Abstract
Segmented filamentous bacterium (SFB) a gram-positive, anaerobic, and intestinal commensal organism directly influences the development of Th17 helper cells in the small intestine of mice. In NOD mice, SFB colonization interferes with the development of type 1 diabetes (T1D), a T-cell–mediated autoimmune disease, suggesting that SFB may influence Th17 cells to inhibit Th1 populations associated with the anti-β-cell immune response. This effect is a serious concern for investigators who use NOD mice for diabetes research because the expected incidence of disease decreases markedly when they are colonized by SFB. A room housing mice for T1D studies at The Jackson Laboratory was determined by fecal PCR testing to have widespread SFB colonization of multiple NOD strains after a steady decline in the incidence of T1D was noted. Rederivation of all NOD-related mouse strains was not feasible; therefore an alternative treatment using antibiotics to eliminate SFB from colonized mice was undertaken. After antibiotic treatment, soiled bedding from NOD mouse strains housed in SFB-free high-health–status production barrier rooms was used to reintroduce the gastrointestinal microbiota. Over the past 16 mo since treating the mice and disinfecting the mouse room, regular PCR testing has shown that no additional SFB colonization of mice has occurred, and the expected incidence of T1D has been reestablished in the offspring of treated mice.
Abbreviations: SFB, segmented filamentous bacterium; T1D, type 1 diabetes
In NOD mice, the principle small-animal model of type 1 diabetes (T1D), the disease is well characterized as a progressive pathology of the pancreas wherein destruction of insulin-secreting pancreatic β cells is initiated and sustained by autoreactive T-cell populations.2,27 Insulitis, the initial phase of diabetes development in young (3- to 5-wk-old) NOD mice, is characterized by a predominately CD4+ T-cell incursion into the pancreas. Importantly, however, CD8+ T cells invade islets prior to the CD4 subset and play an integral role in pancreatic β-cell destruction as insulitis progresses to overt diabetes. Additional leukocyte populations are present within the cellular infiltrates in the pancreas during insulitis and progression to overt diabetes development, including B cells that are critical to the disease process. Nonetheless, the pathogenesis is controlled, and effector mechanisms of cellular destruction are fulfilled by T cells.2,27,35
Segmented filamentous bacterium (SFB) has long been known to scientists as a resident of the murine intestinal tract. Previous histologic and microscopic research recognized the intimate contact between SFB in the ileum of mice and rats and the intestinal epithelial cells of the host.9,10 This intimate contact has several important functions. One is the nourishment of SFB through host-provided nutrients, because genomic studies of SFB have revealed that the organism lacks the genes for biosynthetic pathways that would enable the bacterium to supply its own essential cellular building blocks and nutrients.25,30 Another important function of this contact is the unique and influential association with the host's mucosal immune system,21,22 given that the preferred site of SFB attachment within the rodent ileal epithelium is the cells that overlie Peyer patches. Studies have demonstrated that this level of influence is substantial in that SFB is capable of inducing the same differentiation of the complete T-cell repertoire response in immunocompetent mice that the entire microbiota induces.14 Observations made during the histology and microscopy studies done decades ago led to a proposed SFB life cycle—a proposal largely confirmed by recent genetic studies 25,30 and in vitro cultivation.33
The proposed SFB life cycle begins with the attachment of a morphologic form called the ‘holdfast’ to epithelial cells of the ileum. Thereafter, the holdfast, which interacts directly with host cell plasma membranes, matures distally into segments, which give rise to intracellular daughter cells. Daughter cells develop into pairs of holdfasts, which are released into the local intestinal milieu to initiate new attachments to epithelial cells. Electron microscopy has demonstrated that these intracellular holdfast forms are sometimes converted into spores,7 the form likely to be responsible for transmission between rodents when shed in feces. Thus, the holdfast morphologic form can be derived directly from mature segments and autoinfect mice currently colonized.
In NOD mice, the influence of SFB on the immune response is very important in that colonization of NOD mice by SFB has been shown to generate a robust Th17 cell response coincident with protection of mice from T1D. This effect suggests a role for SFB, and the Th17 cells and IL17 that they generate, in down-regulating the pathogenic mechanism responsible for TID24 rather than enhancing the disease process. Although insulitis occurs in SFB-colonized NOD mice in this model, overt diabetes does not, indicating that at least some T-cell–mediated anti-β-cell mechanisms are intact in these mice.24,27 In addition, silencing IL17 through lentiviral-vector–mediated RNA interference had no effect on the frequency of T1D development in NOD mice, again indicating that Th17 cells and IL17 probably do not contribute to the pathogenesis of T1D.19
Research laboratories using the NOD T1D model routinely monitor their colonies for increasing glucosuria to confirm an intact disease phenotype and as an indicator of exposure of the mice to microbial agents. Immunostimulation of NOD mice by microbial contamination, particularly through infection, interferes with and can abrogate the development of T1D. This effect was first demonstrated when NOD colonies infected with murine viruses either intentionally16,29 or inadvertently38 showed substantial declines in the incidence of diabetes. Furthermore, ample evidence indicates that NOD colonies housed in high-health–status mouse facilities have a higher incidence of T1D than those housed in less sanitary conditions.31 In addition, the mouse gut microbiota has recently been shown to influence the development of T1D. For example, treatment of NOD mice with acidified water,39 antibiotics,5,17 or dietary changes13,15,28,32 alters the NOD gastrointestinal microbiome and affects the progression of T1D pathogenesis. In this regard, although we had a high index of suspicion that SFB was the primary culprit involved in the loss of the diabetic phenotype in our mice, we cannot say with complete certainty that additional microorganisms did not have a role.
