Abstract
Background
HBV DNA is the most important molecular marker in hepatitis B, used to determine treatment indication and monitoring. Most patients require lifelong hepatitis B virus (HBV) management, thus viral load (VL) monitoring may be performed at different laboratories, with different HBV assays, which may result in different VL results. This multicenter study compares the commutability and concordance of results from four different HBV DNA assays: CAP/CTM HBVv2, HPS/CTM HBVv2 and the new cobas 6800/8800 HBV and cobas 4800 HBV assays.
Methods
Across all four assays, HBV limit of detection (LoD) and linearity at lower concentrations were assessed using panels traceable to the World Health Organization international standard, and concordance was investigated at the important medical decision cutoffs 2000 and 20,000 IU/ml, using specimens from HBV-positive patients.
Results
The calculated LoD via a probit curve was 2.7 IU/ml for cobas 6800/8800 HBV, 2.8 IU/ml for cobas 4800 HBV, 9.6 IU/ml for CAP/CTM HBVv2, and 6.2 IU/ml for HPS/CTM HBVv2. The average accuracy was comparable between cobas 6800/8800 HBV, cobas 4800 HBV and CAP/CTM HBVv2 (0.04–0.05 log10 IU/ml), while a slightly lower accuracy was documented for HPS/CTM HBVv2 (−0.16 log10 IU/ml). A total of 211–245 clinical samples were used for a pairwise comparison. Mean paired log differences ranged from −0.17 log10 IU/ml to −0.01 log10 IU/ml. Coefficient of determination was over 98% for all pairs with high overall percent agreement at the 2000 and 20,000 IU/ml cutoffs (from 91.7% to 96.3%). In a subset of samples with VL±0.5 log10 to the 2000 and 20,000 IU/ml thresholds, concordance was still 72% and 82%, respectively.
Conclusions
The new cobas 6800/8800 HBV and 4800 HBV assays show high accuracy in samples with low-level viremia and a high concordance with the established HBV tests, CAP/CTM HBVv2 and HPS/CTM HBVv2, at 2000 and 20,000 IU/ml. Thus, all four HBV assays have high commutability and may be used interchangeably in routine clinical practice.
Keywords: cobas, HBV diagnostics, HBV DNA assay, HBV treatment, hepatitis B virus
Introduction
Hepatitis B virus (HBV) infection is a global health problem. About 20% of the world population have been exposed to the virus.1 However, the majority (>95%) of adults acquiring HBV do not develop chronic hepatitis B (CHB), but achieve permanent immune control, reflected by hepatitis B surface antigen (HBsAg) seroconversion (resolved HBV infection). In contrast, the rate of CHB is far higher if viral transmission occurs during early childhood (20–50%) and even exceeds 90% in those infected perinatally.2,3 Currently, about 240 million people worldwide have CHB.4 Patients with CHB may develop long-term complications of HBV infection, including liver cirrhosis and hepatocellular carcinoma (HCC).2,5–9 Almost 700,000 people die each year due to HBV-related complications.10
However, not all patients with CHB are at high risk of developing HBV-related complications. Thus, it is of high clinical relevance to reliably identify patients with CHB, who are at a considerable risk, and therefore requiring antiviral treatment or close monitoring. Currently, one of the most valuable markers in CHB is quantitative HBV DNA. High levels of plasma HBV DNA have been identified as a major risk factor for the development of liver cirrhosis and HCC.11–14 The suppression of HBV DNA by antiviral treatment significantly reduces the incidence of liver cirrhosis. Consequently, the HBV DNA plasma level plays a central role in all key international HBV guidelines to determine the indication for antiviral therapy. In addition, it is the most important biomarker for monitoring patients during antiviral treatment and to confirm treatment response, particularly if nucleos(t)ide analogues (NUCs) are used.2,5,15
HBV DNA in plasma can be quantitated by nucleic acid amplification technologies, such as polymerase chain reaction (PCR).16–19 In general, the use of real-time PCR methodology is recommended for HBV DNA quantification, primarily due to increased sensitivity and a broader linear range.2,5,15,20
Currently available CHB treatments rarely lead to ‘HBV cure’, but only suppress viral replication or improve immune control. Thus, lifelong HBV management, antiviral therapy, or at least monitoring for the risk of HBV-related complications, is required for the majority of patients. Because of the need for such long-term surveillance, over time, it is not uncommon for different healthcare providers to manage a specific patient’s HBV disease, or that there are different laboratories and/or different systems being used to monitor a specific patient’s viral load. These intralaboratory differences may further be compounded by interlaboratory variations with laboratories transitioning from currently available assays, such as the cobas AmpliPrep/COBAS TaqMan HBV Test, version 2.0 (CAP/CTM HBV v2) and the cobas TaqMan HBV test for use with the High Pure System (CTM/HPS HBV) to newer systems. Recently, two new cobas assays have been approved by the US Food and Drugs Administration: the cobas HBV for use on the cobas 6800/8800 Systems (cobas 6800/8800 HBV) and the cobas 4800 HBV test for use on the cobas 4800 System (cobas 4800 HBV). Currently, there are little or no data demonstrating the commutability or concordance of HBV viral load results among institutions, or among these four cobas HBV assays.
The purpose of this multicenter, international study was to evaluate the clinical performance of and the commutability between the four cobas HBV assays, CAP/CTM HBV v2, CTM/HPS HBV, cobas 6800/8800 HBV and cobas 4800 HBV, which may be routinely used in viral load (VL) monitoring.
