Abstract
Misfolding and aggregation of proteins play an important part in the pathogenesis of several genetic and degenerative diseases. Recent evidence suggests that cells have evolved a pathway that involves sequestration of aggregated proteins into specialized “holding stations” called aggresomes. Here we show that cells regulate inducible NO synthase (iNOS), an important host defense protein, through aggresome formation. iNOS aggresome formation depends on a functional dynein motor and the integrity of the microtubules. The iNOS aggresome represents a “physiologic aggresome” and thus defines a new paradigm for cellular regulation of protein processing. This study indicates that aggresome formation in response to misfolded proteins may merely represent an acceleration of an established physiologic regulatory process for specific proteins whose regulation by aggresome formation is deemed necessary by the cell.
Aggresomes are recently described as discrete cytoplasmic inclusion bodies that form in response to the production of misfolded proteins (1-4). Aggresomes share both morphological and biochemical similarities with inclusion bodies that characterize common neurodegenerative diseases, including amyotrophic lateral sclerosis, Parkinson's disease, and Alzheimer's disease. Aggregated forms of proteins, including mutant forms of cystic fibrosis transmembrane conductance regulator, superoxide dismutase, rhodopsin, T cell receptor α, and presenilin-1, have been shown to localize to the aggresome. Aggresomes form around the microtubule-organizing center (MTOC) near the centrosome by way of dynein-directed retrograde transport of proteins on microtubule tracks. In addition to the aggregated proteins, aggresomes contain ubiquitin, proteasomes, and heat-shock proteins (4, 5).
Nitric oxide (NO), a multifunctional biological messenger, is synthesized from l-arginine by NO synthase (NOS) isoforms (6-8). As a signaling molecule, NO is produced by two constitutive calcium- (Ca2+) dependent isoforms, neuronal NOS (nNOS or NOS1) and endothelial NOS (eNOS or NOSIII). Ca2+-activated calmodulin binds to and transiently activates constitutive NOS dimers. As an agent of inflammation and cell-mediated immunity, NO is produced by a Ca2+-independent cytokine-inducible NOS (iNOS or NOSII) (7, 8). Calmodulin is tightly bound to iNOS even at basal Ca2+ levels (9), and therefore iNOS is notably distinguished from the constitutive isoforms by its prolonged production of a relatively large amount of NO. It has been recognized that overproduction of NO by iNOS may cause tissue damage that outweighs its potential benefit for host defense. iNOS has been implicated in the pathogenesis of many inflammatory syndromes, e.g., asthma, transplant rejection, inflammatory bowel disease, rheumatoid arthritis, and septic shock (10, 11).
Studies of the subcellular localization of endothelial NOS (eNOS) and neuronal NOS (nNOS) provided insight into the biological functions of these isoforms. eNOS is targeted, primarily by acylation, to plasmalemmal caveolae (12), and nNOS is localized in specialized postsynaptic densities (13). In contrast, most studies of iNOS have dealt with the biochemical characterization of purified soluble iNOS (7). The few studies that have dealt with iNOS subcellular localization have provided diverse findings. iNOS has been reported to reside in the cell as a diffuse cytosolic protein or to localize in vesicular or perinuclear structures (14-16). In one study, the perinuclear location was assigned to the Golgi (17). Our study demonstrates that iNOS is expressed initially as a cytosolic protein but is eventually targeted to a perinuclear localization, identified by our data as an aggresome. The latter is hitherto thought to be the site of accumulation of misfolded proteins (1-4). Thus, the iNOS aggresome serves as a prototype for what we term the “physiologic aggresome.” The term physiologic in this context is loosely used to describe aggresome formation not associated with misfolded proteins.
Materials and Methods
Cells were grown on poly(d-lysine)-coated 22-mm glass coverslips to 70% confluence in six-well clusters. Cells were washed with PBS containing 1.2% sucrose and fixed with 4% formaldehyde at room temperature for 10 min. Coverslips were mounted by using the SlowFade Antifade Kit (Molecular Probes) and the blue nuclear chromatin stain DAPI and viewed by using a delta vision deconvolution microscopy system (Applied Precision, Issaquah, WA) equipped with a Zeiss Axiovert microscope. Imaging was performed by using a Zeiss ×100 (1.4 numerical aperture) oil immersion lens, and Z sections were collected at an optical depth of 0.2 μm. Images were optimized by using delta vision deconvolution software (Applied Precision).
Cell culture, transfection, iNOS induction, cell lysis, live cell imaging, and electron microscopy were done as described in refs. 18-20 and in Supporting Text, which is published as supporting information on the PNAS web site.