In view of the influence of SFB on NOD colonies, we have documented the current case study to provide details about the use of antibiotics to remove SFB from the mouse microbiota and the importance of reintroducing microbiota by selective exposure to soiled bedding. Antibiotic treatment of the NOD research colonies eliminated the target bacterial species, thus reestablishing the incidence of diabetes in a colony of NOD mice without requiring additional mice and without incurring the financial expense and time required to rederive the mouse colony.
Materials and Methods
Mice.
Mice used in this research program were from The Jackson Laboratory Research Animal Facility, an AAALAC-approved facility with animal care programs in compliance with the Institute for Laboratory Animal Research's Guide for the Care and Use of Laboratory Animals.18 The study was reviewed and approved by the IACUC. All mice were treated humanely in this effort to eradicate SFB. All of the mice used in this study were on the NOD (nonobese diabetic) background, with the exception of a single strain on the C57Bl/6J background. Immunocompetent mice were housed in duplex microisolation cages with wire-bar lids and filter tops on static racks. Immunodeficient mice were housed in microisolation cages on individually ventilated racks. Mouse housing density varied, with nonbreeding mice group housed at 5 or 6 mice per cage depending on weight; breeding pairs and trios in duplex cages with a maximum of 3 adults and 12 pups (3 wk or younger); and 3 adults and a maximum of 20 pups (3 wk or younger) in weaning cages. Some mice were singly housed, in which case they received cotton squares (Ancare, Bellmore, NY) and Shepard Shacks (Shepard Specialty Papers, Watertown, TN) in their cages. Standard cage bedding was pine shavings; however, cages containing diabetic mice contained Bed-o-Cobs (The Andersons, Maumee, OH).The mice were free of viral antibodies and parasites as determined by routine health monitoring. Several opportunistic bacterial species were present in the mouse population (for example, Pasteurella pneumotropica, Klebsiella oxytoca). The room is presumed positive for Helicobacter spp. and Proteus mirabilis, because these organisms had been detected in the past and no efforts were made to eliminate them. In our experience, none of these organisms is associated with a decline in the incidence of T1D in NOD mouse colonies.
Antibiotic treatment.
Ampicillin (Dava Pharmaceuticals, Ft Lee, NJ) was added to acidified drinking water at a dose of 0.5 mg/mL. Drinking water for mice in this facility is treated by serial filtration to a final filtration at 1.0 μm, exposed to UV light, acidified with hydrochloric acid (pH, 2.50 to 3.0), and autoclave-sterilized before use. To improve palatability, a sweetener (2 mg/mL; Truvia, Cargill, Minneapolis, MN) was added to the water bottles. The antibiotic and sweetener were dissolved in the water by shaking the bottles. The antibiotic was administered for a total of 28 d, with freshly compounded medicated water provided weekly. Results from a pilot study that compare ampicillin with amoxicillin for efficacy against SFB were equivocal; we selected ampicillin because it was more readily available for use in water. Strict policies were enforced for handling the SFB-positive cages that housed mice undergoing antibiotic treatment. Cages were changed weekly in a ventilated change station, and each cage wiped with 1% Virkon S (Dupont, Wilmington, DE) when removed from the shelf and again when removed from the ventilated changing station. Water bottles containing freshly mixed antibiotic solution were added to cages at the weekly cage-change intervals. Mice were observed daily for signs of clinical illness resulting from antibiotic administration. Mice undergoing antibiotic treatment were handled in ventilated changing tables separately from other mice housed the room. Dedicated personnel were assigned to work with the mice undergoing antibiotic treatment. Personal protective equipment consisted of masks, gloves, lab coat, shoe covers, and bonnets.
Treatment of contaminated mouse room.
Concerns regarding residual SFB contamination of the mouse room were eliminated by disinfection of the room with vaporized hydrogen peroxide (VHP), whereas cages and cage racks were autoclave-sterilized. For treatment, the mouse room was emptied and then cleaned with Process NPD (Steris, Mentor, OH). Next, 2 vaporizers (model Z2, Bioquell, Horsham, PA) were used to distribute the hydrogen peroxide vapor throughout the room to a peak concentration of 450 ppm, with a 30-min dwell time. The room was aerated overnight. The following day, 20 biologic indicators (Steris) that were placed throughout the room during hydrogen peroxide treatment were collected. In addition, 32 environmental swabs were obtained and placed in trypticase soy broth; all collected materials were incubated and then evaluated to assess room disinfection. Research instruments and materials that had been in the mouse room were disinfected with a liquid sterilant (SporKlenz, Steris) or removed. Each of these steps—combined with the institution of rigorous husbandry and mouse handling practices including the use of ventilated changing tables, gloves decontaminated with 70% ethanol after handling each cage, masks, bonnets, daily change of lab coats, and designated mouse room entrance and exit policies for research and husbandry personnel—were integral to the establishment and maintenance of SFB-free mouse colonies.
