Abstract
Through a genetic screen using myosin-like protein strains mlp1Δ mlp2Δ and biochemical purification, we identified a complex of eight proteins, each required for growth and DNA repair in Saccharomyces cerevisiae. Among the subunits are Mms21 that contains a putative Siz/PIAS (protein inhibitor of activated signal transducer and activator of transcription) RING domain characteristic of small ubiquitin-like modifier (SUMO) ligases, two structural-maintenance-of-chromosome (Smc) proteins, Smc5 and Smc6, and a protein that contains an ubiquitin ligase signature domain. We show that these proteins colocalized to several distinct nuclear foci. Biochemical and genetic data demonstrated that Mms21 indeed functions as a SUMO ligase and that this activity requires the Siz/PIAS (protein inhibitor of activated signal transducer and activator of transcription) RING domain. The substrates for this SUMO ligase include a subunit of the octameric complex, Smc5, and the DNA repair protein Yku70. We further show that the abolition of the SUMO E3 activity of Mms21 leads to such disparate phenotypes as DNA damage sensitivity, defects in nucleolar integrity and telomere clustering, silencing, and length regulation. We propose that Mms21 sumoylates proteins involved in these diverse processes and that the other members of the complex, particularly Smc5/6, facilitate proper substrate sumoylation by localizing Mms21 to specific chromosomal regions.
Keywords: myosin-like protein, structural maintenance of chromosome, small ubiquitin-like modifier
The budding yeast myosin-like proteins, Mlp1 and Mlp2, like their mammalian homolog TPR (translocated promoter region), contain long coiled-coil domains and form filaments (1, 2). These proteins are associated with the nucleoplasmic side of the nuclear pore complexes (NPCs) and extend from there to the nuclear interior (3, 4). Remarkably, Mlps were found to be associated with only a subset of NPCs: they are excluded from the NPCs juxtaposed to the nucleolus (5, 6). The same biased localization was also observed for a desumoylating enzyme Ulp1 (5), which removes small ubiquitin-like modifier (SUMO) from SUMO-conjugated proteins (7). This correlation suggests that Mlps are involved in anchoring Ulp1. Indeed, the deletion of Mlps or the “localization” domain of Ulp1 yielded displacement of the catalytically active Ulp1 to the nuclear interior (5, 8, 9). This displacement resulted in desumoylation of intranuclear SUMO conjugates that normally are substrates of the other desumoylating enzyme, Ulp2, which is localized to the nuclear interior (5, 8-11). The delocalization of Ulp1 activity yielded two distinct phenotypes: clonal lethality and increased DNA damage sensitivity (5). This finding suggests that a proper maintenance of the sumoylation stasis is important for the cell.
Sumoylation was found to be involved in many cellular processes, such as transcription, nuclear transport, and signal transduction (reviewed in refs. 12-14). The effect of sumoylation on the target proteins can be diverse: it can affect the localization of the targets or regulate the activity or the binding properties of the targets (reviewed in refs. 15 and 16). However, sumoylation does not lead to protein degradation; and in some cases, it can even antagonize ubiquitination and lead to protein stabilization (reviewed in refs. 15 and 16).
Sumoylation, like ubiquitination, is also carried out by the sequential action of three enzymes: an activating enzyme (E1), a conjugating enzyme (E2), and a ligase (E3). In all of the organisms examined so far, a single E1 and E2 but multiple E3s have been detected (reviewed in refs. 15 and 16). Thus, E3s likely are the determinants of substrate specificity. In yeast, two E3s, Siz1 and Siz2, have been identified (17, 18). These proteins and their mammalian homologs, the PIAS (protein inhibitor of activated signal transducer and activator of transcription) proteins, contain the so-called SP-RING (Siz/PIAS RING) domain that is exclusively found in SUMO E3s and is required for the sumoylating activity (reviewed in ref. 19). However, the fact that the deletion of both Siz1 and Siz2 was not lethal, whereas the deletion of E1 or E2 was (16), suggested that additional E3s exist and that they are important for cell growth.
This study was initiated with a synthetic-lethal screen by using mlp1Δ mlp2Δ strains. We found that the deletion of MLPs was synthetically lethal with a mutation in the MMS21 gene. Interestingly, Mms21 contains a SP-RING-like domain, suggesting that it functions as a SUMO E3. Further biochemical and genetic characterization demonstrated that Mms21 is indeed a SUMO E3. In addition, it is part of an octameric complex, which, among others, also contains structural maintenance of chromosome (SMC) proteins, Smc5 and Smc6, and another protein that contains an ubiquitin E3 signature domain. We also present evidence that the Mms21 SUMO E3 activity is required in diverse cellular processes, including DNA repair and nuclear organization.
Materials and Methods
Isolation of the Mms21-Smc5-Smc6 Complex. Yeast cell pellets were washed with TBT buffer (20 mM K·Hepes, pH 7.4/110 mM KAc/2 mM MgCl2/0.1% Tween 20) supplemented with 1 mM DTT, 1:200 dilution of PIC (protease inhibitor mixture, Sigma), 0.36 mg/ml PMSF, and 6 μg/ml pepstatin. Cell pellets were first frozen and then ground vigorously in a mortar and pestle. The cell powder was resuspended in EB buffer (TBT solution supplemented with 250-650 mM NaCl and 0.5% Triton X-100). The resulting extract was centrifuged at 5,000 × g for 10 min, and the supernatant was bound to IgG-coupled Dynabead M-270 Epoxy (Dynal, Great Neck, NY). After washing extensively with EB buffer, protein A (ProA) fusion proteins were eluted with 500 mM NH4OH and 1 mM EDTA. The eluate was dried, resuspended in loading buffer, and subjected to SDS/PAGE analysis.