Results and Discussion
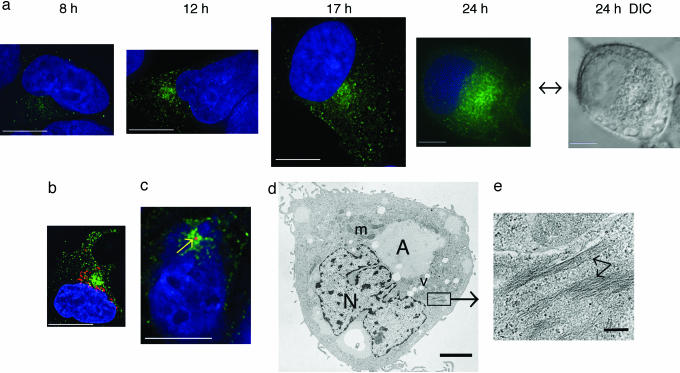
iNOS Subcellular Localization Leads to Identification of iNOS Aggresome. iNOS is expressed in vivo in response to cytokines and inflammatory mediators (18-20). Therefore, we sought to examine the subcellular localization of cytokine-induced iNOS. RT4 is a urinary bladder papilloma cell line that expresses iNOS upon cytokine stimulation (21-23). At varying times after stimulation by a mixture of IFN-γ (100 units/ml), IL-1β (0.5 ng/ml), TNF-α (10 ng/ml), and IL-6 (200 units/ml), RT4 cells were fixed and immunostained by using a monoclonal anti-iNOS antibody. After cytokine stimulation, iNOS was distributed throughout the cytoplasm, mostly as minute vesicles. Over time, iNOS vesicles eventually coalesced into a perinuclear localization (Fig. 1a). To determine whether the iNOS perinuclear accumulation correlates with a cellular palpable structure typical of an aggresome, we used Normaski differential interference contrast (DIC) microscopy in combination with immunofluorescence microscopy. The iNOS perinuclear aggregation detected by immunofluorescence microscopy appeared as a distinct structure when viewed with DIC optics (Fig. 1a; 24-h time point).
Fig. 1.
iNOS subcellular localization in RT4 cells. (a) Cells were stimulated by cytokines to induce iNOS, fixed at various time points, and immunolabeled by using a monoclonal anti-iNOS antibody and a goat anti-mouse conjugated to Alexa Fluor 488 (green). Cells were also stained with DAPI to visualize nuclei (blue). At the 24-h time point, a DIC image is shown. (b) Immunolabeling was done by using a monoclonal antibody against the Golgi resident protein giantin followed by Alexa Fluor 594-conjugated goat anti-mouse antibody (red). Proximity, but no colocalization, of iNOS aggresome to the Golgi is shown. (c) Cells were immunolabeled by using anti-human γ-tubulin antibody and an Alexa Fluor 568-conjugated anti-human secondary antibody. γ-Tubulin labeling appeared as a red dot in the center of iNOS aggresome. [Bars (a-c), 10 μm.] (d) Ultrastructure of iNOS aggresome by transmission EM. A, aggresome; m, mitochondria; N, nucleos; v, vacuole. (Bar, 2 μm.) (e) Higher magnification of section indicated by box in d. Arrows denote intermediate filaments. (Bar, 200 nm.)
The Golgi and the centrosome are the two main cellular organelles in the perinuclear area. In a recent report, the perinuclear distribution of iNOS was interpreted as localization to the Golgi (17). To investigate this possibility, we used an antibody against the Golgi resident protein giantin. iNOS failed to localize with giantin, indicating that iNOS distributes to a compartment distinct from the Golgi (Fig. 1b). Using an anti-body against human γ-tubulin, we found that the iNOS perinuclear accumulation was centered at the MTOC, surrounding the centrosome (Fig. 1c). This observation is highly reminiscent of the described process of aggresome formation associated with misfolded proteins (2, 3). Using transmission EM, the iNOS aggresome appeared as a large perinuclear accumulation (Fig. 1d), which appeared to displace other cellular organelles such as mitochondria. Importantly, the iNOS aggresome was surrounded by intermediate filaments (Fig. 1e), which has been demonstrated as a typical feature for an aggresome (2, 3, 24). These findings suggest that the perinuclear accumulation of iNOS represents an iNOS aggresome that shares some of the features of aggresomes previously thought to be solely associated with misfolded proteins.