PCR analysis for SFB.
Mouse fecal samples were collected from the floors of cages housing each treatment and control group. Samples were stored at −80 °C prior to DNA preparation. Total genomic DNA was extracted by using QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. DNA was eluted with 100 μL DNase–RNase-free water and further diluted to a final nominal concentration of 100 ng/μL as quantified by using a microplate spectrophotometry system (BioTek, Winooski, VT).
The 16S rRNA gene (110 bp) of SFB was amplified by using forward and reverse primers (SFB 726F, 5′ GAC GCT GAG GCA TGA GAG CA 3′; SFB 844R, 5′ GAC GGC ACG GAT TGT TAT TCA 3′).36 Standard SYBR green PCR analysis was performed (CFX96 Touch Real-Time PCR Detection System, BioRad, Hercules, CA; QuantiFast SYBR Green PCR Kit, Qiagen). The cycling parameters were 95 °C for 5 min, followed by 35 cycles of 95 °C for 10 s and 60 °C for 30 s. A melting-curve analysis was included at the end of the 35 cycles, and the quantification cycle was calculated automatically by using instrument software.
Pyrosequencing.
16S rRNA library preparation and pyrosequencing were conducted by MR DNA Lab (Shallowater, TX). A 100-ng aliquot of each sample DNA was used for a 50-μL PCR reaction. Barcoded primers 530F (5′ GTG CCA GCM GCN GCG G 3′) and 1100R (5′ GGG TTN CGN TCG TTG 3′) were used to amplify the 600-bp sequence in variable region V4–V6 of the 16S rRNA genes. HotStar Taq Plus Master Mix Kit (Qiagen) was used for PCR amplification under the following conditions: 94 °C for 3 min followed by 32 cycles of 94 °C for 30 s, 60 °C for 40 s, and 72 °C for 1 min, with a final elongation step at 72 °C for 5 min. Amplicons were purified by using Ampure beads (Agencourt Bioscience, Beverly, MA) to remove smaller fragments. Purified DNA was further measured and combined in equal concentrations. The DNA pool was pyrosequenced (454 FLX Titanium instruments and reagents, Roche Applied Science, Penzburg, Germany) according to the manufacturer's guidelines. Two mock community samples representing genomic DNA from 67 bacterial isolates pooled at even concentrations were processed in parallel with fecal samples to calculate the technical variation due to errors and biases during PCR amplification and sequencing. All sequence metadata are publicly available at the NCBI sequence read archive under bioproject no. PRJNA328251.
Microbiome data analysis.
Sequence data were processed by using a MR DNA Lab (http://mrdnalab.com/) proprietary analysis pipeline. In summary, sequences were depleted of barcodes and primers, and sequences shorter than 150 bp, containing ambiguous base calls, or with homopolymer runs longer than 6 bp were removed. Sequences were denoised and chimeras removed. An average of 3000 sequences per sample was selected according to the highest mean quality score and number of operational taxonomic units generated. Operational taxonomic units were defined by clustering at 97% similarity by using the R-package (http://www.rstudio.org) program pyclust with a bootstrap resampling value of 1000, taxonomically classified by using BLASTn1 analysis against a curated GreenGenes database,11 and compiled to the most relevant taxonomic level according to the percentage identity. Microbial richness, representative taxa, and statistical analyses were performed by using the Quantitative Insights into Microbial Ecology (QIIME) package.6
Bedding transfer.
Soiled bedding taken from NOD mouse colonies housed in maximal barrier rooms in the Jackson Laboratory production facilities (that is, all supplies autoclaved, autoclave-sterilized acidified water, and personnel in sterilized booties, lab coat, gloves, and powered air-purifying respirators, and clean-room–processed scrubs) was added to the cages of each group of mice except the negative controls. Soiled bedding (1.5 cups per cage) was added directly into the normal amount of clean bedding in the cage. Mice were exposed for 2 wk, and soiled bedding was replenished once during the weekly cage change. The maximal barrier rooms from which the soiled bedding was taken are free of opportunistic and pathogenic microorganisms.
Fecal sampling for SFB PCR analysis.
Prior to sampling for this case study, pilot studies confirmed that shed fecal pellets in the bedding of mouse cages were suitable target samples for SFB detection. We chose this strategy rather than collection of fresh fecal samples directly from mice because SFB is a spore-forming anaerobe, and spores were determined as likely to be stable within shed fecal material.
Assessment of T1D development.
