipA, vipD, vipE, and vipF were amplified from L. pneumophila genomic DNA and cloned in the KpnI/XbaI site of pJC141 (9). Plasmids encoding vip-cyaA fusions were electroporated into JR32 or dotA and plated on charcoal yeast extract agar plates containing chloramphenicol (5 μg/ml) (9). Construction of the GFP-tagged carboxypeptidase S (GFP-CPS) plasmid is described elsewhere (36).
Invertase (Inv) Assay. Assays were based on previous methodologies (22). For the qualitative plate assay, NSY01 transformant colonies on SC-Ura/fructose plates were overlaid with agar containing 125 mM sucrose, 100 mM sodium acetate (pH 5.5), 0.5 mM N-ethylmaleimide (NEM), 10 μg/ml horseradish peroxidase, 8 units/ml glucose oxidase, and 2 mM O-dianisidine. The trafficking defect was assessed by the intensity of brownness compared in vector-only strain (white, Vps+) and Vps4E233Q strain (brown, Vps-). The quantitative assay was performed on stationary-phase liquid cultures as previously described (11, 22, 23). Stationary-phase liquid cultures of vip-expressing strains were grown in SC-Ura/fructose and divided into two samples: total Inv activity and exogenous Inv activity. For total Inv activity, yeast were first lysed by four freeze/thaw cycles. Samples were then tested for Inv activity by the addition of sucrose and glucostat reagent. Reactions were stopped by using 6M HCl, and the absorbance at 540 nm was measured. One unit of Inv activity is defined as the amount of enzyme that hydrolyzes sucrose to produce 1 μmol of glucose per minute at 30°C. Values for exogenous and total Inv activity were obtained and used to calculate the percent secreted. To test for a plasmid-dependent Vps- effect, plasmid DNA was isolated from Vps- clones, amplified in Escherichia coli, and used to transform NSY01. Vps- clones were cured of their plasmids by streaking once on yeast extract/peptone/fructose plates, followed by growth on synthetic complete (SCC)/fructose plates supplemented with 5-fluoroorotic acid (5-FOA). 5-FOA-resistant colonies were restreaked on SCC/fructose plates. The qualitative Inv plate assay was performed on these cured strains.
Immunoprecipitations. Cell labeling and immunoprecipitations were performed as described previously (37). Briefly, mid-log-phase (OD600 of ≈0.6) cultures were concentrated to OD600 = 3, and 2 ml was labeled with 23 μl of Tran 35S label (PerkinElmer Life and Analytical Sciences, Boston) for 10 min in SC-Ura medium. Cells were chased with 5 mM methionine, 2 mM cysteine, and 0.2% yeast extract for the indicated times (Fig. 2). Proteins were then precipitated in 10% trichloroacetic acid. Protein pellets were washed twice with ice-cold acetone, dried in vacuo, and processed for immunoprecipitation as described previously (37). Immunoprecipitated proteins were resuspended in sample buffer for resolution by SDS/PAGE, and gels were developed by autoradiography.
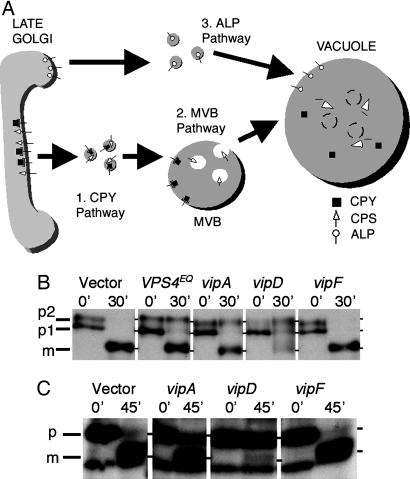
Fig. 2.
vip gene expression inhibits different trafficking pathways in yeast. (A) Golgi membrane-to-vacuole sorting pathways in S. cerevisiae. The CPY and MVB (CPS) pathways diverge at the late endosome, where the latter involves internal vesicle formation. After fusion with the vacuole, MVBs are degraded in the lumen by various enzymes. ALP is sorted via an AP3-dependent pathway that bypasses endosomes. (B and C) Immunoprecipitation of vacuolar proteins in vip-expressing strains. Cells were [35S] methionine-labeled at 26°C with a 10-min pulse, followed by a chase to the indicated time points as described in Materials and Methods. Intracellular CPY (B) and CPS (C) were immunoprecipitated by using the appropriate antibodies, and samples were resolved by SDS/PAGE and visualized by autoradiography. Precursor (p) and mature (m) forms are indicated to the left. (B) For CPY, the ER-modified (p1), Golgi membrane-modified (p2), and mature (m) forms are shown. (C) For CPS, the Golgi membrane-modified (p) and mature (m) forms are shown.