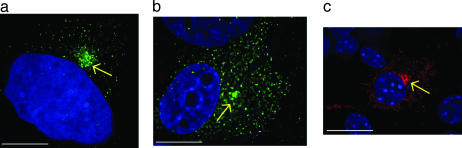
iNOS Aggresome in Various Cell Types. We next sought to determine whether the iNOS perinuclear aggregation was common to a variety of cell types. We studied A172 and A549 human cell lines and a RAW264.7 murine cell line. A172 is a human glioblastoma cell line and was used as a representative of a nonepithelial cell line. A549 is an alveolar type II epithelium lung carcinoma cell line, which produces iNOS upon stimulation by proinflammatory cytokines (21). Thus, these cells serve as a model for lung inflammation milieu. RAW264.7 is a murine macrophage cell line that allowed us to extend our observations to the murine iNOS isoform. A172 and A549 cells were stimulated by a cytokine mixture similar to that used for RT4 cells. RAW264.7 cells were stimulated by LPS and IFN-γ (21, 23). In all cell types examined, iNOS formed an aggresome at the perinuclear area (Fig. 2). However, there were some morphological variations among cell types, consistent with previous reports of cell type specific morphological patterns for aggresome formation (2, 3, 25).
Fig. 2.
iNOS aggresome in various cell types. A172 (a), A549 (b), and RAW264.7 (c) cells were stimulated by cytokine mixture and/or LPS for 18 h to induce iNOS and then fixed and immunolabeled by anti-iNOS antibody followed by goat anti-mouse IgG conjugated to Alexa Fluor 488 (green in a and b) or Alexa Fluor 594 (red in c). Cells were stained with DAPI to visualize nuclei (blue). Arrows indicate the site of iNOS aggresome formation. (Bars, 10 μm.)
Characterization of the iNOS Aggresome in Human Embryonic Kidney (HEK)293 Cells Using iNOS-GFP. To facilitate the study of mechanisms regulating formation of the iNOS aggresome, we constructed a cDNA encoding human iNOS fused to the N terminus of GFP. We generated a stable cell line of HEK293 cells expressing the iNOS-GFP chimera. The expression and activity of iNOS-GFP in this cell line were confirmed by Western blot analysis and iNOS activity assays, respectively (data not shown). HEK293 cells were used because they do not express endogenous NOS genes, and they have been used for iNOS biochemical characterization (18, 23). Three general patterns of iNOS-GFP subcellular localization were observed. In some cells, iNOS-GFP was diffuse and cytoplasmic. In other cells, cytoplasmic iNOS-GFP was diffuse, but perinuclear accumulations were evident. In a third category of cells, iNOS-GFP was present exclusively as a perinuclear accumulation that varied in size between 1 and 5 μm (data not shown). To investigate whether the cellular heterogeneity of iNOS localization represented a cellular trafficking pathway for iNOS, we performed transient transfection of HEK293 cells using cDNA of iNOS-GFP. After transfection, cells were fixed and evaluated for iNOS-GFP expression at time points ranging from 6 to 96 h. At early time points, iNOS appeared mostly as a diffuse cytosolic protein, whereas at later time points, iNOS appeared in a perinuclear distribution that eventually became the predominant localization for iNOS (data not shown).
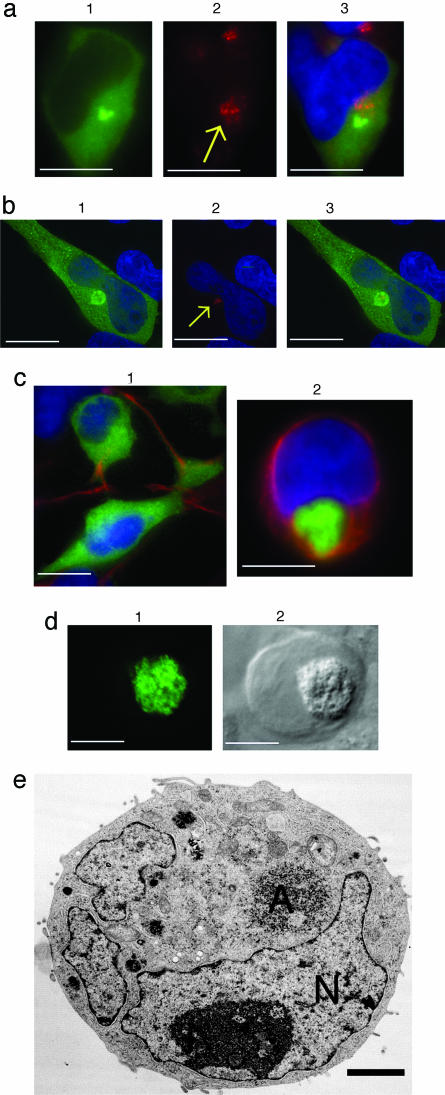
We examined HEK293 cells stably expressing iNOS-GFP by immunostaining with an antibody against the Golgi resident protein, golgin 97. iNOS perinuclear accumulation was close to, but quite distinct from, the Golgi (Fig. 3a). We investigated whether the iNOS aggresome localized with the centrosome at the MTOC using an antibody to human γ-tubulin. The iNOS aggresome was centered at the MTOC, surrounding the centrosome (Fig. 3b).