T1D status was assessed by weekly monitoring of glycosuria (Ames Diastix, Bayer, Diagnostics Division, Elkhart, IN), with disease onset defined by 2 consecutive readings of 3 or greater.
Background History of Colony
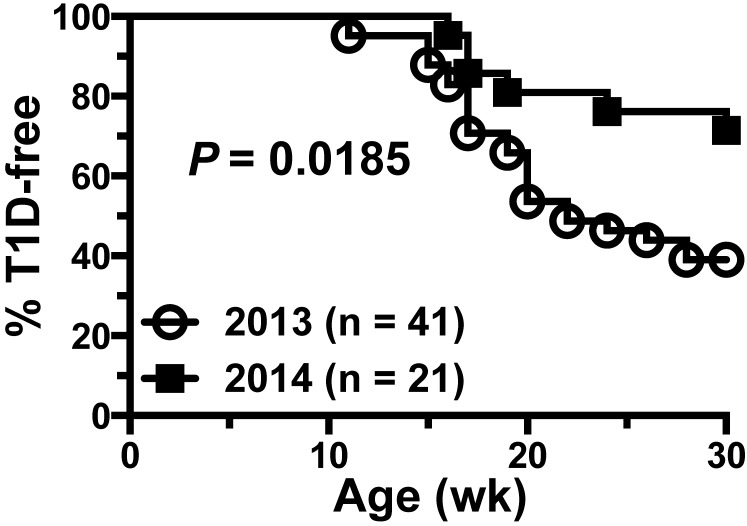
The acquisition of mice from an SFB-positive room occurred between 1 and 2 y prior to this investigation. Mice were sent from an SFB-positive mouse room in the Research Animal Facility to an animal room in the same facility housing approximately 55 mouse strains on the NOD background that were used in T1D research. The SFB-positive mouse room from which the mice originated housed mice for autoimmune disease research other than T1D and for which SFB is essential for initiation of the Th17-mediated immune responses under study. Laboratory personnel from the diabetes research group noted a decline in the incidence of disease in groups of standard NOD/ShiLtDvs mice serving as putative disease-susceptible controls in ongoing experiments (Figure 1). This discovery initiated an investigation of potential causes for the change. Quarterly routine health monitoring of this mouse room during that same period revealed no changes to the organisms normally found within the mice housed therein. The principle investigator of the T1D research group requested that The Jackson Laboratory Diagnostic Laboratory test some mice for the presence of SFB in light of a recent publication24 that demonstrated an association between SFB and protection from diabetes in female NOD mice. The current report describes the steps taken to identify and eliminate SFB from mouse colonies within the affected mouse room, repopulation of the microbiota, and the subsequent management of the room after antibiotic treatment of the mice and disinfection of the room.
Figure 1.
Historical diabetes data for mice in the SFB-contaminated room. NOD/ShiLtDvs mice that serve as putative disease-susceptible groups in ongoing experiments received weekly intraperitoneal injections of an IgG1 isotype-control antibody prior to SFB (2013, n = 41) and during SFB contamination (2014, n = 21). T1D incidence curves show a significant decrease (P = 0.0185, Mantel–Cox log-rank test) in incidence from approximately 60% to 30% by 30 wk of age. These mice were positive controls for ongoing antibody-based therapy studies and received an irrelevant IgG1 isotype control antibody that does not alter T1D development.
At the request of the principal investigator, initial SFB PCR testing of mice imported from the SFB-positive mouse room as well as a survey of the entire T1D research mouse room were undertaken by the in-house diagnostic laboratory. Fecal samples from cages of an initial cohort of tested mice showed a 49% positive rate for SFB, whereas as subsequent survey of all boxes showed a 71% SFB-positive frequency, and there was no specific pattern of positive cages within the room (Table 1). Discussions between the veterinary staff and the T1D research group verified that contamination of the research mice with SFB was severely affecting the T1D research program and that a strategy was required to deal with this issue.
Table 1.
Fecal PCR testing results before and after antibiotic treatment for SFB
| Test results |
||||
| No. of cages tested | Date tested | No. of cages positive | No. of cages negative | % of cages positive |
| Before treatment | ||||
| 214b | 11 Sep 2014 | 105 | 109 | 49 |
| 554a | 22 Sep 2014 | 394 | 160 | 71 |
| After treatment | ||||
| 675a | 30 Dec 2014 | 0 | 675 | 0 |
| 127c | 2 Feb 2015 | 0 | 127 | 0 |
| 788a | 12 Apr 2016 | 0 | 788 | 0 |
Fecal samples were collected from the cage bottom and their DNA tested for SFB by PCR analysis.
Includes all cages in the room.
Includes mouse strains originally obtained from an SFB-positive room.
Includes samples acquired randomly from 18% of cages in room.
Eradication Effort
The chosen strategy was to eliminate this organism from the mouse population through antibiotic treatment rather than rederivation of all contaminated mouse strains. The proposed strategy was to 1) reduce the size of contaminated colonies to fewer breeding pairs by euthanizing SFB-positive mice, 2) treat the remaining mice with antibiotics to eliminate SFB, 3) establish the gastrointestinal flora of antibiotic-treated mice with fecal material from SFB-free mice of the same genetic background from high health-status barriers in the production facilities, 4) decontaminate the mouse room and equipment, and 5) revise policies for entering, exiting, and working within the mouse room.