Fluorescence Microscopy. Cells expressing GFP-CPS were grown to an OD600 of 0.6 and labeled with the fluorescent lipophilic dye N-[3-triethylammoniumpropyl]-4-[p-diethylaminophenylhexatrienyl] pyridinium dibromide (FM4-64; Molecular Probes) as previously described (38). Cells were visualized by using a Zeiss Axiovert S1002TV inverted fluorescent microscope equipped with fluorescein isothiocyanate (FITC) and rhodamine filters. Images were captured with a Photometrics (Tucson, AZ) camera and processed with deltavision deconvolution software (Applied Precision, Seattle) and adobe photoshop 8.0 (Adobe Systems, San Jose, CA). All observations are based on the examination of at least 100 cells, and representative fields are shown.
cAMP Measurement. cAMP measurement was performed as previously described (9). Briefly, 4 × 106 J774 cells in RPMI medium 1640 (Cellgro; Mediatech, Washington, DC) with 10% normal human serum were infected with L. pneumophila strains by using a multiplicity of infection (moi) of 50 and incubated at 37°C for 30 min. After removal of supernatant, lysates were extracted by the addition of cold 2.5% perchloric acid for 10 min, followed by neutralization with KOH. cAMP levels were measured by using the cAMP Biotrak Enzymeimmunoassay System (Amersham Pharmacia Biosciences).
Results
Pathogen Effector Protein Screening in Yeast (PEPSY) Screening Identifies L. pneumophila Genes That Inhibit Vacuolar Traffic. The methodology used to identify a VPS phenotype (11, 12, 19, 22) takes advantage of the properties of a hybrid protein resulting from a fusion between Inv and carboxypeptidase Y (CPY). The SUC2 gene encodes an Inv (β-d-fructofuranoside fructohydrolase) that is normally localized at the cell surface. The PRC1 gene encodes CPY, a vacuolar hydrolase that undergoes posttranslational modifications in the Golgi body, followed by trafficking to the late endosome and sorting to the lysosome-like yeast vacuole. Because the sorting signals for CPY lie in the N-terminal region, where the first 50 aa of CPY are fused to the 433 C-terminal amino acids of Inv, the CPY-Inv hybrid protein traffics from the ER to the Golgi body and then to the vacuole via the late endosome. Because the CPY-Inv hybrid is sequestered in the vacuole and cannot reach the cell surface, the cell cannot hydrolyze exogenous sucrose. However, when normal trafficking of CPY-Inv is perturbed, cargo vesicles are blocked from reaching the vacuole, resulting in missorting of the hybrid protein and subsequent spillover into vesicles destined for the cell surface. This aberrant secretion of the CPY-Inv hybrid allows the cell to hydrolyze exogenous sucrose. This phenotype can be scored at the colony level by a simple agar overlay containing sucrose and chromogenic reagents that indicate glucose production by a brown precipitate. A wild-type Vps+ phenotype is scored as a white colony, whereas a Vps- phenotype is scored as a brown colony (22).