Immunoprecipitation and Immunoblotting. Yeast strains containing chromosomal Myc-tagged Yku70, Smc5, or Pol30 were grown to early log phase and treated with 0.3% methyl methanesulfonate (MMS) for 2 h. Yeast lysates were prepared as described (20). The tagged proteins were immunoprecipitated from the yeast lysate by using agarose conjugated with anti-Myc antibody (Sigma). After stringent washing, the bound proteins were eluted by using the SDS/PAGE loading dye. The eluate was then subjected to SDS/PAGE. Standard protocols were followed in all immunoblotting analyses. Antibodies used in immunoblotting analyses were: anti-SUMO antibody (20), anti-Myc (Sigma), anti-Yku70 (21), anti-T7 (Novagen), and anti-pol30 (22).
Yeast Strains, Plasmid Construction, and Synthetic Lethal Screen. For yeast strains, plasmid construction, and synthetic-lethal screen see Table 1 and Supporting Materials and Methods, which are published as supporting information on the PNAS web site.
Other Techniques. Recombinant proteins were expressed in Escherichia coli and purified by Ni-NTA chromatography as described by Johnson and Gupta (17). Protein preparations were further purified by using Superdex 200 chromatography. The expression and purification of the Yku70-Yku80 complex were carried out as described (23). In vitro sumoylation assay was performed following the same protocol as in ref. 17. Yeast telomere length measurement was carried out as described (24). Cells were prepared and images were taken as described (25).
Results
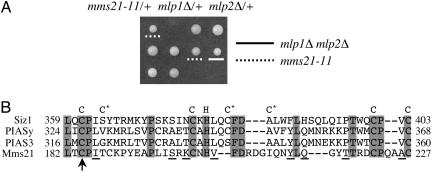
A Mutation in the MMS21 Gene Is Synthetic-Lethal with mlp1Δ mlp2Δ. To identify proteins functionally related to Mlps, we used transposon-mediated mutagenesis to find genes that are required for the survival of mlp1Δ mlp2Δ cells. One mutant isolated in this screen exhibited synthetic lethality with the mlp1Δ mlp2Δ double, but not with either mlp1Δ or mlp2Δ single mutants, as demonstrated by plasmid shuffle experiments (data not shown) and tetrad analysis (Fig. 1A). Cloning and sequence analysis showed that this mutant contained a transposon insertion in the MMS21 ORF at 552 nt. The MMS21 gene was previously identified in a screen for genes affecting resistance to DNA-damaging agent MMS (26). The insertion recovered in our screen results in a truncated protein lacking the C-terminal region. This region resembles the SP-RING domain that is exclusively found in one type of SUMO E3s (19) (Fig. 1B and see below). We refer to this mutation as mms21-11 hereafter.
Fig. 1.
The deletion of the SP-RING domain of Mms21 is synthetic-lethal with mlp1Δ mlp2Δ.(A) A diploid strain heterozygous for mms21-11, mlp1Δ, and mlp2Δ was sporulated and dissected. Three tetrads are shown. mlp1Δ mlp2Δ and mms21-11 spore clones are indicated. The genotype of inviable spores was deduced from those of sister spore clones and is mms21-11 mlp1Δ mlp2Δ.(B) Alignment of the SP-RING domains of Siz1, mammalian PIAS enzymes, and Mms21. The eight signature Cys (C) and His (H) residues are indicated above the alignment. Cysteines marked by * vary among different proteins or change to other amino acids. Identical amino acids in the alignment are shaded in gray, and conserved amino acids are underlined. The arrow indicates a transposon insertion in mms21-11.
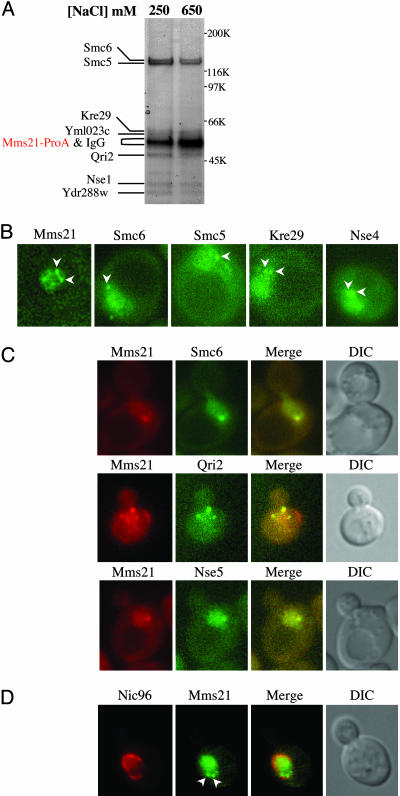
Mms21 Associates with Smc5, Smc6, and Five Other Proteins in Vivo. As a step toward understanding the function of Mms21, we attempted to identify its binding partners. We tagged the MMS21 gene with a ProA module at its chromosomal locus. The resulting fusion protein was fully functional as indicated from its WT-like phenotype (data not shown). A cell extract was prepared, and Mms21-ProA and seven associated proteins were isolated by affinity purification (Fig. 2A). MS analyses revealed two of these proteins to be Smc5 and Smc6/Rhc18. To obtain additional evidence for this complex, we tagged SMC5 and SMC6 with ProA at their chromosomal loci. Again, these fusion proteins were fully functional (data not shown). Purification of Smc5-ProA or Smc6-ProA revealed that each of them also associates with seven other proteins (Fig. 7, which is published as supporting information on the PNAS web site). The apparent molecular mass of each of these proteins was in agreement with that in the Mms21-ProA pull-down experiment. Moreover, the same set of proteins was copurified in all three pull-down experiments at both moderate and high salt conditions (Fig. 2A), indicating they form a stable complex.