Materials and methods
Study sites and ethical statement
This was an international multicenter study, with sites located at Hannover Medical School (Hanover, Germany), Covance Central Laboratory Services (Geneva, Switzerland), and Department of Laboratory Medicine, Korea University, Anam Hospital (Seoul, South Korea).
This study was conducted in compliance with the International Conference on Harmonisation (ICH) Good Clinical Practice (GCP) Guidelines, the Declaration of Helsinki, and applicable local regulations. This study was approved by the ethics committee (EC) of Hannover Medical School, which waived the need for written informed consent. This was a nonsignificant risk study. All specimens were de-identified leftover or prospectively collected specimens. The study was noninterventional, nonobservational, and no results were used for patient care.
Sample selection and HBV DNA quantification
All experiments were performed in International Standard Organization (ISO) 9001:2008-certified or ISO 17,025:2005-accredited laboratories.
The following four generations of the Roche cobas HBV assay were utilized: cobas 6800/8800 HBV (cobas HBV test for use on the cobas 6800/8800 Systems; Roche Diagnostics, Pleasanton, CA, USA), cobas 4800 HBV (cobas HBV test for use on the cobas 4800 Systems; Roche Diagnostics, Pleasanton, CA, USA), CAP/CTM HBV v2 (COBAS AmpliPrep/COBAS TaqMan HBV v2 test; Roche Diagnostics, Pleasanton, CA, USA), and HPS/CTM HBV v2 (COBAS TaqMan HBV test; Roche Diagnostics, Pleasanton, CA, USA for use with the High Pure System).21–24
All four cobas HBV assays are quantitative nucleic acid tests that enable the detection and quantification of HBV DNA in EDTA plasma or serum of HBV-infected patients. Assays (cobas 6800 HBV, cobas 4800 HBV, CAP/CTM HBV v2, and HPS/CTM v2) were used according to the manufacturer’s instructions.21–24 Assay and system characteristics are described in Table 1. All assays are calibrated to the World Health Organization (WHO) International Standard for HBV DNA, and results are reported in IU/ml.
Table 1.
Lower limit of quantitation, upper limit of quantitation, sample input volume, and processing volume of the four cobas HBV assays*.
| Platform* | Quantitative standard control (type) |
Linear range | Workflow | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LLOQ (IU/ml) |
ULOQ (IU/ml) |
Sample processing volume (µl) |
Type of sample prep | Maximum walk away time (h) |
Sample processing time$ (h) | Hands on time per 8 h shift (h) | Detection time (h) |
Interventions per 8 h (n) |
Time to first results [h (n)] | Throughput in 8 h (n) |
||
| HPS/CTM | Competitive | 29 | 1.1E + 08 | 500 | Manual | 3 | 2 | 2.5 | 3 | N/A | 5 (48) | 96 |
| CAP/CTM | Competitive | 20 | 1.7E + 08 | 650 | Automated | 2.5 | 2 | 0.5 | 3 | 4 | 5.5 (96) | 96 |
| cobas 4800 | Noncompetitive | 10 | 1.0E + 09 | 200 or 400 | Automated | 2.7 | 3 | 0.5 | 1.5 | 5 | <5 (96) | 192 |
| cobas 6800 | Noncompetitive | 10 | 1.0E + 09 | 200 or 500 | Automated | 8 | 1.5 | 0.5 | 1.5 | 3 | <3.5 (96) | 384 |
| cobas 8800 | Noncompetitive | 10 | 1.0E + 09 | 200 or 500 | Automated | 4 | 1.5 | 1 | 1.5 | 4‡ | <3.5 (96) | 960 |
Corresponding assays for each platform: COBAS TaqMan HBV test for use with the High Pure System (HPS/CTM); COBAS AmpliPrep/COBAS TaqMan HBV v2 test using the COBAS AmpliPrep Instrument for automated specimen processing and the COBAS TaqMan Analyzer (CAP/CTM); cobas HBV test for use on the cobas 4800 System (cobas 4800); cobas HBV test for use on the cobas 6800/8800 Systems (cobas 6800/8800).
Sample processing time includes sample extraction and purification and polymerase chain reaction setup.
Extra waste removal step compared with cobas 6800.
HBV, hepatitis B virus; LLOQ, lower limit of quantitation; ULOQ, upper limit of quantitation.
Assay performance was evaluated using the following two approaches.
Comparison of analytical performance of four assays using the third WHO HBV standard panel
A seven-member panel was prepared from the third WHO International Standard Material for HBV. In accordance with the original concentrations assigned by the National Institute for Biological Standards and Control, the WHO material was diluted into HBV-negative human EDTA plasma in specified concentrations (see Supplemental Table S1). The HBV DNA results from the four assays were compared to evaluate analytical sensitivity and linearity at lower concentrations.
Assay correlation using clinical specimens
HBV DNA test results were compared by using specimens from patients with HBV-positive test results who were (a) on therapy and suppressed; (b) at or near clinically relevant decision points; and (c) not on treatment or nonsuppressed, in which specimen VLs spanned across the linear range of the four assays. All patient specimens that were included in this comparison were obtained from frozen plasma that remained, from specimens that were previously tested with the CAP/CTM HBV v2 assay, in previous clinical studies. The specimens were selected based on the volume and viral load necessary to cover the assay linear range, in a balanced way, for this study.