After population reduction in the research mouse colonies, remaining mice were transferred to a mouse room maintained as SFB-positive for antibiotic treatment and to enable decontamination of their room of origin. This action was based on the need to keep these mice out of SFB-negative rooms.
As part of the strategy to determine the effectiveness of antibiotics in eliminating SFB and to examine the effect of antibiotic treatment on the mouse microbiota, most (but not all) potentially infected mouse cages were treated with antibiotics and exposed to soiled bedding from SFB-negative mice. Some cages of SFB positive mice were not given antibiotics or soiled bedding to serve as controls for antibiotic efficacy. In addition, a few cages of antibiotic-treated mice were not exposed to soiled bedding, to assess the changes in gut microflora resulting from treatment.
Colony Evaluation after Eradication
After 4 consecutive weeks of antibiotic administration, mice were rested for 1 wk to allow for a decline in residual tissue antibiotic concentrations. The mice then were changed into clean cages under ventilated changing stations in preparation for transport back to their room of origin. Just prior to removal from the room in which the mice were treated, each cage was wiped down with 1% Virkon S (Dupont), the cages were placed onto a transport rack in the adjacent hallway, and the racks were covered with a plastic hood. The racks were immediately wheeled to the hallway outside their room of origin, which had been decontaminated with vaporized hydrogen peroxide and wiped down with SporKlenz (Steris). Cages were wiped down again with Virkon S as they were placed onto racks within the recently decontaminated mouse room. After the mice had adapted to their room of origin for one week fecal samples were taken from every mouse cage in the room. PCR testing of the samples revealed that all the cages were devoid of SFB (Table 1, 30 Dec 2014 test date). Note that more cages were sampled after culling and antibiotic treatment than at the outset of the investigation, due to the previously described redistribution of breeding pairs after mice were culled to reduce the population of contaminated animals.
After SFB PCR testing, 2 rounds of 1-wk exposure to soiled bedding from SFB-negative mice were implemented to provide gastrointestinal microbiota to all except a few cages of antibiotic-treated mice. All cages were changed to fresh bedding and water bottles after the second round of exposure. One week after the final exposure to soiled bedding, fecal samples were taken from at least one cage from each mouse strain. Fecal pellets were used for SFB PCR analysis and for sequencing of the gastrointestinal microbiome of the mice. All sampled cages were negative for SFB (Table 1, 2 Feb 2015 sampling date). A follow-up assessment of the SFB status of the mice was performed 14 mo later (Table 1, 12 Apr 2016 sampling date), with all cages in the room still PCR-negative for SFB.
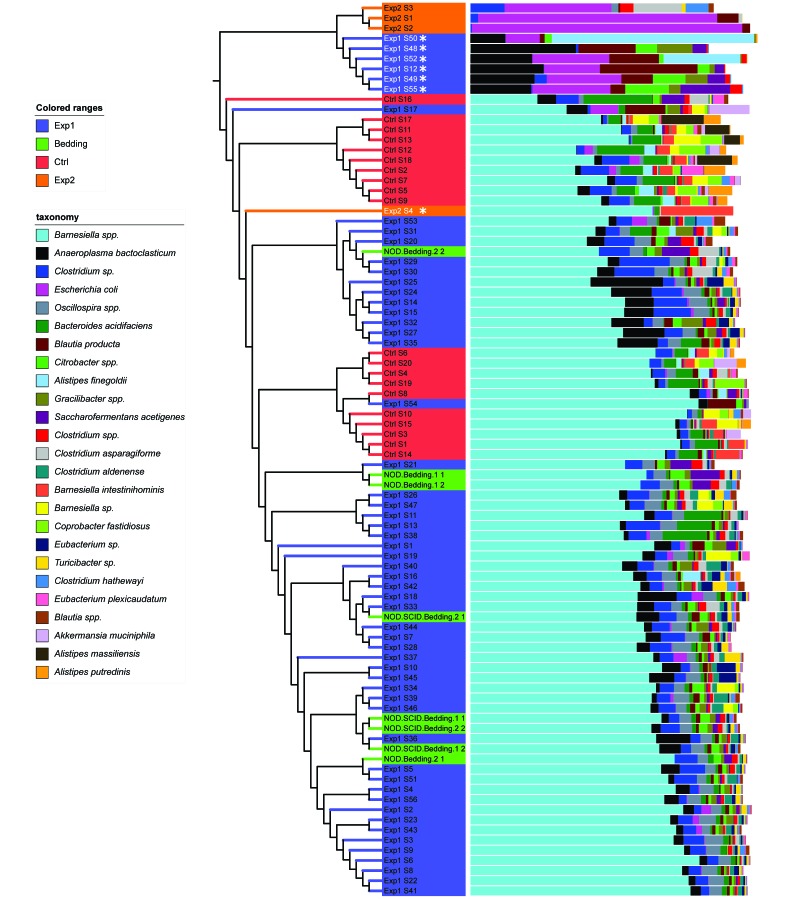
Overall microbiome data, presented in a multivalued bar chart associated with a phylogenetic tree (Figure 2), demonstrate that samples formed 2 distinct clusters: one including all samples from control, bedding-donor, and antibiotic-treated mice exposed to SFB-free soiled bedding (except 6 samples), with Barnesiella being dominant, and the other including all but one sample from antibiotic-treated mice that were not exposed to soiled bedding, with Escherichia being dominant.