Fig. 2.
The Mms21-Smc5-Smc6 complex is composed of eight subunits and forms perinuclear centers. (A) Mms21-ProA was affinity-purified from yeast at the indicated salt concentrations. Copurifying proteins were resolved by SDS/PAGE and stained with Coomassie blue. Individual bands were excised and identified by MS. The identity of each protein is indicated. (B) Subunits of the Mms21-Smc5-Smc6 complex form nuclear foci. Each subunit shown was tagged by YFP at its chromosomal locus. Examples of live-cell fluorescence images are shown for the proteins indicated. Arrowheads mark foci formed by each protein. (C and D) Mms21 foci colocalize with those formed by other subunits of the complex (C), and they are juxtaposed to the nuclear envelope delineated by Nic96 (D). In C, cells contain Mms21-CFP and one of the YFP-tagged subunits as indicated. In D, cells contain Nic96-CFP and Mms21-YFP. Representative live-cell fluorescence images of CFP and YFP fusion proteins as well as their merged pictures are shown. CFP fusion proteins are pseudocolored as red and YFP fusion proteins are green. Arrowheads indicate Mms21-YFP foci in D.
Along with Mms21, Smc5, and Smc6, the other five proteins were identified by MS to be Nse1, Qri2, Kre29, and the protein products encoded by ORF YDR288w and YML023c (Fig. 2A). Based on sequence analysis, five or six subunits of the complex have homologs in fission yeast and humans, respectively (Table 2, which is published as supporting information on the PNAS web site), suggesting that this complex is evolutionarily conserved. Interestingly, Nse1 possesses a RING finger motif and therefore might function as an ubiquitin ligase, as previously noted for the Schizosaccharomyces pombe homolog (27). Following the naming of the non-SMC elements (Nse), we propose to name Ydr288w and Yml023c as Nse3 and Nse4, respectively. During the course of this study, a proteomic analysis reported Mms21 being associated with Nse4, Qir2, Smc5, and Smc6, but not with Nse5 and Kre29 (28). Because Mms21 copurified with all seven proteins even at 650 mM salt, it is likely that all eight proteins identified in this study form one complex. In this article, we refer to the octameric complex as the Mms21-Smc5-Smc6 complex.
Cell imaging using yellow fluorescent protein (YFP) or ProA-tagged subunits of the complex revealed that each of the eight proteins formed a few scattered nuclear foci in addition to a diffuse nuclear localization (examples shown in Fig. 2 B and C). Moreover, Mms21-cyan fluorescent protein (CFP) colocalized with YFP-tagged components of the complex at their nuclear foci (examples shown in Fig. 2C). This result supported our biochemical purification data and suggested that the eight proteins function together in vivo. Colocalization studies using a CFP-tagged nucleoporin, Nic96, and YFP-tagged-Mms21 showed that the Mms21 foci are right next to the nuclear envelope (Fig. 2D). These results demonstrated that the Mms21-Smc5-Smc6 complex forms perinuclear centers.
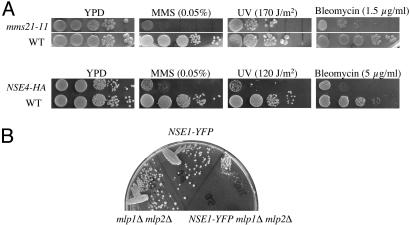
Consistent with the aforementioned biochemical and cell biological data, defects in different subunits of the Mms21-Smc5-Smc6 complex led to similar phenotype. First, the deletion of each gene resulted in lethality (data not shown). Second, mms21-11 and hypomorphic alleles of other components of the complex conferred sensitivities to various DNA-damaging agents, including MMS, UV, and the γ-irradiation-mimicking drug bleomycin (examples shown in Fig. 3A). These results extended previous reports on a subset of the subunits (29-31) by demonstrating that all of them are required for growth and repair. Lastly, like mms21-11, several hypomorphic alleles of the other subunits exhibit synthetic interactions with mlp1Δ mlp2Δ (Fig. 3B and data not shown), indicating that other members of the Mms21-Smc5-Smc6 complex also functionally interacted with Mlps.
Fig. 3.
Defects of the subunits of the Mms21-Smc5-Smc6 complex lead to similar phenotype. (A) Strains containing mms21-11 (Upper) or hemagglutinin-tagged NSE4 (Lower) exhibit sensitivity to different DNA damaging agents. Mid-log phase, yeast extract/peptone/dextrose (YPD)-grown cells were spotted in 10-fold serial dilutions (from 105 to 10 cells) on YPD plates or plates containing the indicated amount of DNA-damaging agents. One spotted YPD plate was treated with UV light. All plates were incubated at 30°C for 3 days. (B) A hypomorphic allele of Nse1 exhibits synthetic growth defect when combined with mlp1Δ mlp2Δ. mlp1Δ mlp2Δ, NSE1-YFP, and NSE1-YFP mlp1Δ mlp2Δ strains were streaked out on a YPD plate and grown at 30°C for 3 days. The mlp1Δ mlp2Δ NSE1-YFP strain grew significantly slower than either the mlp1Δ mlp2Δ or Nse1-YFP strains.