Statistics
All statistical analyses were carried out using SAS/STAT software (SAS Systems).25 All confidence intervals (CIs) were constructed using a 95% confidence level. For both objectives, outlier testing was conducted using the method of Studentized residuals.
Assay comparison using the third WHO HBV panel
HBV limit of detection (LoD), used as a measure of analytical sensitivity, was defined as the lowest concentration at which at least 95% of samples gave positive results in the respective assay. LoD was calculated using a probit curve fit to the lowest panel members. Associated 95% CIs for the LoD were constructed and used to compare each assay. HBV DNA concordance analysis was performed by comparing the mean log10 difference of the observed titers for each panel member for each assay pair. Supportive statistics including 95% CI for the mean log10 differences were calculated. Linearity was assessed by comparing the mean log10 of the observed titers for each WHO standard panel member to the predicted value from an ordinary least squares (OLS) regression.
Assay correlation using clinical specimens
For pairwise assay comparisons, Deming regression analyses were performed. The coefficient of determination was calculated, as well as the 95% CI for the slopes and intercept. Bland–Altman bias plots were constructed to evaluate bias between the mean differences of the assay comparisons. If bias was deemed constant and linear, an overall mean bias estimate was computed at the medically relevant decision points of 2000 and 20,000 IU/ml.2,15 Overall percent agreement (OPA), a measure of agreement between assays, across pairwise comparisons for the clinically relevant cutoffs of 2000 and 20,000 IU/ml respectively were calculated. To calculate the OPA, the total number of times in which the assays agree is divided by the total number of readings or classifications made.26
To further analyze the robustness of VL results at clinically relevant decision points, a subset of samples with viral loads ±0.5 log10 to the clinical important cutoffs 2000 and 20,000 IU/ml were selected. These medical decision points were established using AMPLICOR HBV MONITOR Test and corroborated with the HPS/CTM HBV v2 assay in the tenofovir pivotal registration study.27–29 Thus, for this subanalysis, we used samples with HBV DNA levels ±0.5 log10 for 2000 and 20,000 IU/ml using the HPS/CTM HBVv2 assay results as the ‘reference population’ from which to partition samples by viral load.
Results
Assay comparison using the third WHO HBV standard panel
The number of replicates of each panel member tested, per assay, and the number of valid tests are summarized in Supplemental Table S1. Of the 1202 replicates tested, 1170 gave a valid result (97.3%).
From the 1170 valid test results, the LoD estimated, via a probit curve, was 2.7 IU/ml for cobas 6800/8800 HBV, 2.8 IU/ml for cobas 4800 HBV, 9.6 IU/ml for CAP/CTM HBV v2, and 6.2 IU/ml for HPS/CTM HBV v2 (Table 2).
Table 2.
Limit of detection, accuracy and standard deviation of four cobas HBV assays* (N = 1170).
| Nominal titer (IU/ml) |
cobas 6800/8800 | cobas 4800 HBV | CAP/CTM HBV v2 | HPS/CTM HBV v2 | |
|---|---|---|---|---|---|
| (IU/ml) | (log10 IU/ml) | % (#) | % (#) | % (#) | % (#) |
| 2.000 | 3.3 | 100 (42/42) | 100 (40/40) | 100 (40/40) | 100 (40/40) |
| 200 | 2.3 | 100 (48/48) | 100 (40/40) | 100 (40/40) | 100 (40/40) |
| 60 | 1.78 | 100 (48/48) | 100 (40/40) | 100 (40/40) | 100 (40/40) |
| 20 | 1.3 | 100 (48/48) | 100 (40/40) | 100 (40/40) | 100 (40/40) |
| 10 | 1 | 100 (48/48) | 100 (40/40) | 95.0 (38/40) | 97.5 (39/40) |
| 5 | 0.7 | 100 (48/48) | 100 (40/40) | 77.5 (31/40) | 95.0 (38/40) |
| 2,5 | 0.4 | 83.3 (40/48) | 75.0 (30/40) | 47.5 (19/40) | 60.0 (24/40) |
|
LoD$via PROBIT
95% hit rate (IU/ml) (95%CI) |
2.7 (nc–nc) |
2.8 (nc–nc) |
9.6 (7.3–16.5) |
6.2 (4.8–10.8) |
|
|
LoD ⩾ 95% hit rate
(IU/ml) |
5 | 5 | 10 | 5 | |
|
Average accuracy‡
(log10 IU/ml) |
0.05 | 0.04 | 0.04 | –0.16 | |
|
Standard deviation (log10 IU/ml)
(min, max) |
(0.04, 0.14) | (0.06, 0.24) | (0.05, 0.16) | (0.07, 0.15) | |
cobas 6800/8800 HBV, cobas HBV test for use on the cobas 6800/8800 Systems; cobas 4800 HBV, cobas HBV test for use on the cobas 4800 Systems; CAP/CTM HBV v2, COBAS AmpliPrep/COBAS TaqMan HBV v2 test; and HPS/CTM HBV v2, COBAS TaqMan HBV test for use with the High Pure System.
LoD was calculated using panel members with concentration ranging from 2.5 to 2000 IU/ml.
Average accuracy and standard deviation were calculated using panel members with nominal concentration within each assay linear range.
CI, confidence interval; HBV, hepatitis B virus; LoD, limit of detection; nc, noncalculable.