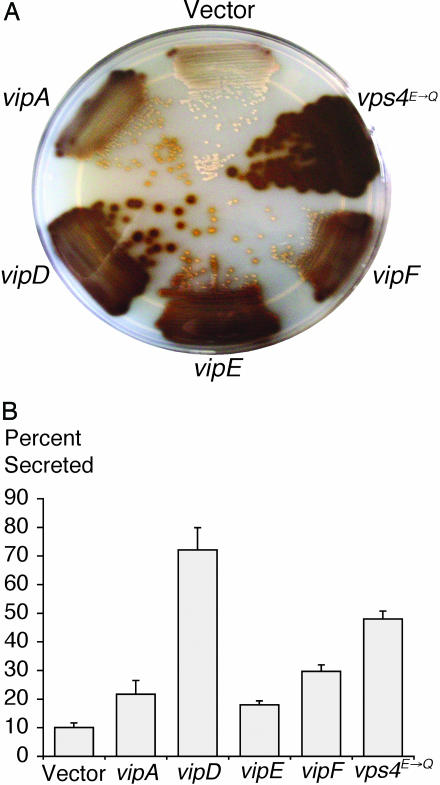
To look for L. pneumophila proteins that would perturb VPS, we screened for bacterial genes capable of inducing a Vps- phenotype in yeast. A random library of L. pneumophila genomic DNA was generated by a Sau3AI partial digest. Fragments were cloned into the URA3+ 2 μ yeast expression vector pNS00. Gene expression from inserted DNA in this plasmid is under the control of the strong constitutive ADH1 promoter. Plasmid pools were transformed into the diploid Ura- indicator yeast strain NSY01, which expresses the CPY-Inv hybrid protein. Transformants were plated on SC-Ura plates with fructose as the sole carbon source. We screened ≈80,000 colonies by overlaying with agar containing glucose detection reagents, and those that turned brown were restreaked and retested (Fig. 1A). CPY-Inv secretion was also quantified in liquid cultures (Fig. 1B). Plasmid DNA was isolated from brown Vps- colonies and retransformed into NSY01. In parallel, the Vps- yeast strains were cured of the plasmids, and their Vps phenotype was retested. Candidates in which the Vps phenotype was linked to the plasmid by both retransformation and curing were kept for further analysis. The DNA sequences of the inserts in these plasmids were determined and analyzed by using the L. pneumophila genome database (24). The positive control in our assays was the expression of a dominant negative allele of the AAA ATPase Vps4p. Mutation of a conserved glutamate at position 233 of Vps4p abolishes its VPS function in a dominant negative manner (21). We identified four independent clones that induce a Vps- phenotype in yeast. The genes carried by the clones were designated vipA, vip D, vipE, and vipF.
Fig. 1.
vip gene expression causes CPY-Inv secretion in yeast. (A) Qualitative Inv assay on NSY01 strains transformed with plasmids carrying empty vector, VPS4E233Q, vipA, vipD, vipE, and vipF clones. A SC-Ura/fructose plate was overlaid with top agar containing chromogenic reagents that resulted in a brown precipitate upon secretion of the CPY-Inv hybrid protein. (B) Quantitative Inv assay on vip clones. Inv activity in total cultures and in culture supernatants was measured as described in Materials and Methods. Values for exogenous and total Inv activity were obtained and used to calculate the percent secreted. Assays were done in triplicate at least twice.
Protein Similarities and Properties. The fragment expressing vipA contained two other ORFs downstream of vipA, but mutational analysis determined that only removal of the vipA coding region abolished CPY-Inv secretion (data not shown). We cloned the full-length vipA gene into pNS00, and the corresponding NSY01 strain exhibited CPY-Inv secretion similar to that of the original Sau3AI vipA fragment (data not shown). The sequence of VipA is predicted to have a coiled-coil region but exhibits no extensive homologies to known proteins. The fragment designated vipD contained an 848-bp sequence encoding a C-terminal region of the VipD protein, where the initiating methionine is at residue 341. This results in a polypeptide encompassing residues 341-613 of the full-length VipD. The full-length gene encodes a patatin domain and shows strong homology to the Pseudomonas aeruginosa type III translocated effector, ExoU. The patatin domain also showed considerable homology to eukaryotic phospholipase A genes. However, the vipD fragment that results in a Vps- phenotype does not include the catalytic patatin domain. Interestingly, when cloned into pNS00, the full-length vipD gene also caused a Vps- phenotype but missecreted only 24 ± 3% of total CPY-Inv (data not shown), significantly less than the 72 ± 8% missecreted by the fragment encoding the C-terminal region (Fig. 1B). Expression of full-length VipD in yeast did not result in any detectable toxicity (data not shown). The vipE clone contained a single ORF that exhibited no extensive homology to other genes. The vipF clone contained a single gene that exhibited homologies to genes that encode acetyltransferases (e scores between e-4 and e-24; data not shown).