Figure 2.
Hierarchical clustering and microbial community composition. Samples were clustered by using the pvclust program, with Euclidean distances based on summarized tables of operational taxonomic units. Microbial community composition is displayed as relative abundance at the genus level. Except for samples Exp1 S12, 48, 49, 50, 52, and 55 (font color white marked with an asterisk), all samples from antibiotic-treated and microbiome-reconstituted mice (Exp1 group) are clustered together with those from control (Ctrl) and SFB-negative bedding donors. Except for sample Exp2 S4, all samples from antibiotic-treated but nonreconstituted mice (Exp2 group) are in a single cluster.
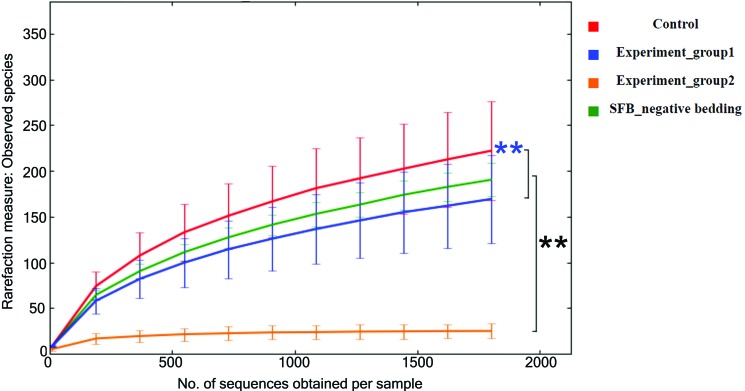
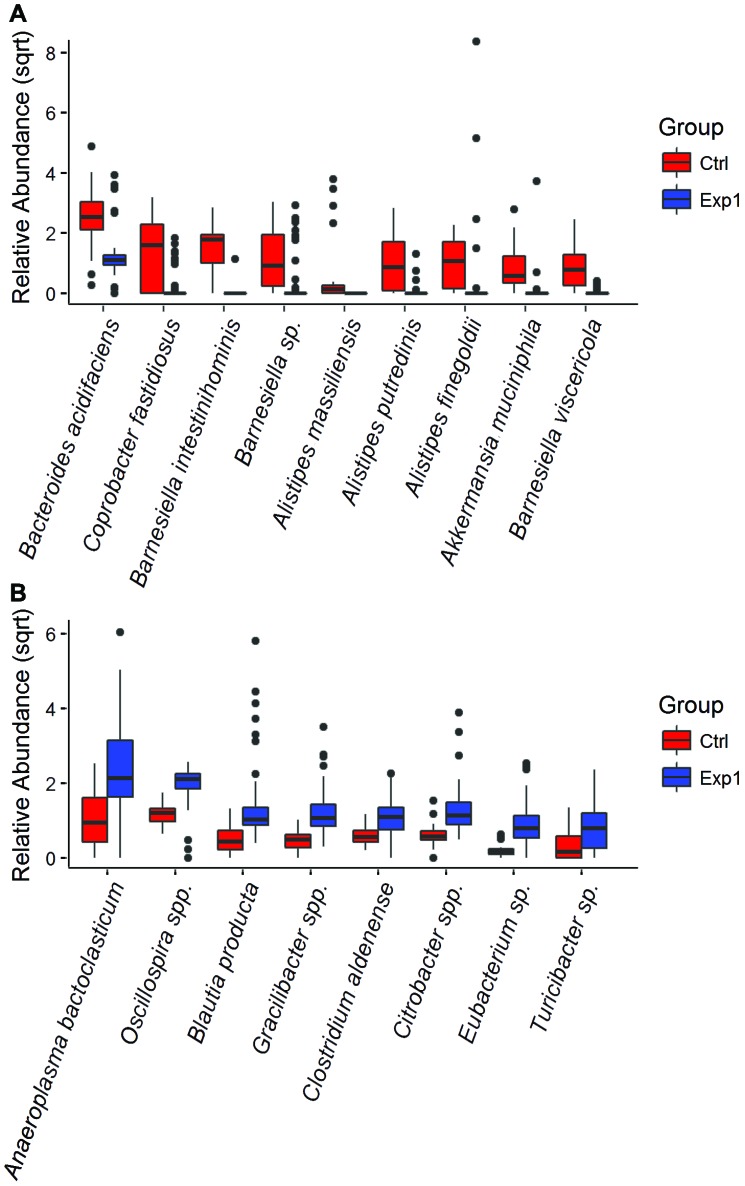
Rarefaction curves (Figure 3) are close to plateauing, indicating that a sequencing depth of 1804 reads per sample is sufficient to estimate the microbial richness present in each experimental group. Similar numbers of species were recovered from antibiotic- treated mice exposed to SFB-free soiled bedding and SFB-negative, nonantibiotic-treated donors of the soiled bedding, whereas antibiotic-treated mice that were not exposed to soiled bedding had significantly (P < 0.05) decreased microbial richness than both other groups. The results of both the clustering and rarefaction analyses suggest that antibiotics markedly decreased the species richness of the gut microbiota and that SFB-free soiled bedding can be used as a reliable source to repopulate the gastrointestinal microbiota after antibiotic treatment. Microbial diversity was greater (P < 0.05) in the control group than in mice with reconstituted microbiotas, most likely due to the 6 mice in which soiled bedding failed to restore the microbiome (Figure 2). In addition, 10 representative taxa differed significantly (P < 0.05) between control and reconstituted groups (Figure 4), with 4 of these taxa having higher abundance in control mice and the other 6 being higher in reconstituted mice.