All assays achieved a 100% hit rate if HBV DNA concentration was 20 IU/ml or higher and a hit rate of at least 95% if viral load was 10 IU/ml. Even samples with a viral load of 2.5 IU/ml were identified as HBV DNA positive in the majority of cases (Table 2).
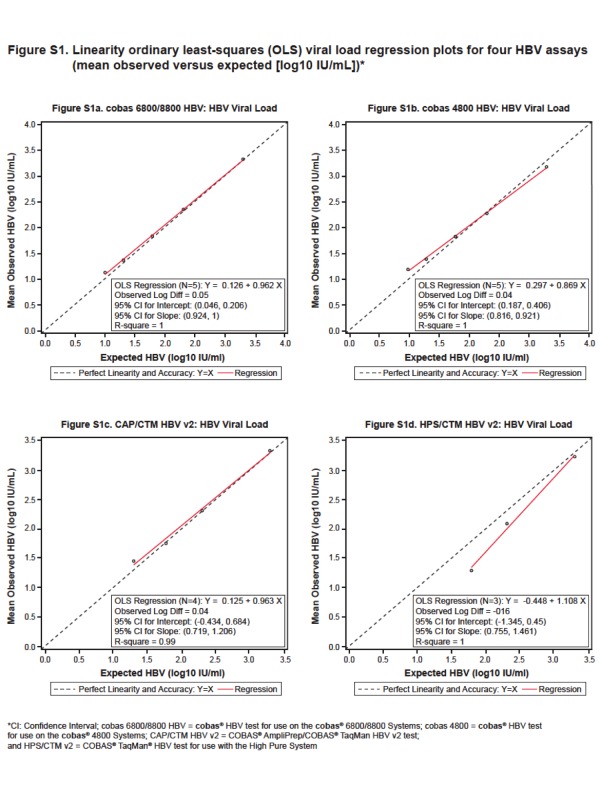
The average accuracy calculated using panel members with nominal concentration within each assay linear range was comparable between cobas 6800/8800 HBV, cobas 4800 HBV and CAP/CTM HBV v2 (0.04–0.05 log10 IU/ml), while a slightly lower accuracy was documented for the HPS/CTM HBV v2 (−0.16 log10 IU/ml) (Table 2). The observed mean log10 IU/ml titers from each of the five panel members (2000, 200, 60, 20, and 10 IU/ml) were plotted against the nominal HBV DNA log10 IU/ml concentrations. Individual accuracy values (mean log10 observed – log10 nominal) ranged from −0.50 to 0.20 log10 IU/ml across the four assays whereas linearity [observed mean (log10 IU/ml) – linearized (log10 IU/ml)] ranged from −0.35 to 0.16 log10 IU/ml (Supplemental Table S2).
The linearity OLS regression plots for cobas 6800/8800 HBV, cobas 4800 HBV, CAP/CTM HBV v2, and HPS/CTM HBV v2 are shown in Supplementary Figure S1.
Assay comparison using clinical specimens
Overall correlation between the four assays
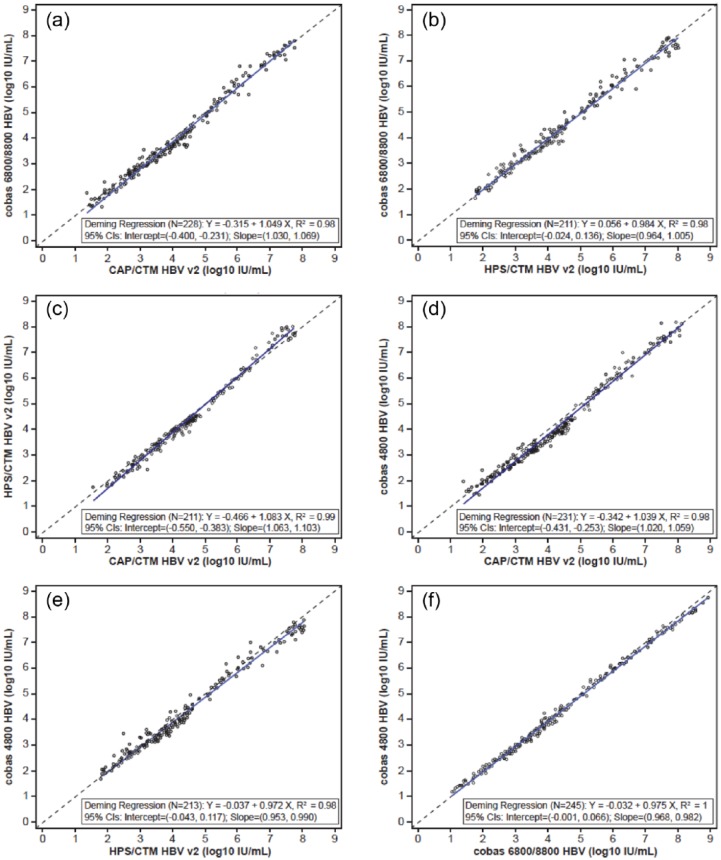
A total of 211–245 clinical samples were tested with all four assays and the results within the linear range were used in pairwise method comparisons. Across all pairs of assays, the mean paired log difference ranged from −0.17 to −0.01 log10 IU/ml (Supplemental Table S3). The coefficient of determination was at least 98% for all pairs of assays, demonstrating a strong correlation between the four assays (Supplemental Table S3, Figure 1).
Figure 1.
Deming regression plot of viral loads (IU/ml) for four hepatitis B virus (HBV) assays.