Characterization of Trafficking Pathways Affected in Yeast by vip Expression. To characterize the defects in trafficking produced by vip gene expression, we examined the proteolytic maturation of proteins that are sorted to the vacuole via the three major pathways in yeast (Fig. 2A): (i) the CPY sorting pathway (13), (ii) the multivesicular body (MVB) pathway (25), and (iii) the alkaline phosphatase (ALP) pathway (26). CPY is a soluble hydrolase that traffics to the vacuole via endosomes in a receptor-mediated fashion and does not require all components of the MVB pathway. CPS, however, is a transmembrane protein that is sorted into MVB vesicles at the late endosome before final delivery and maturation at the vacuole. Finally, ALP, a transmembrane protein residing in the vacuolar membrane, bypasses endosomes entirely and reaches the vacuole by the AP3 adaptor-mediated sorting pathway (27, 28).
Processing of these proteins in the Golgi body and the vacuole results in precursor and mature forms of different molecular weights. To examine maturation in yeast strains expressing different Vip proteins, we 35S-labeled nascent proteins by pulse-chase and determined the extent of CPS, CPY, and ALP processing by immunoprecipitation and SDS/PAGE (Fig. 2 B and C; data not shown for ALP). VipA expression resulted in partial accumulation in p2 CPY (Golgi form), indicating a block in trafficking between the Golgi body and the late endosome and/or between the late endosome and the vacuole. VipA expression also caused a partial accumulation of precursor CPS but no detectable effect on ALP. Strains expressing the VipD C-terminal fragment caused a more severe defect in processing: there was a partial kinetic delay in the conversion of the p1 (ER) form to the p2 (Golgi) form, and most of the intracellular CPY accumulated in the p2 form, indicating a defect in Golgi body-to-vacuole trafficking. Furthermore, given the strong CPY-Inv secretion exhibited by this strain, it is likely that the majority of the p2 CPY is secreted into the growth medium and therefore was not detected on the gel. Similarly, expression of the VipD C-terminal region inhibited nearly all CPS and ALP processing (Fig. 2C; data not shown for ALP). In contrast, VipF expression did not cause a detectable alteration in the processing of any of the three proteins.
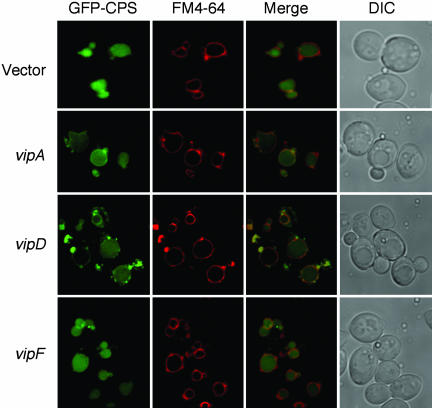
We also analyzed the localization of GFP-CPS in the Vip-expressing strains by fluorescence microscopy (Fig. 3). Although the GFP-CPS protein is normally localized in the vacuole lumen, VipA expression resulted in partial mislocalization to the membrane. Expression of the VipD C-terminal region resulted in a more striking phenotype, where some cells showed localization of the GFP-CPS protein exclusively in the vacuole membrane and others showed partial membrane localization. Interestingly, all cells exhibited an accumulation of GFP-CPS on punctate structures on or near the vacuole membrane. These structures are reminiscent of an aberrant endosomal membrane compartment observed in class E vps mutants (29), wherein MVB sorting is impaired (30). As in the case of the processing assays, VipF had no discernible effect on GFP-CPS localization.
Fig. 3.
Steady-state localization of a GFP-CPS fusion in vip-expressing strains. Cells were harvested in mid-log phase and labeled with N-[3-triethylammoniumpropyl]-4-[p-diethylaminophenylhexatrienyl] pyridinium dibromide (FM4-64) to visualize the vacuole membrane (as described in Materials and Methods). Fluorescence was observed and imaged by using the deltavision system (Applied Precision). DIC, differential interference contrast microscopy (Nomarski optics).