Fig. 3.
Characterization of the iNOS aggresome in HEK293 cells stably expressing human iNOS-GFP. (a) Lack of colocalization of iNOS and Golgi. Cells (1) were immunostained with anti-human golgin 97, followed by Alexa Fluor 594-conjugated goat anti-mouse antibody. Arrow denotes the Golgi (2). Overlap of the two images with a nuclear DAPI staining, is shown (3). (b) Cells (1) were immunostained with anti-human γ-tubulin; arrow points to the centrosome (2). Overlap of the two images is shown (3). (c) Vimentin localization. Cells were immunostained by antivimentin antibody (red). (1) Vimentin is seen as a rope-like filament; (2) vimentin appears as a cage surrounding iNOS-GFP at its perinuclear localization. (d) Characterization of iNOS aggresome by DIC. Cells (1) were evaluated by DIC (2). [Bars (a-d), 10 μm.] (e) Ultrastructure of iNOS aggresome by transmission EM. Cells were incubated in the presence of 10 μm of the proteasome inhibitor MG132 for 18 h and examined with a JEOL 200CX electron microscope. A, aggresome; N, nucleus. (Bar, 2 μm.)
A consistent feature associated with aggresome formation is the displacement of the intermediate filament proteins from their normal cellular distribution. Type III intermediate filaments such as vimentin normally display an extended cytoplasmic distribution of rope-like 8- to 10-nm filaments. In aggresome-containing cells, vimentin is radically redistributed to form a cage-like structure wrapped around the exterior of the inclusion. The molecular events that lead to vimentin rearrangements are not understood. Furthermore, the function of this vimentin cage is unclear. However, a cage of collapsed intermediate filaments might contribute to the stability of aggresomes. Whether it is related to aggresome formation directly or indirectly, intermediate filament collapse is a robust marker for aggresome formation (2, 3, 24). To determine whether the iNOS aggresome is associated with vimentin collapse, we immunostained HEK293 cells expressing iNOS-GFP with an antivimentin antibody. In cells in which iNOS was mostly cytosolic, vimentin had a normal extended cytoplasmic rope-like distribution (Fig. 3c1). By contrast, in cells exhibiting perinuclear accumulation of iNOS, vimentin collapsed to form a cage around iNOS (Fig. 3c2). These observations represent typical features of aggresomes and thus further establish cardinal features of the iNOS aggresome. We then confirmed that the iNOS-GFP aggresome, as detected by fluorescence microscopy, correlated with a distinct structure as visualized by DIC (Fig. 3d) and by EM (Fig. 3e). In these cells, which were treated with proteasome inhibitor MG132 (10 μM) for 18 h, iNOS aggresome appeared as a large accumulation of closely packed electron-dense particles. The EM ultrastructure of iNOS aggresome is consistent with published data of aggresomes associated with misfolded proteins (2, 3).
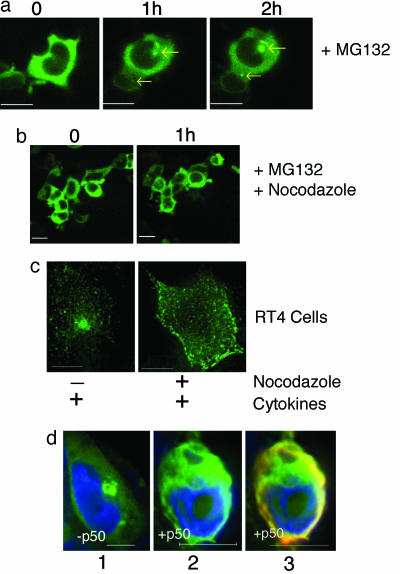
Effect of Proteasomal Inhibition and Microtubular Disruption on iNOS Aggresome. Two additional cardinal characteristics of aggresome formation include its dependency on microtubule integrity and its accelerated formation after proteasomal inhibition (2, 3, 24). To verify whether the iNOS aggresome exhibits these two characteristics, we studied the effect of proteasomal inhibitors and microtubule depolymerizing agents on iNOS aggresome formation in live HEK293 cells stably expressing iNOS-GFP. After proteasomal inhibition by MG132, iNOS aggresomes formed by 1 h and were increased in size at 2 h (Fig. 4a). In parallel, experiments using the microtubular depolymerizing agent nocodazole (30 μM), microtubule disruption prevented iNOS aggresome formation (Fig. 4b). These data suggest similarities among mechanisms of iNOS aggresome formation and previously described aggresome formation associated with misfolded proteins.