(a) Deming regression plot for cobas 6800/8800 HBV versus CAP/CTM HBV v2 (n = 228);
(b) Deming regression plot for cobas 6800/8800 HBV versus HPS/CTM HBV v2 (n = 211);
(c) Deming regression plot for HPT/CTM HBV v2 versus CAP/CTM HBV v2 (n = 211);
(d) Deming regression plot for cobas 4800 HBV versus CAP/CTM HBV v2 (n = 231);
(e) Deming regression plot for cobas 4800 HBV versus HPS/CTM HBV v2 (n = 213);
(f) Deming regression plot for cobas 4800 HBV versus cobas 6800/8800 HBV (n = 245). CI, confidence interval.
OPA between assays
Table 3 shows the OPA between the HPS/CTM HBV v2 and the cobas 6800/8800 HBV across pairwise comparisons for the clinically relevant cutoffs of 2000 and 20,000 IU/ml, respectively. For all pairwise comparisons at the 2000 IU/ml cutoff, the OPA between the four assays ranged from 91.7% to 96.3%, and the 95% CIs ranged from 88.0% to 98.2%. Similar results were found at the 20,000 IU/ml cutoff. The OPA ranged from 93.0% to 97.7% across all pairwise comparisons, and the 95% CI ranged from 89.5% to 99.1%.
Table 3.
Overall concordance in the classification at the 2000 IU/ml and 20,000 IU/ml medical decision points between HPS/CTM HBV v2 and cobas 6800/8800 HBV assays*.
| Concordance between HPS and c6800 | c6800/8800 HBV | Total | OPA (95% CI) |
||
|---|---|---|---|---|---|
| <2000 IU/ml | ⩾2000 IU/ml | ||||
| 2000 IU/ml cutoff | |||||
| HPS/CTM HBV v2 | <2000 IU/ml | 134 | 8 | 142 | 95.3% (92.3–97.4%) |
| ⩾2000 IU/ml | 6 | 152 | 158 | ||
| Total | 140 | 160 | 300 | ||
| Concordance between HPS and c6800/8800 | c6800/8800 HBV | Total | OPA (95% CI) |
||
| <20,000 IU/ml | ⩾20,000 IU/ml | ||||
| 20,000 IU/ml cutoff | |||||
| HPS/CTM HBV v2 | <20,000 IU/ml | 202 | 3 | 205 | 97% (94.4–98.6%) |
| ⩾20,000 IU/ml | 6 | 89 | 95 | ||
| Total | 208 | 92 | 300 | ||
cobas 6800/8800 HBV, cobas HBV test for use on the cobas 6800/8800 Systems; HPS/CTM HBV v2, COBAS TaqMan HBV test for use with the High Pure System.
CI, confidence interval; HBV, hepatitis B virus; OPA, overall percent agreement.
To further investigate concordance of clinical results in samples with viral loads close to the medically relevant decision points of 2000 and 20,000 IU/ml, we defined a subset of data that were within 0.5 log10 of these two cutoffs and conducted a post hoc analysis. Specifically, if the HPS/CTM HBV v2 value fell within 0.5 log10 of the medical decision points of either 2000 IU/ml (3.3 log10) or 20,000 IU/ml (4.3 log10), the pairwise comparisons were used. Using these criteria, 171 of the 899 possible pairwise comparisons to HPS/CTM HBV v2 data fell within 0.5 log10 of the 2000 IU/ml threshold, and 174 of the 899 possible pairwise comparisons fell within 0.5 log10 of the 20,000 IU/ml threshold. A total of 265 out of the 345 pairwise results were concordant across the thresholds representing an overall concordance of 76.8% (range 71.9–81.6%). Although the sample sizes were smaller, for the paired comparisons, at 20,000 IU/ml, the highest rate of concordance was between the cobas 6800 HBV and HPS/CTM HBV v2 at 84.5%, and the lowest rate of concordance was 77.6%, between CAP/CTM HBV v2 and HPS/CTM HBV v2. At 2000 IU/ml, the highest rate of concordance was between CAP/CTM HBV v2 and HPS/CTM HBV v2 at 78.9% and the lowest rate of concordance was 59.6% between cobas 4800 HBV and HPS/CTM HBV v2.
Further information and full pairwise comparisons of all tests can be found in the supplemental material, Tables S4–S11.
Discussion
Adequate management of patients with CHB is highly dependent on HBV DNA quantification. However, different assay characteristics or local differences may potentially alter VL results and therefore influence clinical decisions. In this international multicenter study, we showed that the new cobas 6800/8800 and cobas 4800 HBV assays have a high sensitivity and accuracy. Importantly, both new assays not only demonstrated a high concordance to one another, but also to the two already established and routinely used HBV tests, cobas CAP/CTM HBV v2 and HPS/CTM v2.
In this study, all four commercially available cobas HBV tests were analyzed with respect to their performance and concordance. This study was divided into two parts. In the first part of the study, the performance of the four assays in low viremic samples was assessed using the WHO standard to create samples with different, predefined HBV DNA concentrations. All assays demonstrated a high level of concordance in this low viremic setting. In the second part of the study, clinical samples were tested. Pairwise assessments of the four HBV assays also demonstrated a comparable performance and high concordance between the four assays, in a routine clinical setting.