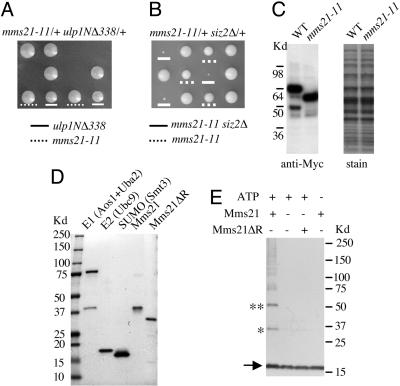
Mms21 Genetically Interacts with Enzymes of the Sumoylation Path-way. Recently, we found that Mlps are required to anchor and stabilize the desumoylating enzyme Ulp1 at NPCs (5). Deleting Mlps or the localization domain of Ulp1 can both delocalize Ulp1 from the NPCs and subsequently lead to unphysiological desumoylation (5, 8, 9). Given that mms21-11 lacks the putative SUMO E3 signature domain, we suspected that the genetic interactions of Mlps with mms21-11 or other subunits of the Mms21-Smc5-Smc6 complex might be related to the function of Mlps in anchoring Ulp1. A straightforward test of this idea is to determine whether deleting Ulp1 localization domain (residues 1-403) can mimic mlp1Δ mlp2Δ in causing synthetic lethality with mms21-11. We examined the genetic interaction between a N-terminal deletion construct, ulp1NΔ338 (residues 1-338 deleted) and mms21-11. As shown in Fig. 4A, whereas mms21-11 and ulp1NΔ338 single mutants grew slightly slower, mms21-11 and ulp1NΔ338 double mutants were inviable. This experiment suggested that the synthetic lethality of mlp1Δ mlp2Δ and mms21-11 is related to the impaired Ulp1 functions.
Fig. 4.
Mms21 functions in sumoylation processes. (A and B) mms21-11 is synthetic-lethal with ulp1NΔ338 and synthetic-sick with siz2Δ. Diploid strains heterozygous for mms21-11 and ulp1NΔ338 (A) or mms21-11 and siz2Δ (B) were sporulated and dissected. Three tetrads are shown for each case. In A, mms21-11 and ulp1NΔ338 spore clones are indicated. The genotype of inviable spores was deduced from those of sister spore clones and is mms21-11 ulp1NΔ338. In B, mms21-11 and mms21-11 siz2Δ spore clones are indicated. siz2Δ and WT spore clones grew equally well. (C) mms21-11 does not affect the level of the mutated protein. Mms21 (WT) or mms21-11 was tagged with Myc tags at their corresponding chromosomal loci. Total yeast lysate was prepared from these cells, separated by SDS/PAGE, and analyzed by immunoblotting using anti-Myc antibody. The nitrocellulose membranes were stained by amidoblack before blotting to check equivalent loading. (D) Yeast SUMO E1 (Aos1/Uba2), E2 (Ubc9), SUMO (Smt3), full-length Mms21, and Mms21 lacking the SP-RING domain (Mms21ΔR) were purified from E. coli via chromatography on Ni-NTA and gel-filtration columns. Protein preparations were subjected to SDS/PAGE analysis to assess purity. (E) Mms21 promotes di-SUMO and tri-SUMO formation in a SP-RING domain-dependent manner. All reactions contain E1 (110 ng), E2 (80 ng), and SUMO (800 ng), and they were carried out at 30°C for 30 min. Additional components included in the reactions are indicated above the gel. Reactions were analyzed by SDS/PAGE and immunoblotting with anti-SUMO antibody. The arrow indicates the position of free SUMO. * and ** depict di-SUMO and tri-SUMO, respectively.
The genetic interaction with ulp1NΔ338 raised the possibility that mms21-11 is defective in sumoylation and thus can cause lethality when combined with unregulated desumoylation. To explore this possibility, we examined whether mms21-11 also exhibited synergistic interaction with SUMO E3 mutants. In yeast, the two known SUMO E3s, Siz1 and Siz2, are responsible for most, but not all SUMO conjugates (17, 18). Consistently, the deletion of Siz1 and Siz2 is viable, whereas the deletion of SUMO, SUMO E1, or E2 is lethal (16). We found that mms21-11 siz1Δ or mms21-11 siz2Δ mutants grew very poorly (Fig. 4B and data not shown). Considering that the mms21-11 protein was expressed at WT level (Fig. 4C) and supported cell growth (unlike the deletion of MMS21), its synergistic interaction with Siz1/2 is likely caused by a defect in sumoylation.
Mms21 Exhibits Sumoylating E3 Activity in Vitro. To further investigate a role of Mms21 in sumoylation, we examined whether it can promote the formation of di-SUMO and tri-SUMO in vitro, because known SUMO E3s have been shown to exhibit such activity (17, 32, 33). As shown in Fig. 4D, sumoylating E1, E2, SUMO, Mms21, and Mms21ΔR (Mms21 without the SP-RING domain) were expressed from E. coli and purified. The addition of the full-length Mms21 to the reaction containing E1, E2, and SUMO promoted the formation of di-SUMO and tri-SUMO (Fig. 4E, *). In contrast, the truncated Mms21 protein, Mms21ΔR, did not support the formation of di-SUMO or tri-SUMO (Fig. 4E). These results show that Mms21 exhibits SUMO E3 activity in vitro and that the SP-RING domain is required for this activity. Taken together, our in vivo and in vitro data strongly support the notion that Mms21 can function as a sumoylating E3.