Figure 3.
Comparison of microbial richness of gut microbiota. The number (mean ± 1 SD) of microbial species present was estimated by using collated α diversity values at rarefaction depth of 1804 (the least number of sequences obtained among all samples). The antibiotic-treated but soiled-bedding–unexposed (experiment 2) group has significantly (black asterisks, P < 0.05) lower microbial richness than the other 3 groups. The control group has significantly greater (blue asterisks, P < 0.05) species richness than antibiotic-treated reconstituted mice (experiment 1 group; blue asterisks). Statistical analysis was performed within QIIME by using nonparametric 2-sample t tests through 999 Monte Carlo permutations.
Figure 4.
Boxplots of relative abundance of representative genera in control mice (Ctrl) and antibiotic-treated microbiota-reconstituted mice (Exp1). Only the taxa with an abundance greater than 1% are plotted, with P < 0.05. (A) Taxa with higher abundance in control mice. (B) Taxa with higher abundance in the Exp1 group. Statistical tests were performed within QIIME by using nonparametric ANOVA with Bonferroni correction for 999 permutations.
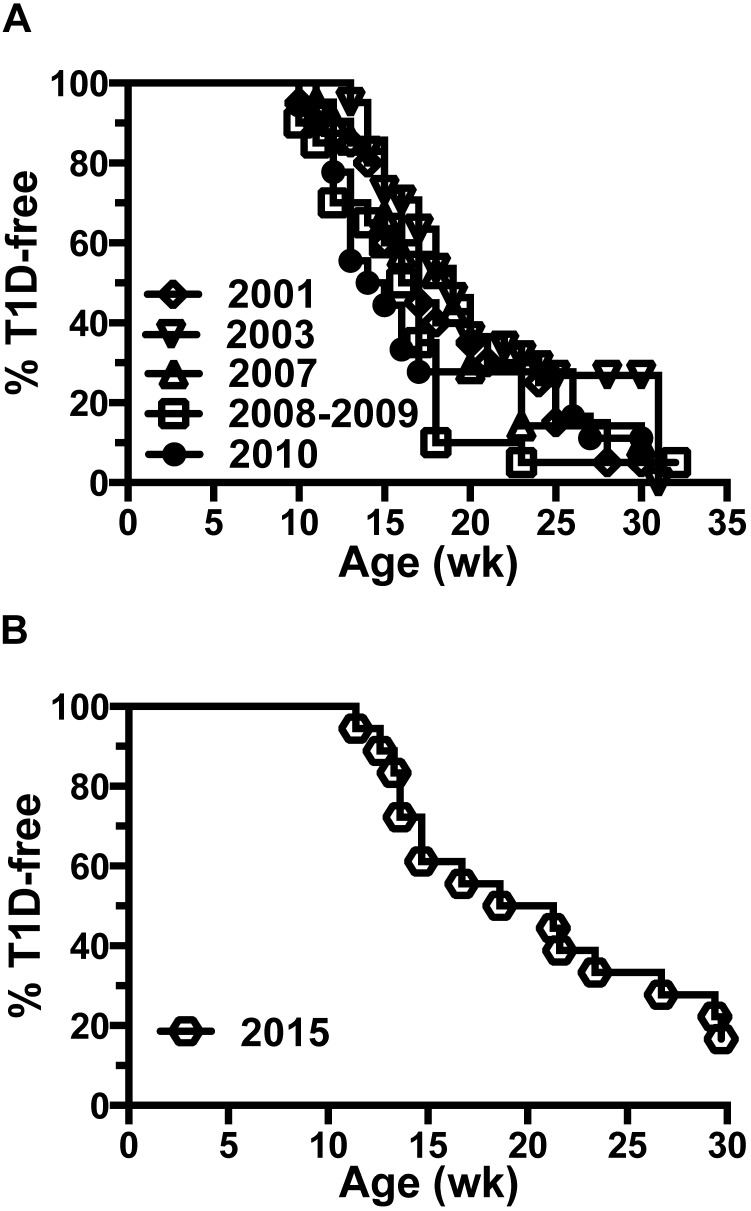
Historical T1D incidence curves for the NOD colony prior to SFB introduction (Figure 5 A) and afterward (Figure 5 B) show a return to similar incidence by 30 wk of age in reconstituted mice.