Fig. 4.
Effect of proteasomal inhibition and microtubular disruption on iNOS aggresome. (a) Live cell imaging of HEK293 cells stably expressing human iNOS-GFP and incubated with 10 μM of the proteasome inhibitor MG132 for 0-2 h. Arrows indicate the early formation of iNOS aggresomes at 1 h and their increase in size at 2 h. (b) Parallel experiments were done in which cells were concomitantly treated with 30 μM of the microtubule depolymerizing agent nocodazole. Microtubule disruption by nocodazole prevented iNOS aggresome formation. (c) RT4 cells were incubated for 18 h with a cytokine mixture to induce iNOS and in the absence or presence of nocodozole (10 μM). Cells were fixed and evaluated for iNOS expression by immunofluorescence by using an anti-iNOS antibody (green). (d) Overexpression of p50/dynamitin inhibits iNOS aggresome formation. HEK293 cells were transfected with plasmids encoding iNOS-GFP combined in a 1:2 molar ratio with either FLAG-tagged p50/dynamitin or with an empty vector. Twenty-four hours after transfection, cells were incubated with 10 μM of the proteasome inhibitor MG132 for 7 h to accelerate iNOS aggresome formation. Cells were then fixed and stained with DAPI to visualize nuclei (blue). Cells expressing iNOS-GFP were further examined for the coexpression of p50/dynamitin by using anti-body to the FLAG epitope (red). Representative images are shown for cells expressing iNOS-GFP in the absence (1) or presence (2) of p50/dynamitin. The overlap between iNOS-GFP and p50/dynamitin is also shown (3). iNOS aggresome was present in 63% of cells expressing iNOS-GFP only, compared with 16% of cells expressing both iNOS-GFP and p50/dynamitin. (Bars, 10 μm.)
We then examined the microtubule dependence for iNOS aggresome formation in RT4 cells expressing cytokine-induced iNOS. In RT4 cells stimulated by cytokines to produce iNOS and simultaneously treated with nocodazole (10 μM), iNOS failed to reach its perinuclear target as an aggresome and remained in minute vesicles scattered around the cytoplasm (Fig. 4c). These data demonstrate the requirement of a microtubule transport mechanism for iNOS aggresome formation by endogenous cytokine-induced iNOS, similar to the observation shown above for transfected iNOS.
iNOS Aggresome Formation Depends on Functional Dynein/Dynactin Motor. The inhibition of iNOS aggresome formation by nocodazole treatment indicated that a microtubule-dependent motor is involved in its formation. The minus-end-directed transport processes require dynein, which is typically associated with dynactin. The dynein-dynactin associated complex has been shown to be involved in aggresome formation (3). Dynein-dynactin-associated minus-end motor activity can be experimentally inhibited by overexpressing the p50/dynamitin component of the dynactin complex (3). To test whether targeting of iNOS to the aggresome depends on an intact dynein-dynactin complex, HEK293 cells were transfected with plasmids encoding iNOS-GFP combined in a 1:2 molar ratio with either FLAG-tagged p50/dynamitin or an empty vector. Twenty-four hours after transfection, cells were incubated with 10 μM of the proteasome inhibitor MG132 for 7 h to accelerate iNOS aggresome formation. Cells expressing iNOS-GFP were examined for the coexpression of p50/dynamitin by using antibody to the FLAG epitope. iNOS aggresome was present in 63% of cells expressing iNOS-GFP only (237 cells counted), compared with 16% of cells expressing both iNOS-GFP and p50/dynamitin (177 cells counted). Representative images are shown in Fig. 4d. These results indicate that targeting of iNOS to the aggresome at the MTOC is powered by the microtubule motor complex of dynein-dynactin.