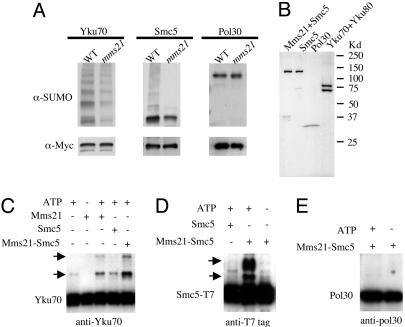
Mms21 Is Required for Sumoylating Smc5 and Yku70 in Vivo and in Vitro. To identify physiological substrates for the SUMO E3 activity of Mms21, we undertook a “candidate-screen” approach. The observed DNA damage sensitivity of mms21-11 suggested that some of Mms21's substrates may be involved in DNA repair. We thus examined the sumoylation status of several repair proteins after MMS treatment in mms21-11 and WT strains. To avoid potential artifacts caused by overexpression, candidate proteins were tagged at their chromosomal loci with the Myc epitope. In addition, to be able to detect the small amount of endogenous sumoylated proteins (<1% of the unmodified forms) (16), we performed Western blot analysis of the immunoprecipitated proteins. Among the examined proteins, Smc5 and Yku70 showed dramatically decreased levels of sumoylation in mms21-11 mutants, whereas the amounts of their unmodified forms were apparently unchanged (Fig. 5A). Both proteins showed polysumoylated forms that were more pronounced in the case of Yku70 and the decrease affected all polysumoylated forms (Fig. 5A). These effects are specific, as sumoylation of other repair proteins, such as Pol30, was unchanged in mms21-11 strains (Fig. 5A).
Fig. 5.
Mms21 is required for sumoylation of Yku70 and Smc5 in vivo and in vitro.(A) Mms21 is required for sumoylation of Yku70 and Smc5 in vivo.WTor mms21-11 cells containing chromosomal Myc-tagged Yku70, Smc5, or Pol30 were grown to early log phase and treated with 0.3% MMS for 2 h. Tagged proteins were affinity-purified from the total lysate by using anti-Myc-conjugated agarose. The eluate was subjected to SDS/PAGE and Western blot analysis using anti-SUMO antibody to detect the sumoylated forms of the protein. The membranes were subsequently stripped and reprobed with anti-Myc antibody to detect the unmodified forms. (B) The Mms21-Smc5 complex, Smc5, and Pol30 were purified from E. coli by chromatography on Ni-NTA and gel-filtration columns. The Yku70-Yku80 complex was purified from yeast by chromatography on a Ni-NTA and a Mono-Q column. Protein preparations were subjected to SDS/PAGE analysis to assess purity. (C-E) Mms21 stimulates sumoylation of Yku70 and Smc5 in vitro. All in vitro sumoylation reactions contain E1, E2, and SUMO and were carried out at 30°C for 30 min. Additional components included in the reactions are indicated above the gel. The reactions were analyzed by SDS/PAGE and immunoblotting using antibodies specific to the target proteins. In C, all reactions contain the Yku70-Yku80 complex, and a monoclonal anti-Yku70 antibody was used in the immunoblotting analysis. Arrows indicate the positions of mono- and di-SUMO-Yku70. In D, Smc5 was detected in the immunoblotting analysis by using anti-T7 antibody as Smc5 was fused to the T7 tag. Arrows indicate the positions of mono- and di-SUMO-Smc5. In E, an anti-Pol30 antibody was used in the immunoblotting analysis. Only the unmodified form of Pol30 was detected.
We further examined whether Mms21 can stimulate sumoylation of Smc5 and Yku70 in vitro. To this end, we purified these proteins and the control Pol30 (Fig. 5B). We also coexpressed Mms21 and Smc5 and found that the two proteins can be purified as a complex (Fig. 5B). As shown in Fig. 5C, the addition of purified Mms21 (Fig. 4D) promoted the formation of SUMO-Yku70 and di-SUMO-Yku70. The stimulation of SUMO-Yku70 and di-SUMO-YKu70 formation was more efficient with the addition of the Mms21-Smc5 complex, even though the concentration of Mms21 was only 15% of that when Mms21 was added alone (Fig. 5C). Smc5 alone had no sumoylating activity (Fig. 5C). The much higher activity of the Mms21-Smc5 complex was most likely caused by the stabilization of Mms21 by Smc5 (data not shown). Moreover, Smc5 was also sumoylated in an Mms21-dependent manner (Fig. 5D). In contrast, neither Mms21 nor Mms21-Smc5 promoted sumoylation of Pol30 (Fig. 5E). These results suggest that Mms21 can promote sumoylation of these specific substrates in vitro.
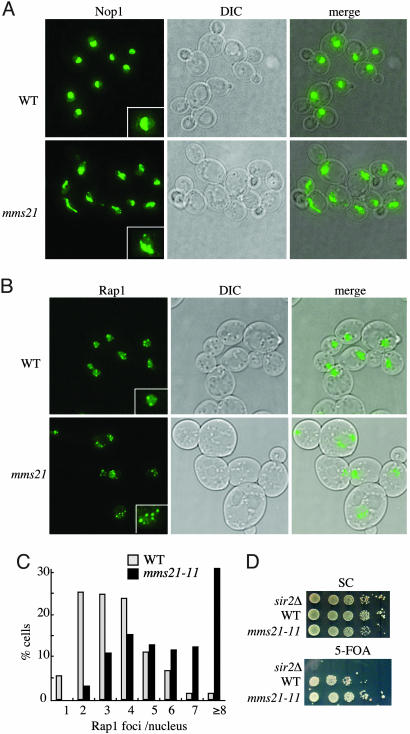
The Sumoylating Activity of Mms21 Is Required for Nucleolar Integrity and Telomere Functions. Next, we tested whether the sumoylating activity of Mms21 is also required in other cellular processes besides DNA damage response. The protein complex composed of Smc5/6 has been implicated in nuclear organization, as the human Smc5 is associated with DNA (34) and that deletions of a subset of the yeast subunits lead to fragmented nuclei (27, 30, 35, 36). Therefore, we examined the effect of mms21-11 on the integrity of specific nuclear structures, such as centromeres, telomeres, and the nucleolus. In a haploid budding yeast cell, the 16 centromeres form one to two foci, whereas the 32 telomeres form a few clusters at the nuclear periphery (37, 38). In addition, the yeast nucleolus assumes a half-moon shape adjacent to the nuclear envelope. To test whether mms21-11 affects any of these nuclear structures, we tagged proteins shown to mark each structure with YFP or CFP and carried out live-cell imaging across the whole nucleus.