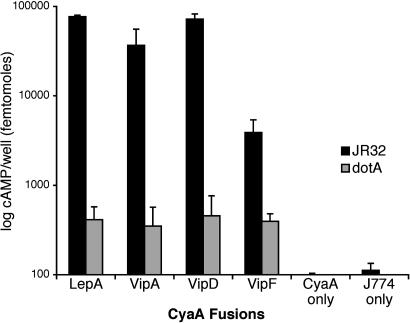
Legionella Translocates the VipA, VipD, and VipF Proteins into Macrophages via the Icm/Dot TFBSS. If the Vip proteins are bona fide effectors, they should be substrates of the Icm/Dot TFBSS. To determine whether they are translocated into host cells by L. pneumophila, we used a reporter that has been used to study the translocation of many bacterial effectors into eukaryotic cells. This reporter, the cyaA sequence of Bordetella pertussis, encodes an adenylate cyclase that is activated by eukaryotic calmodulin. Thus, increased intracellular cAMP levels after infection by strains expressing effector-cyclase hybrids is indicative of CyaA translocation from bacteria into the host cytosol (31). The vipA, vipD, vipE, and vipF genes were cloned into pJC141, a reporter plasmid derived from pMMB207C that is used for creating upstream translational fusions to cyaA (9). Plasmids containing the Vip-CyaA hybrids were transformed into the wild-type JR32 strain and a dotA mutant in which the TFBSS is inactive. Because the VipE-CyaA hybrid was unstable and had low intrinsic adenylate cyclase activity, it was not analyzed further. J774 macrophages were infected with L. pneumophila expressing the Vip-CyaA hybrid proteins or unfused CyaA (Fig. 4). The levels of cAMP produced by the strains containing the Vip-CyaA hybrids resembled the levels produced by the positive control LepA-CyaA, a previously characterized translocated effector hybrid (9). The differences in cAMP levels produced by infection by wild-type and dotA strains were between 10- and 100-fold, similar to values obtained with other effectors (9), indicating that a functional Icm/Dot TFBSS is required for translocation. Constructs expressing only CyaA resulted in very low levels of cAMP, indicating that increased cAMP levels required the presence of the Vip sequences. We conclude that the VipA, VipD, and VipF proteins that produce trafficking defects in yeast are also substrates of the Icm/Dot TFBSS.
Fig. 4.
Cyclase translocation assays. J774 macrophages were seeded into 96-well tissue culture plates and infected with L. pneumophila strains for 30 min. After the removal of the supernatant, cells were lysed with cold 2.5% perchloric acid and neutralized with KOH. cAMP levels measured as in previous studies (9) by using the cAMP Biotrak Enzymeimmunoassay System (Amersham Pharmacia Biosciences).
Discussion
L. pneumophila modulates host vesicle traffic to create a protective, replicative vacuole. This is likely accomplished by injecting effector proteins into the host via the Icm/Dot TFBSS. Effectors modulate phagosome trafficking by unknown mechanisms. Despite the identification of numerous Icm/Dot substrates, all appear to be dispensable for proper phagosome trafficking and intracellular growth. It has been proposed that L. pneumophila possesses a high degree of functional redundancy and species specificity in effector function (9, 10, 24). We sought to directly identify effectors that can interact with endosomal trafficking pathways. We took advantage of the CPY-Inv assay, previously used to identify the vps genes (11, 12) that encode components of these pathways in both lower and higher eukaryotes. Using pathogen effector protein screening in yeast (PEPSY), we identified four Sau3AI fragments and characterized three genes: vipA, vipD, and vipF, whose expression in yeast results in mistrafficking of vacuolar proteins. Using calmodulin-activated adenylate cyclase as a reporter, we determined that L. pneumophila indeed translocates the VipA, VipD, and VipF proteins via the Icm/Dot TFBSS into host macrophages.