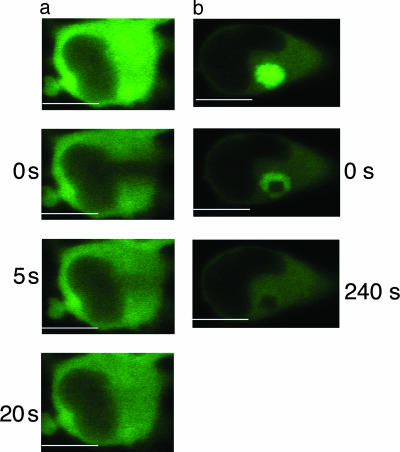
Determination of iNOS Mobility in Cytosolic and Perinuclear Compartments. The intense fluorescence of iNOS-GFP at its perinuclear location suggested that this accumulation might contain a relatively large concentration and/or an aggregated mass of iNOS-GFP. This “crowdedness” could impose restriction on iNOS mobility. To test this hypothesis, we used fluorescence recovery after photobleaching to determine the mobility of iNOS (26). Recovery of iNOS fluorescence in the cytosolic compartment was relatively fast, with a half life of 5.1 ± 0.3 s (Fig. 5a). However, for perinuclear iNOS, there was no significant recovery of fluorescence up to 240 s, indicating that iNOS mobility was extremely limited within this compartment (Fig. 5b). These data suggest that iNOS in the perinuclear area may be aggregated and thus an inactivated protein.
Fig. 5.
Determination of iNOS mobility in cytosolic and perinuclear compartments. Live HEK293 cells stably expressing human iNOS-GFP were transferred to a prewarmed live-cell chamber, and fluorescence recovery after photobleaching was performed by using laser-scanning confocal microscope at wavelength of 488 nm. Recovery of iNOS-GFP fluorescence in the cytosolic compartment was relatively fast, with a calculated half life of 5.1 ± 0.3 s; n = 3 (a). However, for the iNOS perinuclear accumulation, there was no significant recovery of fluorescence up to 240 s (b). (Bars, 10 μm.)
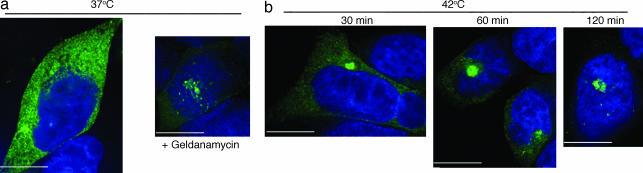
iNOS Aggresome Acceleration by Heat Shock and hsp 90 Inhibition. To evaluate the effect of cellular stress on iNOS aggresome formation, we subjected HEK293 cells expressing iNOS-GFP to chaperone inhibition or heat shock. hsp 90 is a major cellular chaperone involved in protein folding (27). Inhibition of hsp 90 by incubating cells in the presence of the specific inhibitor geldanamycin (10 μM) led to acceleration of iNOS aggresome formation (Fig. 6). In heat-shock experiments, cells were initially monitored at 37°C, followed by incubation at 42°C (28). Increasing the temperature to 42°C accelerated iNOS aggresome formation that was evident in the majority of cells within 30 min of the temperature shift. These results indicate that cellular stress in the form of heat shock or inhibition of chaperone function exerts a regulatory role on iNOS aggresome formation.
Fig. 6.
iNOS aggresome acceleration by heat shock and hsp 90 inhibition. (a) HEK293 cells stably expressing iNOS-GFP were incubated at 37°C with or without the hsp 90 inhibitor geldanamycin (10 μM; 3 h). (b) Cells were incubated at 42°C and evaluated by fluorescence microscopy as above. (Bars, 10 μm.)
The current literature suggests that aggresomes represent a cellular response to the formation of misfolded and aggregated proteins. Previous studies have clearly defined several cardinal features that were found to be common characteristics of cellular aggresomes (1-3). These features include: (i) aggresomes occur in response to misfolded proteins; (ii) aggresome formation is microtubule-dependent; (iii) aggresomes form around or near the MTOC; (iv) aggresome formation is associated with collapse of intermediate filament forming a cage around the aggresome; and finally, (v) aggresome formation is accelerated by proteasomal inhibition. Our study demonstrates that iNOS perinuclear accumulation meets all of the criteria listed above, except that iNOS is not considered to be a misfolded protein. The concern inherently associated with ectopic expression of proteins such as iNOS-GFP could not explain iNOS aggresome, because iNOS aggresome was a common feature for endogenous iNOS expressed in various cell types (Figs. 1, 2, and 4c). Thus, the inevitable conclusion from our data is that the iNOS aggresome represents a new paradigm by extending the role of aggresomes to the regulation of normal functional cellular proteins. This mechanism endows the aggresome with additional regulatory functions and permits cells to rapidly eliminate proteins from the active cellular environment by sweeping them toward the MTOC, forming an aggresome. This strategy may help cells avoid potential cellular injury. This protective mechanism may be particularly important with specific proteins whose overproduction could be associated with cell toxicity, e.g., iNOS. In addition to serving a sequestration role, it is possible that the aggresome provides a staging ground for the incorporation of proteins into autophagic structures, perhaps by facilitating interactions with endosomes and lysosomes that are also delivered by microtubules to the same region of the cell (4, 29).