We observed no apparent changes of centromere clustering in mms21-11 cells (data not shown). However, we did see abnormalities in the nucleolar structure and telomere foci in mms21-11 cells. Whereas the nucleoli in WT cells are compact and form a half-moon shape in 3D-rendered images, the nucleoli of mms21-11 cells were more spread out and irregularly shaped. In addition, ≈8% of the mms21-11 cells contained fragmented nucleoli (Fig. 6A). Moreover, the number of telomere foci was increased in mms21-11 cells, indicating a defect in clustering (Fig. 6B). Quantitation of the data showed that ≈75% (74/98) of WT cells contained 2-4 telomere foci (Fig. 6C), whereas telomere foci increased to >5 in ≈70% (59/85) and to >10 in ≈9% (8/85) mms21-11 cells (Fig. 6C). To test whether defects in telomere clustering were related to impaired telomere functions, we examined the telomere length and silencing (telomere position effect). We found that telomere length was slightly increased in mms21-11 cells (Fig. 8, which is published as supporting information on the PNAS web site). In addition, mms21-11 led to an enhanced silencing of a reporter gene (URA3) that was placed in the telomeric region (Fig. 6D). Together, these observations suggest that the sumoylating activity of Mms21 is required for the proper organization of telomeres and the nucleolus.
Fig. 6.
mms21-11 affects nucleolus structure and telomere clustering. (A and B) WT and mms21-11 cells containing Nop1-CFP (a nucleolar marker) (A) or Rap1-YFP (a telomere marker) (B) were grown to early log phase. Fluorescence images of live cells were taken at 10 Z-sections (step size, 0.4 μm). Maximum projections of all Z-sections, differential interference contrast images, and the merge of the two are shown. (Insets) Enlarged images of single nuclei are shown. (C) The number of Rap1-YFP foci of 72 WT and 85 mms21-11 cells were quantified based on the projections. (D) Telomere position effect increases in mms21-11 strains. WT and mms21-11 cells containing reporters for telomere silencing (VR TEL:URA3) (44) were spotted in 10-fold serial dilutions on plates that have 5-fluoroorotic acid (5-FOA) (counter selective for Ura3) or lack 5-FOA [synthetic complete (SC) medium, for control]. Partial repression of URA3 expression in WT strains allows some growth on 5-FOA. Complete abolition of silencing in the sir2Δ strain results in the failure to grow on 5-FOA (44). Increased silencing in the mms21-11 strain has the opposite effect to that of sir2Δ.
Discussion
We have identified a SUMO E3, Mms21, in yeast, the third such enzyme in this organism. Like some known SUMO E3s, this E3 contains a SP-RING-like motif. But unlike the two known SUMO E3s, Siz1 and Siz2, which function as single proteins, we found that this additional SUMO E3 is part of a nuclear, octameric complex. Interestingly, the subunits of this complex colocalize at several perinuclear foci. Other subunits of this complex include two SMC proteins, Smc5 and Smc6, and Nse1 that possesses a RING finger motif and therefore might have ubiquitin ligase activity. Hence this nuclear complex potentially combines several functions: sumoylation, ubiquitination, and chromosome organization. The function of the other members of the octameric complex is currently not known. Yet they also serve essential functions because we found that deletion of any of the eight subunits is lethal.
We demonstrated that deletion of the SP-RING motif of Mms21 (Mms21-11) is not lethal but that it abolishes SUMO E3 activity, confirming that this motif is essential for this activity. We detected three apparently disparate phenotype in mms21-11 strains: increased sensitivity to DNA damaging agents, defects in nucleolar structure and in the clustering, and silencing as well as length regulation of telomeres. We propose therefore that these phenotypes are the result of defective sumoylation of proteins that are involved in these processes.
In search of such sumoylation substrates we found two proteins. Their sumoylation was dramatically decreased in mms21-11 mutants after DNA damage. One is the Smc5 subunit of the octameric complex, and the other is Yku70 that functions in nonhomologous end joining repair (39). The observed decrease of sumoylation in mms21-11 cells is likely to be direct because we showed that these two proteins can also be sumoylated in vitro by recombinant Mms21. Interestingly, Smc5 directly binds to Mms21 and stimulates the sumoylating activity of Mms21. Although we do not know how Mms21 recognizes Yku70 or other yet to be identified substrates in vivo, we envision that the other members of the octameric complex play an important role. A clue for targeting Mms21 may be its association with Smc5/6. Smc5 and Smc6 belong to the SMC protein family, members of which have ATPase activity and form filaments via their long coiled-coil domains (40-42). The other two pairs of the SMC family have been proposed to form rings that enclose either two chromatin fibers (cohesin) or bring together different regions of the same chromatin fiber (condensin) (41, 43). Two or three non-SMC proteins stabilize the clamps of cohesin or condensin, respectively (41, 43). We propose that the Smc5/6 pair may similarly enclose/connect chromatin fiber(s) with the six non-SMC proteins helping to stabilize and localize the complex to specific regions of the chromatin fiber. A selective localization would then allow Mms21 to sumoylate proteins associated with such chromatin regions. This sumoylation may then serve to displace or recruit other proteins to that site. The disparate phenotype that we observed in the mms21-11 strain therefore may result from a failure to efficiently sumoylate these target proteins.