The data generated from assay comparisons, using the third WHO HBV standard panel, allowed for the determination of the LoD. All four assays achieved a LoD up to 10 IU/ml. Interestingly, the new cobas 6800/8800 and cobas 4800 HBV assays showed a slightly improved sensitivity, with a LoD of 2.7–5 IU/ml (depending on the applied statistical methodology) compared with the older cobas HBV tests.
Improved assay sensitivity may possibly offer some potential clinical value, in the current era of NUC therapies and also for future HBV treatment strategies. Currently, a complete virological response to NUC therapy is defined as undetectable HBV DNA by a sensitive PCR assay. However, several patients may show residual viremia during regular treatment monitoring.2,5,30 Most recently, Kim and colleagues have published data suggesting that patients under long-term NUC treatment who still have low levels of HBV DNA, even below the limit of quantification, have a higher risk for HCC compared with patients with an undetectable HBV DNA result.31 A higher assay sensitivity may increase the number of patients identified with low levels of viremia, and potentially affect their clinical management, such as HCC screening, or possibly a change in the patient’s current antiviral treatment regimen.
Another potential impact of assay sensitivity could be the prevention of HBV reactivation during strong immunosuppressive treatments (i.e. rituximab). HBV reactivation usually occurs in HBsAg-positive patients but may also happen in patients with an anti-HBc only status or even in those who already have achieved HBsAg seroconversion.32,33 The use of prophylaxis versus preemptive antiviral treatment in HBsAg-negative, anti-HBc-positive patients is still a matter of debate and some practitioners consider close monitoring for HBV DNA as sufficient strategy under these circumstances and initiation of HBV therapy may only be required if HBV DNA is detected. Future studies are required to assess whether HBV assays with higher sensitivities can improve the prediction of HBV reactivation in this setting and help to identify patients who require antiviral treatment. A higher sensitivity must not impair specificity. Specificity has not been assessed in this study but has been intensively studied for each assay during the respective registration trials. Each of the here studied assays showed a specificity of 100%.21–24
A particular focus of our study was on the accuracy and concordance around important clinical decision points, namely the 2000 and 20,000 IU/ml thresholds. Clinical concordance, when switching platforms, is the most important metric by which to judge equivalency of results. Current European Association for the Study of the Liver guidelines recommend a liver biopsy in patients with a VL of 2000–20,000 IU/ml and an alanine aminotransferase level over two times the upper limit of normal. Patients with a VL less than 2000 IU/ml do not require antiviral treatment, whereas patients with an HBV DNA over 20,000 IU/ml are supposed to start treatment without the need for a biopsy result.2 As such, considerable differences or discordance between assays could potentially lead to significantly higher, or lower, HBV DNA quantification, which could result in a different clinical management decision, depending upon the assay that is used. In other words, decisions for the therapeutic or diagnostic management of a patient would not exclusively be based upon the patient’s status but, potentially, on the assay that is used. According to our data, the concordance between all four cobas assays was in general high, including at the 2000 IU/ml cutoff, which ranged from 91.7% to 96.3% across all pairwise comparisons. Thus, patient classification and clinical decisions for liver biopsy or initiation of antiviral HBV treatment should be the same in almost all patients, regardless of which of the four assays is used.
Not surprisingly, the risk for discordant results increased if the HBV DNA result was close to the 2000 or the 20,000 IU/ml cutoff, as documented in our post hoc analysis of a subset of samples. While we realize the smaller sample size of the subset limits the power of the analysis, it does suggest that the concordance at these particular cutoffs are never 100%. It is also important to realize there is intra-assay variability, and a result close to a medical decision point, if repeated, can generate a second result, on the opposite side of the cutoff, potentially leading to different medical decisions. Similar observations have been made for clinically important viral load cutoffs in hepatitis C virus infection.34,35 These observations suggest medical decision making should not be based exclusively upon a single viral load result, but on an established trend, observed over time or multiple clinical and serological parameters.
In conclusion, we showed that the new cobas 6800/8800 HBV and 4800 HBV assays show a high accuracy, in samples with low-level viremia, and also a high concordance with the established HBV tests, CAP/CTM HBV v2 and HPS/CTM HBV v2, at the clinically important decision cutoffs of 2000 and 20,000 IU/ml. Thus, all four HBV assays have high commutability and may safely be used interchangeably in routine clinical practice. In addition, the higher sensitivity of the two newer cobas assays, cobas 6800/8800 HBV and cobas 4800 HBV, may offer a potential additive clinical value that requires further study.
Supplementary Material
Acknowledgments
The authors would like to thank the Clinical Operations, Data Management, and Biostatistics Teams at Roche Molecular Systems in Pleasanton, California for their help. Specifically, Jasmine Lau, Kevin Luk, Matthew Lin, Keerthi Bodinaidu, Jessie Canchola, Smitha Krishnamurthy, Varun Sama, Marizen Cunanan, Trisha Zeni, and Pari Hemyari. Medical writing support was funded by Roche Molecular systems and furnished by Sue Currie PhD and Amy Keller of Health Interactions, Inc.
Footnotes
Funding: This study was supported, in part, by Roche Molecular Systems, through financial, instrumentation, and reagent contributions, as well as support with the protocol design, sample procurement, data analyses, and manuscript preparation.