Improperly folded, i.e., unfolded or misfolded, proteins are prone to aggregation. This mechanism may account for the formation of aggresome associated with misfolded proteins. Could this mechanism also contribute to our discovered iNOS physiologic aggresome? A speculative, yet intriguing, hypothesis is that cells may induce protein unfolding on purpose, leading to its aggregation, such that it would qualify for transport to the aggresome. This mechanism could serve as a regulatory mechanism to sweep iNOS out of circulation until further degradation takes place. In fact, unfolding of proteins is a physiologic function required for protein degradation by the proteasome. To allow the target protein to be threaded through the relatively narrow hole of the 20S core unit of the proteasome, the protein has to be unfolded. This task is done by the regulatory 19S unit located at the lid of the proteasome (30).
In contrast to the constitutive NOS isoforms, which are tightly controlled by the level of intracellular Ca2+, iNOS is Ca2+-independent and is thought to be primarily regulated at the transcriptional level (7, 8). We have recently shown that cells exert posttranslational regulation on iNOS activity by maintaining a relatively rapid rate of iNOS turnover (31). Cells may use the aggresome pathway as a rapid method to inactivate iNOS by sequestering it to the perinuclear localization. Thus cells have a dual strategy for regulating iNOS activity. First, persistent calmodulin binding to iNOS regardless of intracellular levels of Ca2+ ensures that, once iNOS is expressed, it is continuously active. This results in the production of large amounts of NO that are essential for host defense. Second, cells redistribute iNOS to the aggresome, thereby ensuring that, once the need for NO is abated, its production is rapidly terminated. This strategy may be designed to avoid injury to host cells. The process of aggresome formation is accelerated under conditions perceived by cells as stress or imbalance. These conditions include the inability of cells to degrade proteins (e.g., modeled by proteasomal inhibition) or to fold protein efficiently (e.g., modeled by heat shock or chaperone inhibitors).
Our data indicate that the iNOS physiologic aggresome shares certain features with what were previously described as pathologic aggresomes associated with misfolded proteins. Thus, the pathologic aggresomes may merely represent an acceleration of an established physiologic regulatory process for specific proteins whose regulation by aggresome formation is deemed necessary by the cell. The regulation of aggresome formation appears to be linked to the cellular capacity to degrade proteins in a timely manner. Whenever cells sense that this capacity is likely to be exceeded due to the generation of either a misfolded protein or a large amount of certain proteins, they trigger aggresome formation. These data further our understanding of a process crucial in the pathogenesis of several genetic and degenerative diseases.
Supplementary Material
Acknowledgments
We thank William Brinkely (Baylor College of Medicine) for the γ-tubulin antibody and for useful discussions, Li-YuanYu-Lee (Baylor College of Medicine) for the p50/dynamitin plasmid, and Margarita Moczygemba (Baylor College of Medicine) for critical review of the manuscript. This work was supported by the National Heart Lung and Blood Institute, the National Institute of Allergy and Infectious Diseases, the American Heart Association, and the American Lung Association.
Author contributions: K.E.K., A.R.B., R.H.M., D.L.S., and N.T.E. designed research; K.E.K., A.R.B., and D.L.S. performed research; K.E.K., A.R.B., R.H.M., and N.T.E. analyzed data; and K.E.K., A.R.B., and N.T.E. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: iNOS, inducible NO synthase; HEK, human embryonic kidney; DIC, Normaski differential interference contrast; MTOC, microtubule-organizing center.