Sumoylation is a reversible process. Our previous study showed that one of the desumoylating enzymes, Ulp1, is anchored at NPCs via Mlps (5). Deletion of Mlps or the localization domain of Ulp1 led to delocalization of this enzyme to the nucleoplasm and caused unphysiological desumoylation of intranuclear proteins (5, 8, 9). Our finding that deletion of either MLPs or the localization domain of Ulp1 was synthetically lethal with mms21-11 indicates that a proper balance of sumoylation and desumoylation is crucial for cell survival. Moreover, we observed that mms21-11 was synthetically “sick” with the deletion of Siz1 or Siz2, suggesting that different SUMO E3s may share overlapping substrate specificities. Indeed, sumoylation of Yku70 and Smc5, although strongly reduced in mms21-11 strains, was not completely abolished, indicating that Siz1/2 or other, not yet identified E3s, can substitute for Mms21.
Our results here have established that the SUMO ligase activity of Mms21 is required for DNA repair and chromatin organization. A systematic identification of the entire inventory of substrates of this SUMO E3 may help in understanding the disparate phenotype that we describe here. Moreover, the viability of the mms21-11 cells suggests that Mms21 has additional functions that need to be identified. Finally, the functions of the other subunits remain to be investigated. For example, does the Nse1 subunit possesses ubiquitin ligase activity or do the Smc5/6 subunits form a ring around chromatin fibers? What are the functions of the other subunits? Such studies should shed more light on how this nuclear, multiprotein complex functions in DNA repair and chromatin organization.
Supplementary Material
Acknowledgments
We thank Zhonghui Huang for excellent technical assistance, Erica Johnson (Thomas Jefferson University, Philadelphia) for many reagents and helpful discussions, and Mark Longtine (Oklahoma State University, Stillwater), Wei Xiao (University of Saskatchewan, Saskatoon, Canada), David Shore (University of Geneva, Geneva), Stefan Jentsch (Max Planck Institute of Biochemistry, Munich), Heidi Feldmann (University of Munich, Munich), Alan Tomkinson (University of Maryland, College Park), Rodney Rothstein (Columbia University, New York), and The National Center for Research Resources' Yeast Resource Center (University of Washington, Seattle) for reagents. This work was supported by a Damon Runyon-Walter Winchell Postdoctoral Fellowship (to X.Z.).
Author contributions: X.Z. designed research; X.Z. performed research; X.Z. and G.B. contributed new reagents/analytic tools; X.Z. analyzed data; and X.Z. and G.B. wrote the paper.
Abbreviations: Mlp, myosin-like protein; PIAS, protein inhibitor of activated signal transducer and activator of transcription; SP-RING, Siz/PIAS RING; SUMO, small ubiquitin-like modifier; SMC, structural maintenance of chromosome; NPC, nuclear pore complex; MMS, methyl methanesulfonate; ProA, protein A; YFP, yellow fluorescent protein; CFP, cyan fluorescent protein; YPD, yeast extract/peptone/dextrose; 5-FOA, 5-fluoroorotic acid.
See Commentary on page 4661.
References
- 1.Kolling, R., Nguyen, T., Chen, E. Y. & Botstein, D. (1993) Mol. Gen. Genet. 237, 359-369. [DOI] [PubMed] [Google Scholar]
- 2.Kosova, B., Pante, N., Rollenhagen, C., Podtelejnikov, A., Mann, M., Aebi, U. & Hurt, E. (2000) J. Biol. Chem. 275, 343-350. [DOI] [PubMed] [Google Scholar]
- 3.Strambio-de-Castillia, C., Blobel, G. & Rout, M. P. (1999) J. Cell Biol. 144, 839-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galy, V., Olivo-Marin, J. C., Scherthan, H., Doye, V., Rascalou, N. & Nehrbass, U. (2000) Nature 403, 108-112. [DOI] [PubMed] [Google Scholar]
- 5.Zhao, X., Wu, C.-Y. & Blobel, G. (2004) J. Cell Biol. 167, 605-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galy, V., Gadal, O., Fromont-Racine, M., Romano, A., Jacquier, A. & Nehrbass, U. (2004) Cell 116, 63-73. [DOI] [PubMed] [Google Scholar]
- 7.Li, S. J. & Hochstrasser, M. (1999) Nature 398, 246-251. [DOI] [PubMed] [Google Scholar]
- 8.Panse, V. G., Kuster, B., Gerstberger, T. & Hurt, E. (2003) Nat. Cell Biol. 5, 21-27. [DOI] [PubMed] [Google Scholar]
- 9.Li, S. J. & Hochstrasser, M. (2003) J. Cell Biol. 160, 1069-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwienhorst, I., Johnson, E. S. & Dohmen, R. J. (2000) Mol. Gen. Genet. 263, 771-786. [DOI] [PubMed] [Google Scholar]