At present, it is difficult to identify the precise nature of the defects caused by the Vip proteins or their direct targets. However, some information can be gleaned from the Vip protein sequences. VipA contains a relatively large coiled-coil region, as do previously documented L. pneumophila effectors such as LepA, LepB, LidA, SidC, SidE, SdeA, SdeB, and SdhB. Coiled coils are highly versatile structures involved in protein interactions, and several trafficking components, such as SNAREs, EEA1, and Uso1p (32, 33), possess coiled coils. We speculate that L. pneumophila effectors may interact with similar host proteins via their coiled-coil regions. The N-terminal domain of VipD contains a patatin domain that is found in the P. aeruginosa effector ExoU, which has lipolytic activity that is lethal to eukaryotic cells. However, this activity depends on activation by unknown eukaryotic cell-derived factors (34-36). The VipD patatin domain also bears strong homology to eukaryotic phospholipase A2 proteins that generate the signaling lipid arachidonic acid, which is also the precursor for other important lipid molecules such as leukotrienes and prostaglandins. Recent work from various laboratories has also implicated a role for phospholipase A2 in Golgi membrane dynamics, ER-to-Golgi membrane traffic, and even phagosome biogenesis (37-39). However, the fragment isolated from the Vps screen encodes only the C-terminal domain and lacks the patatin domain. This C-terminal region does not bear any sequence or structural homology to known proteins or motifs, yet it causes the strongest defect in vacuolar traffic, even when compared with the dominant negative Vps4pE233Q allele. Expression of the VipD C-terminal region significantly delayed the trafficking of CPY from the ER to the Golgi membrane and from the Golgi membrane to the vacuole. The trafficking of CPS and ALP to the vacuole was also delayed. When we expressed the full-length VipD protein, it resulted in a less severe defect in CPY-Inv trafficking, which could be caused by differential stability or expression levels between the two forms of VipD or possibly by the N-terminal domain's negatively regulating the C-terminal domain. It is also possible that the C-terminal region targets or titrates a limiting host trafficking component more efficiently than the full-length protein. In support of the latter model, it has been hypothesized that the C terminus of ExoU (which also excludes the patatin domain) interacts with a eukaryotic factor necessary for ExoU function and cytotoxicity (40, 41). Unlike ExoU, expression of full-length VipD was not lethal in yeast but was sufficient to cause a Vps- phenotype. VipF is a protein that has an acetyltransferase domain. Despite its ability to cause mistrafficking of CPY-Inv, pulse-chase experiments and fluorescence microscopy did not reveal a detectable phenotype. However, it is important to note that several of the original vps mutants also exhibited stronger defects in CPY-Inv sorting than did the wild-type protein; a likely explanation for this observation is that the fusion protein is sorted less efficiently than CPY because it is not recognized as well by the CPY receptor Vps10p (12).
Yeast has been used as a tool to understand bacterial virulence by many groups and was the subject of a recent review (42). However, these studies used yeast to characterize known virulence factors. We wanted to use yeast as a gene discovery tool to identify effectors that inhibit vesicle traffic. We were encouraged by earlier work showing that many bacterial effectors normally involved in mammalian infection retain their biological function in yeast (42, 43). Furthermore, L. pneumophila infects and modulates host traffic in species from different kingdoms. This may be due to interactions between effectors and host targets that are highly conserved. There may be other host-specific effectors, such as LepA and LepB, which appear to function in protozoa but not in mammalian hosts (9). The fact that we recovered Icm/Dot translocated effectors that interfere with organelle trafficking in yeast supports the idea that some effector-host interactions are conserved. The three Vip proteins do not share common structures, nor do they appear to inhibit organelle trafficking by the same mechanisms. This may reflect multiple strategies that L. pneumophila effectors use to intersect with the host trafficking pathways. We found four other patatin domain-containing VipD paralogs in the L. pneumophila genome database, lpg1227, lpg2410, lpg1426, and lpg0670. Interestingly, they share strong homology among their N-terminal patatin-like domains but not in their C-terminal regions. There may be additional vip genes that we did not find because of the lack of a proximal Sau3AI site or because of lethality in yeast. For example, lepB expression is lethal in yeast and tolerated only under low-copy, repressing conditions (data not shown). Furthermore, it has been shown that expression of lepA in yeast results in a Vps- phenotype (C. D. Pericone, N.S., and H.A.S., unpublished data).
It should be possible to use other yeast tools such as suppressor analysis to identify host factors that either dampen or enhance the Vip-dependent Vps- phenotype. The pathogen effector protein screening in yeast (PEPSY) approach can also be applied to other pathogens that modulate host cell trafficking such as Salmonella typhimurium and Mycobacterium tuberculosis and less characterized pathogens such as Coxiella burnetii and Brucella abortus. This strategy provides additional approaches to identify virulence factors and study their mode of action, thus contributing to the understanding of host-pathogen interactions during intracellular survival and multiplication.