Conflict of interest statement: BM has received speaking or consulting fees from Abbott, AbbVie, Bristol Myers Squibb, Fujirebio Europe and Falk, Janssen-Cilaq, and Roche; research support from Abbott Molecular and Roche; travel grants from Bristol Myers Squibb, Gilead, and Janssen-Cilaq. MC received speaker or consulting fees from AbbVie, BMS, Falk, Gilead, Janssen-Cilag, Merck/MSD, Roche Pharma, Roche, and Novartis; research support from Fujirebio, Gilead, Merck/MSD, and Roche. JV has received advisory board or teaching fees from Abbott, AbbVie, Bristol-Myers Squibb, Gilead, Medtronic, and Roche. CV has received grants or research support from Abbott, Gilead, Janssen, Quintiles, Roche, and Siemens; speaking or consulting fees from Abbott, AbbVie, Achillion, Bristol-Myers Squibb, Gilead, Janssen, Merck/MSD, and Roche. MPM has received grants and personal fees from Bristol Myers Squibb, Boehringer Ingelheim, Gilead, GlaxoSmithKline, Janssen-Cilaq, MSD/Merck, Novartis, and Roche. YC and JYS have received research support from Roche. HW has received speaking or consulting fees from Abbott, AbbVie, Siemens, Bristol Myers Squibb, Janssen-Cilaq, Elger, Novartis, and Roche; research support from Abbott Molecular, Bristol Myers Squibb, Janssen-Cilaq, Elger, Novartis, and Roche. EM, COS, MN, and EP are employed by Roche. BB, PL, and VMT have no interests to declare.
Contributor Information
Benjamin Maasoumy, Department of Gastroenterology, Hepatology and Endocrinology, Hannover Medical School, Carl-Neuberg-Strasse 1, 30625 Hannover, Germany.
Birgit Bremer, Hannover Medical School, Hanover, Germany.
Patrick Lehmann, Hannover Medical School, Hanover, Germany.
Ed G. Marins, Roche Molecular Systems Inc., Roche Diagnostics, Pleasanton, CA, USA
Véronique Michel-Treil, Covance Central Laboratory Services SA, Geneva, Switzerland.
Christian O. Simon, Roche Molecular Systems Inc., Roche Diagnostics, Pleasanton, CA, USA
Merlin Njoya, Roche Molecular Systems Inc., Roche Diagnostics, Pleasanton, CA, USA.
Markus Cornberg, Hannover Medical School, Hanover, Germany.
Ellen Paxinos, Roche Molecular Systems Inc., Roche Diagnostics, Pleasanton, CA, USA.
Michael P. Manns, Hannover Medical School, Hanover, Germany
Johannes Vermehren, University Hospital, Frankfurt, Frankfurt am Main, Germany.
Christoph Sarrazin, University Hospital, Frankfurt, Frankfurt am Main, Germany.
Ji Yeon Sohn, Department of Laboratory Medicine, National Cancer Center, Goyang, Republic of Korea.
Yunjung Cho, Korea University College of Medicine, Seoul, Republic of Korea.
Heiner Wedemeyer, Hannover Medical School, Hanover, Germany.
References
- 1. Merrill R, Hunter BD. Seroprevalence of markers for hepatitis B viral infection. Int J Infect Dis 2011; 15: e78–e121. [DOI] [PubMed] [Google Scholar]
- 2. European Association for the Study of the Liver. EASL clinical practice guidelines: management of chronic hepatitis B infection. J Hepatol 2012; 57: 167–185. [DOI] [PubMed] [Google Scholar]
- 3. McMahon BJ, Alward WL, Hall DB, et al. Acute hepatitis B virus infection: relation of age to the clinical expression of disease and subsequent development of the carrier state. J Infect Dis 1985; 151: 599–603. [DOI] [PubMed] [Google Scholar]
- 4. Ott JJ, Stevens GA, Groeger J, et al. Global epidemiology of hepatitis B virus infection: new estimates of age-specific HBsAG seroprevalence and endemicity. Vaccine 2012; 30: 2212–2219. [DOI] [PubMed] [Google Scholar]
- 5. Terrault NA, Bzowej NH, Chang KM, et al. AASLD guidelines for treatment of chronic hepatitis B. Hepatology 2016; 63: 261–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dienstag JL. Hepatitis B virus infection. N Engl J Med 2008; 359: 1486–1500. [DOI] [PubMed] [Google Scholar]
- 7. Liaw YF. Natural history of chronic hepatitis B virus infection and long-term outcome under treatment. Liver Int 2009; 29: 100–107. [DOI] [PubMed] [Google Scholar]
- 8. Fattovich G, Bortolotti F, Donato F. Natural history of chronic hepatitis B: special emphasis on disease progression and prognostic factors. J Hepatol 2008; 48: 335–352. [DOI] [PubMed] [Google Scholar]
- 9. But DY, Lai CL, Yuen MF. Natural history of hepatitis-related hepatocellular carcinoma. World J Gastroenterol 2008; 14: 1652–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Global Burden of Disease Collaborators. Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 2015; 385: 117–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen CJ, Yang HI, Su J, et al. Risk of hepatocellular carcinoma across a biological gradient of serum hepatitis B virus DNA level. JAMA 2006; 295: 65–73. [DOI] [PubMed] [Google Scholar]
- 12. Wu CF, Yu MW, Lin CL, et al. Long-term tracking of hepatitis B viral load and the relationship with risk for hepatocellular carcinoma in men. Carcinogenesis 2008; 29: 106–112. [DOI] [PubMed] [Google Scholar]