- 11.Li, S. J. & Hochstrasser, M. (2000) Mol. Cell. Biol. 20, 2367-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melchior, F. (2000) Annu. Rev. Cell Dev. Biol. 16, 591-626. [DOI] [PubMed] [Google Scholar]
- 13.Muller, S., Hoege, C., Pyrowolakis, G. & Jentsch, S. (2001) Nat. Rev. Mol. Cell Biol. 2, 202-210. [DOI] [PubMed] [Google Scholar]
- 14.Seeler, J. S. & Dejean, A. (2003) Nat. Rev. Mol. Cell. Biol. 4, 690-699. [DOI] [PubMed] [Google Scholar]
- 15.Gill, G. (2004) Genes Dev. 18, 2046-2059. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, E. S. (2004) Annu. Rev. Biochem. 73, 355-382. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, E. S. & Gupta, A. A. (2001) Cell 106, 735-744. [DOI] [PubMed] [Google Scholar]
- 18.Takahashi, Y., Kahyo, T., Toh, E. A., Yasuda, H. & Kikuchi, Y. (2001) J. Biol. Chem. 276, 48973-48977. [DOI] [PubMed] [Google Scholar]
- 19.Hochstrasser, M. (2001) Cell 107, 5-8. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, E. S. & Blobel, G. (1999) J. Cell Biol. 147, 981-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Driller, L., Wellinger, R. J., Larrivee, M., Kremmer, E., Jaklin, S. & Feldmann, H. M. (2000) J. Biol. Chem. 275, 24921-24927. [DOI] [PubMed] [Google Scholar]
- 22.Hoege, C., Pfander, B., Moldovan, G. L., Pyrowolakis, G. & Jentsch, S. (2002) Nature 419, 135-141. [DOI] [PubMed] [Google Scholar]
- 23.Chen, L., Trujillo, K., Ramos, W., Sung, P. & Tomkinson, A. E. (2001) Mol. Cell 8, 1105-1115. [DOI] [PubMed] [Google Scholar]
- 24.Ritchie, K. B., Mallory, J. C. & Petes, T. D. (1999) Mol. Cell. Biol. 19, 6065-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lisby, M., Rothstein, R. & Mortensen, U. H. (2001) Proc. Natl. Acad. Sci. USA 98, 8276-8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prakash, S. & Prakash, L. (1977) Genetics 87, 229-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDonald, W. H., Pavlova, Y., Yates, J. R. J. & Boddy, M. N. (2003) J. Biol. Chem. 278, 45460-45467. [DOI] [PubMed] [Google Scholar]
- 28.Hazbun, T. R., Malmstrom, L., Anderson, S., Graczyk, B. J., Fox, B., Riffle, M., Sundin, B. A., Aranda, J. D., McDonald, W. H., Chiu, C. H., et al. (2003) Mol. Cell 12, 1353-1365. [DOI] [PubMed] [Google Scholar]
- 29.Montelone, B. A., Prakash, S. & Prakash, L. (1981) Curr. Genet. 4, 223-232. [DOI] [PubMed] [Google Scholar]
- 30.Fujioka, Y., Kimata, Y., Nomaguchi, K., Watanabe, K. & Kohno, K. (2002) J. Biol. Chem. 277, 21585-21591. [DOI] [PubMed] [Google Scholar]
- 31.Onoda, F., Takeda, M., Seki, M., Maeda, D., Tajima, J., Ui, A., Yagi, H. & Enomoto, T. (2004) DNA Repair (Amst.) 3, 429-439. [DOI] [PubMed] [Google Scholar]
- 32.Tatham, M. H., Jaffray, E., Vaughan, O. A., Desterro, J. M., Botting, C. H., Naismith, J. H. & Hay, R. T. (2001) J. Biol. Chem. 276, 35368-35374. [DOI] [PubMed] [Google Scholar]
- 33.Bencsath, K. P., Podgorski, M. S., Pagala, V. R., Slaughter, C. A. & Schulman, B. A. (2002) J. Biol. Chem. 277, 47938-47945. [DOI] [PubMed] [Google Scholar]
- 34.Taylor, E. M., Moghraby, J. S., Lees, J. H., Smit, B., Moens, P. B. & Lehmann, A. R. (2001) Mol. Biol. Cell 12, 1583-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lehmann, A. R., Walicka, M., Griffiths, D. J., Murray, J. M., Watts, F. Z., McCready, S. & Carr, A. M. (1995) Mol. Cell. Biol. 15, 7067-7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fousteri, M. I. & Lehmann, A. R. (2000) EMBO J. 19, 1691-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gotta, M., Laroche, T., Formenton, A., Maillet, L., Scherthan, H. & Gasser, S. M. (1996) J. Cell Biol. 134, 1349-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin, Q. W., Fuchs, J. & Loidl, J. (2000) J. Cell Sci. 113, 1903-1912. [DOI] [PubMed] [Google Scholar]
- 39.Lewis, L. K. & Resnick, M. A. (2000) Mutat. Res. 451, 71-89. [DOI] [PubMed] [Google Scholar]
- 40.Jessberger, R. (2002) Nat. Rev. Mol. Cell. Biol. 3, 767-778. [DOI] [PubMed] [Google Scholar]
- 41.Hirano, T. (2002) Genes Dev. 16, 399-414. [DOI] [PubMed] [Google Scholar]
- 42.Hagstrom, K. A. & Meyer, B. J. (2003) Nat. Rev. Genet. 4, 520-534. [DOI] [PubMed] [Google Scholar]
- 43.Haering, C. H. & Nasmyth, K. (2003) BioEssays 25, 1178-1191. [DOI] [PubMed] [Google Scholar]
- 44.Roy, N. & Runge, K. W. (2000) Curr. Biol. 10, 111-114. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.