Acknowledgments
We thank Marian Carlson and Aaron Mitchell for advice and materials, and Chris Pericone and Jonathan Dworkin for helpful comments on the manuscript. This work was supported by National Institutes of Health Grants AI23549 (to H.A.S.) and CA58689 (to S.D.E.). S.D.E. is an investigator of the Howard Hughes Medical Institute.
Author contributions: N.S., S.D.E., and H.A.S. designed research; N.S. and J.A.E. performed research; N.S. and J.A.E. contributed new reagents/analytic tools; N.S., S.D.E., and H.A.S. analyzed data; and N.S. wrote the paper.
Abbreviations: ER, endoplasmic reticulum; TFBSS, type IVB secretion system; VPS, vacuole protein sorting; Vip, VPS inhibitor protein; SC-Ura, synthetic complete medium without uracil; CPS, carboxypeptidase S; GFP-CPS, GFP-tagged CPS; Inv, invertase; CPY, carboxypeptidase Y; MVB, multivesicular body; ALP, alkaline phosphatase.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AY779515, AY779516, AY779517, and AY779518 (vipA, vipD, vipE, and vipF, respectively)].
References
- 1.Cianciotto, N. P. (2001) Int. J. Med. Microbiol. 291, 331-343. [DOI] [PubMed] [Google Scholar]
- 2.Horwitz, M. A. & Silverstein, S. C. (1983) J. Clin. Invest. 71, 15-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horwitz, M. A. & Maxfield, F. R. (1984) J. Cell Biol. 99, 1936-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kagan, J. C. & Roy, C. R. (2002) Nat. Cell Biol. 4, 945-954. [DOI] [PubMed] [Google Scholar]
- 5.Vogel, J. P., Andrews, H. L., Wong, S. K. & Isberg, R. R. (1998) Science 279, 873-876. [DOI] [PubMed] [Google Scholar]
- 6.Segal, G., Purcell, M. & Shuman, H. A. (1998) Proc. Natl. Acad. Sci. USA 95, 1669-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conover, G. M., Derre, I., Vogel, J. P. & Isberg, R. R. (2003) Mol. Microbiol. 48, 305-321. [DOI] [PubMed] [Google Scholar]
- 8.Nagai, H., Kagan, J. C., Zhu, X., Kahn, R. A. & Roy, C. R. (2002) Science 295, 679-682. [DOI] [PubMed] [Google Scholar]
- 9.Chen, J., Suwwan de Felipe, K., Clarke, M., Lu, H., Anderson, O. R., Segal, G. & Shuman, H. A. (2004) Science 303, 1358-1361. [DOI] [PubMed] [Google Scholar]
- 10.Luo, Z. Q. & Isberg, R. R. (2004) Proc. Natl. Acad. Sci. USA 101, 841-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bankaitis, V. A., Johnson, L. M. & Emr, S. D. (1986) Proc. Natl. Acad. Sci. USA 83, 9075-9079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robinson, J. S., Klionsky, D. J., Banta, L. M. & Emr, S. D. (1988) Mol. Cell. Biol. 8, 4936-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bryant, N. J. & Stevens, T. H. (1998) Microbiol. Mol. Biol. Rev. 62, 230-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siddhanta, U., McIlroy, J., Shah, A., Zhang, Y. & Backer, J. M. (1998) J. Cell Biol. 143, 1647-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fratti, R. A., Backer, J. M., Gruenberg, J., Corvera, S. & Deretic, V. (2001) J. Cell Biol. 154, 631-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Babst, M., Odorizzi, G., Estepa, E. J. & Emr, S. D. (2000) Traffic 1, 248-258. [DOI] [PubMed] [Google Scholar]