- 13. Chen CJ, Yang HI, Iloeje UH. Hepatitis B virus DNA levels and outcomes in chronic hepatitis B. Hepatology 2009; 49: S72–S84. [DOI] [PubMed] [Google Scholar]
- 14. Chen CF, Lee WC, Yang H, et al. Serum levels of HBV DNA and alanine aminotransferase determine risk for hepatocellular carcinoma. Gastroenterology 2011; 141: 1240–1248. [DOI] [PubMed] [Google Scholar]
- 15. Sarin SK, Kumar M, Lau GK, et al. Asian-Pacific clinical practice guidelines on the management of hepatitis B: a 2015 update. Hepatol Int 2016; 10: 1–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sorrell MF, Belongia EA, Costa J, et al. National Institutes of Health Consensus Development Conference Statement: management of hepatitis B. Ann Intern Med 2009; 150: 104–110. [DOI] [PubMed] [Google Scholar]
- 17. Longo MC, Berninger MS, Hartley JL. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene 1990; 93: 125–128. [DOI] [PubMed] [Google Scholar]
- 18. Higuchi R, Dollinger G, Walsh PS, et al. Simultaneous amplification and detection of specific DNA sequences. Bio/technology 1992; 10: 413–417. [DOI] [PubMed] [Google Scholar]
- 19. Heid CA, Stevens J, Livak KJ, et al. Real time quantitative PCR. Genome Res 1996; 6: 986–994. [DOI] [PubMed] [Google Scholar]
- 20. Saldanha J, Gerlich W, Lelie N, et al. An international collaborative study to establish a World Health Organization international standard for hepatitis B virus DNA nucleic acid amplification techniques. Vox Sanguinis 2001; 80: 63–71. [DOI] [PubMed] [Google Scholar]
- 21. Roche Molecular Diagnostics. cobas® AmpliPrep/COBAS® TaqMan® HBV Test, v2.0, https://molecular.roche.com/assays/cobas-ampliprep-cobas-taqman-hbv-test-v2/ (accessed 31 May 2017).
- 22. Roche Molecular Diagnostics. cobas® TaqMan® HBV Test for use with the High Pure System, https://molecular.roche.com/assays/cobas-taqman-hbv-test-for-use-with-the-high-pure-system/ (accessed 31 May 2017).
- 23. Roche Molecular Diagnostics. cobas® HBV Quantitative nucleic acid test for use on the cobas® 4800 System, https://molecular.roche.com/assays/cobas-hbv-for-use-on-the-cobas-4800-system (accessed 31 May 2017).
- 24. Roche Molecular Diagnostics. cobas® HBV Quantitative nucleic acid test for use on the cobas® 6800/8800 Systems, https://molecular.roche.com/assays/cobas-hbv-for-use-on-the-cobas-6800-8800-system (accessed 31 May 2017).
- 25. SAS Institute, Inc. SAS System for Windows software. Cary, NC, 2014. [Google Scholar]
- 26. Food and Drug Administration. Statistical guidance on reporting results from studies evaluating diagnostic tests, https://www.fda.gov/RegulatoryInformation/Guidances/ucm071148.htm (2007, accessed 29 April 2017).
- 27. Manesis EK, Papetheodondis GV, Sevastianos V, et al. Significance of hepatitis B viremia levels determined by a quantitative polymerase chain reaction assay in patients with hepatitis B and antigen-negative chronic hepatitis B virus infection. Am J Gastroenterol 2003; 98: 2261–2267. [DOI] [PubMed] [Google Scholar]
- 28. Papatheodoridis GV, Manesis EK, Manolakopoulos S, et al. Is there a meaningful serum HBV DNA cut-off level for therapeutic decisions in HBeAg-negative chronic hepatitis B virus infection? Hepatology 2008; 48: 1451–1459. [DOI] [PubMed] [Google Scholar]
- 29. Marcellin P, Heathcote EJ, Buti M, et al. Tenofovir disoproxil fumarate versus adefovir dipivoxil for chronic hepatitis B. N Engl J Med 2008; 359: 2442–2455. [DOI] [PubMed] [Google Scholar]
- 30. Maier M, Liebert UG, Wittekind C, et al. Clinical relevance of minimal residual viremia during long-term therapy with nucleos(t)ide analogues in patients with chronic hepatitis B. PLoS One 2013; 8: e67481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim JH, Sinn DH, Kang W, et al. Low-level viremia and the increased risk of hepatocellular carcinoma in patients receiving entecavir treatment. Hepatology. Epub ahead of print 5 November 2016. [DOI] [PubMed] [Google Scholar]
- 32. Hwang JP, Lok AS. Management of patients with hepatitis B who require immunosuppressive therapy. Nat Rev Gatroenterol Hepatol 2014; 11: 209–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shouval D, Shibolet O. Immunosuppression and HBV reactivation. Semin Liver Dis 2013; 33: 167–177. [DOI] [PubMed] [Google Scholar]
- 34. Vermehren J, Maasoumy B, Maan R, et al. Applicability of hepatitis C virus RNA viral load thresholds for 8-week treatments in patients with chronic hepatitis C virus genotype 1 infection. Clin Infect Dis 2016; 62: 1228–1234. [DOI] [PubMed] [Google Scholar]
- 35. Maasoumy B, Cobb B, Brember B, et al. Detection of low HCV viraemia by repeated HCV RNA testing predicts treatment failure to triple therapy with telaprevir. Aliment Pharmacol Ther. 2014; 39: 85–92. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.