References
- 1.Kawaguchi, Y. Kovacs, J. J., McLaurin, A., Vance, J. M., Ito, A. & Yao, T. P. (2003) Cell 115, 727-738. [DOI] [PubMed] [Google Scholar]
- 2.Johnston, J. A., Ward, C. L. & Kopito, R. R. (1998) J. Cell Biol. 143, 1883-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garcia-Mata, R., Bebok, Z., Sorscher, E. J. & Sztul, E. S. (1999) J. Cell Biol. 146, 1239-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kopito, R. R. (2000) Trends Cell Biol. 10, 524-530. [DOI] [PubMed] [Google Scholar]
- 5.Wigley, W. C., Fabunmi, R. P., Lee, M. G., Marino, C. R., Muallem, S., DeMartino, G. N. & Thomas, P. J. (1999) J. Cell Biol. 145, 481-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ignarro, L. J., Buga, G. M., Wood, K. S., Byrns, R. E. & Chaudhuri, G. (1987) Proc. Natl. Acad. Sci. USA 84, 9265-9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stuehr, D. J. (1999) Biochim. Biophys. Acta 1411, 217-230. [DOI] [PubMed] [Google Scholar]
- 8.Xie, Q.-W., Cho, H. J., Calaycay, J., Mumford, R. A., Swiderek, K. M., Lee, T. D., Ding, A., Troso, T. & Nathan, C. (1992) Science 256, 225-228. [DOI] [PubMed] [Google Scholar]
- 9.Cho, H. J., Xie, Q.-W., Calaycay, J., Mumford, R. A., Swiderek, K. M., Lee, T. D. & Nathan, C. (1992) J. Exp. Med. 176, 599-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nathan, C. (1997) J. Clin. Invest. 100, 2417-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo, F. H., Comhair, S. A. A., Zheng, S., Dweik, R. A., Eissa, N. T., Thomassen, M. J., Calhoun, W. & Erzurum, S. C. (2000) J. Immunol. 164, 5970-5980. [DOI] [PubMed] [Google Scholar]
- 12.Shaul, P. W. (2002) Annu. Rev. Physiol. 64, 749-774. [DOI] [PubMed] [Google Scholar]
- 13.Brenman, J. E., Chao, D. S., Gee, S. H., McGee, A. W., Craven, S. E., Santillano, D. R., Wu, Z., Huang, F., Xia, H., Peters, M. F., et al. (1996) Cell 84, 757-767. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt, H. H., Warner, T. D., Nakane, M., Förstermann, U. & Murad, F. (1992) Mol. Pharmacol. 41, 615-624. [PubMed] [Google Scholar]
- 15.Vodovotz, Y., Russell, D., Xie, Q.-W., Bogdan, C. & Nathan, C. (1995) J. Immunol. 154, 2914-2925. [PubMed] [Google Scholar]
- 16.Wheeler, M. A. Smith, S. D., García-Cardeña, G., Nathan, C. F., Weiss, R. M. & Sessa, W. C. (1997) J. Clin. Invest. 99, 110-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Webb, J. L., Harvey, M. W., Holden, D. W. & Evans, T. J. (2001) Infect. Immun. 69, 6391-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eissa, N. T., Yuan, J., Haggerty, C. M., Choo, E. K. & Moss, J. (1998) Proc. Natl. Acad. Sci. USA 95, 7625-7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eissa, N.T., Haggerty, C. M., Palmer, C. D., Patton, W. & Moss, J. (2001) Am. J. Respir. Cell Mol. Biol. 24, 616-620. [DOI] [PubMed] [Google Scholar]
- 20.Kolodziejski, P. J., Musial, A., Koo, J. S. & Eissa, N. T. (2002) Proc. Natl. Acad. Sci. USA 99, 12315-12320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eissa, N. T., Strauss, A. J., Haggerty, C. M., Choo, E. K., Chu, S. C. & Moss, J. (1996) J. Biol. Chem. 271, 27184-27187. [DOI] [PubMed] [Google Scholar]
- 22.Musial, A. & Eissa, N. T. (2001) J. Biol. Chem. 276, 24268-24273. [DOI] [PubMed] [Google Scholar]
- 23.Kolodziejski, P. J., Rashid, M. B. & Eissa, N. T. (2003) Proc. Natl. Acad. Sci. USA 100, 14263-14268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Earl, R. T., Mangiapane, E. H., Billett, E. E. & Mayer, R. J. (1987) Biochem. J. 241, 809-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia-Mata, R., Gao, Y. S. & Sztul, E. (2002) Traffic 3, 388-396. [DOI] [PubMed] [Google Scholar]
- 26.Stenoien, D. L., Mielke, M. & Mancini, M. A. (2002) Nat. Cell Biol. 4, 806-810. [DOI] [PubMed] [Google Scholar]
- 27.Yoshida, M. & Xia, Y. (2003) J. Biol. Chem. 278, 36953-36958. [DOI] [PubMed] [Google Scholar]
- 28.Hsu, A. L., Murphy, C. T. & Kenyon, C. (2003) Science 300, 1142-1145. [DOI] [PubMed] [Google Scholar]
- 29.Matteoni, R. & Kreis, T. E. (1987) J. Cell Biol. 105, 1253-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sauer, R. T., Bolon, D. N., Burton, B. M., Burton, R. E., Flynn, J. M., Grant, R. A., Hersch, G. L., Joshi, S. A., Kenniston, J. A., Levchenko, I., et al. (2004) Cell 119, 9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolodziejski, P. J., Koo, J. S. & Eissa, N. T. (2004) Proc. Natl. Acad. Sci. USA 101, 18141-18146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.