- 17.Yoshimori, T., Yamagata, F., Yamamoto, A., Mizushima, N., Kabeya, Y., Nara, A., Miwako, I., Ohashi, M., Ohsumi, M. & Ohsumi, Y. (2000) Mol. Biol. Cell 11, 747-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poupon, V., Stewart, A., Gray, S. R., Piper, R. C. & Luzio, J. P. (2003) Mol. Biol. Cell 14, 4015-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horazdovsky, B. F., Busch, G. R. & Emr, S. D. (1994) EMBO J. 13, 1297-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herskowitz, I. & Jensen, R. E. (1991) Methods Enzymol. 194, 132-146. [DOI] [PubMed] [Google Scholar]
- 21.Babst, M., Wendland, B., Estepa, E. J. & Emr, S. D. (1998) EMBO J. 17, 2982-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darsow, T., Odorizzi, G. & Emr, S. D. (2000) Methods Enzymol. 327, 95-106. [DOI] [PubMed] [Google Scholar]
- 23.Klionsky, D. J., Banta, L. M. & Emr, S. D. (1988) Mol. Cell. Biol. 8, 2105-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chien, M., Morozova, I., Shi, S., Sheng, H., Chen, J., Gomez, S. M., Asamani, G., Hill, K., Nuara, J., Feder, M., et al. (2004) Science 305, 1966-1968. [DOI] [PubMed] [Google Scholar]
- 25.Katzmann, D. J., Odorizzi, G. & Emr, S. D. (2002) Nat. Rev. Mol. Cell Biol. 3, 893-905. [DOI] [PubMed] [Google Scholar]
- 26.Odorizzi, G., Cowles, C. R. & Emr, S. D. (1998) Trends Cell Biol. 8, 282-288. [DOI] [PubMed] [Google Scholar]
- 27.Stepp, J. D., Huang, K. & Lemmon, S. K. (1997) J. Cell Biol. 139, 1761-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piper, R. C., Bryant, N. J. & Stevens, T. H. (1997) J. Cell Biol. 138, 531-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raymond, C. K., Howald-Stevenson, I., Vater, C. A. & Stevens, T. H. (1992) Mol. Biol. Cell 3, 1389-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Odorizzi, G., Babst, M. & Emr, S. D. (1998) Cell 95, 847-858. [DOI] [PubMed] [Google Scholar]
- 31.Ladant, D. & Ullmann, A. (1999) Trends Microbiol. 7, 172-176. [DOI] [PubMed] [Google Scholar]
- 32.Burkhard, P., Stetefeld, J. & Strelkov, S. V. (2001) Trends Cell Biol. 11, 82-88. [DOI] [PubMed] [Google Scholar]
- 33.Gillingham, A. K. & Munro, S. (2003) Biochim. Biophys. Acta 1641, 71-85. [DOI] [PubMed] [Google Scholar]
- 34.Phillips, R. M., Six, D. A., Dennis, E. A. & Ghosh, P. (2003) J. Biol. Chem. 278, 41326-41332. [DOI] [PubMed] [Google Scholar]
- 35.Rabin, S. D. & Hauser, A. R. (2003) Infect. Immun. 71, 4144-4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sato, H., Frank, D. W., Hillard, C. J., Feix, J. B., Pankhaniya, R. R., Moriyama, K., Finck-Barbancon, V., Buchaklian, A., Lei, M., Long, R. M., et al. (2003) EMBO J. 22, 2959-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Figueiredo, P., Doody, A., Polizotto, R. S., Drecktrah, D., Wood, S., Banta, M., Strang, M. S. & Brown, W. J. (2001) J. Biol. Chem. 276, 47361-47370. [DOI] [PubMed] [Google Scholar]
- 38.Kuroiwa, N., Nakamura, M., Tagaya, M. & Takatsuki, A. (2001) Biochem. Biophys. Res. Commun. 281, 582-588. [DOI] [PubMed] [Google Scholar]
- 39.Girotti, M., Evans, J. H., Burke, D. & Leslie, C. C. (2004) J. Biol. Chem. 279, 19113-19121. [DOI] [PubMed] [Google Scholar]
- 40.Sato, H. & Frank, D. W. (2004) Mol. Microbiol. 53, 1279-1290. [DOI] [PubMed] [Google Scholar]
- 41.Rabin, S. D. & Hauser, A. R. (2005) Infect. Immun. 73, 573-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valdivia, R. H. (2004) Eukaryotic Cell 3, 827-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lesser, C. F. & Miller, S. I. (2001) EMBO J. 20, 1840-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]