Figure 5.
Return to normal T1D incidence after SFB elimination and microbiome reconstitution. (A) Incidence curves for T1D development in female NOD/ShiLtDvs mice in 2001 (n = 20), 2003 (n = 43), 2007 (n = 21), 2008–2009 (n = 20), and 2010 (n = 18) showing approximately 75% to 95% incidence by 30 wk of age. (B) After eliminating SFB from the NOD colony in 2015, T1D development in female mice returned to an incidence (approximately 85% by 30 wk of age; n = 18) consistent with that historically observed.
Discussion
The importance of this case study is the demonstration that antibiotic treatment followed by reconstitution of the gastrointestinal microbiota offers an option other than rederivation to eliminate SFB from contaminated NOD mice and induce a return to the expected T1D phenotype in offspring of treated mice. Antibiotic use in mice must be carefully considered with regard to the variable effects on microbiota and, by extension, the reliability of the mouse phenotype and health. For this reason, we are currently examining other options for elimination of SFB from mice by using shorter and different routes of exposure to spare the elimination of gut microbiota to the greatest extent possible. Before the DNA-sequencing era, studies of antibiotic effects on the gastrointestinal microflora showed that roxithromycin and erythromycin eliminated SFB from mice and, according to standard tests at the time, had little effect on the microbial ecology of the mouse gut.23
Two recent studies using next-generation sequencing methods to assess the integrity of the intestinal microbiota have yielded discordant results after antibiotic treatment of NOD mice. The first study5 showed significantly increased T1D incidence in male mice, whereas the female NOD mice cohort demonstrated a robust T1D incidence regardless of antibiotic treatment. In the cited study,5 2 different treatment groups received different antibiotic regimens: vancomycin, which primarily affects gram-positive bacteria, and a mixture of streptomycin, colistin, and ampicillin, which targets both gram-positive and gram-negative bacteria. Both treatment groups showed substantial effects regarding the number of species and species composition of the gut microbiota. The second study,17 which used a different antibiotic mixture (neomycin, polymyxin B, and streptomycin) to specifically target gram-negative bacteria in NOD mice, demonstrated protection from T1D rather than an increased disease incidence. In both studies, the authors pointed out the critical importance of the timing of antibiotic administration, suggesting that prenatal administration is the most effective treatment time, given that the effect on the intestinal microbiota and subsequently T1D, is the most evident then.
A third antibiotic study just published26 also used early administration of antibiotics to mice to mimic human exposure starting in early childhood and the influence of this treatment on T1D development. Two antibiotic regimens were used: penicillin V at a subtherapeutic dose starting in late pregnancy and throughout the life of the offspring, and tylosin at the therapeutic dose level for mice and pulsed over 3 doses after birth, thereby recapitulating antibiotic use in young children. Similar to the previous studies,5,17 the pulsed therapeutic dose of antibiotics in the recent publication23 markedly affected the microbiome of the mice and initiated a concomitant increase in the incidence of T1D. However, the subtherapeutic dose was insufficient to induce a noteworthy microbiome or immunologic effect. The antibiotics used in each of the 3 cited studies5,15,23 are all likely to have impaired the viability of SFB, but none specifically identified SFB as having a role in the outcome of their experiments. It is important to note that each of the studies described the influence of antibiotics on the gut microbiota and, consequently, on the immune response. However, the precise immunologic mechanisms affected by changes to the microbiota and the means by which they influenced the development of T1D have not been fully determined.
The different antibiotics used in the previous and current studies likely was the primary cause of the divergent outcomes on the microbiota and on T1D development, yet other factors—including differences in microbiota and mouse health at each institution—were equally likely to contribute. In addition, studies in rat models of T1D have demonstrated that antibiotic administration to diabetes-prone rats directly influences disease incidence.4,34 Furthermore, the administration of acidified water affects the microbiota and influences T1D incidence of NOD mice,39 as do modified diets, in which the absence of wheat and barley significantly delays the onset of diabetes in this strain.28,32
As described earlier, the incidence of T1D in NOD colonies is variable and dependent on changes in gut microbiota. In this regard, we specifically addressed the issue of antibiotic-induced microbiota changes in the NOD mice by reconstituting the microbiota of treated mice through exposure to fecal microbiota from similar mouse strains on the NOD background, by using bedding from high health-status mouse colonies. Ideally, we would have matched the microbiota from each of the mouse strains in the SFB-contaminated colonies, but doing so was not logistically possible. The effectiveness of this reconstitution in restoring T1D was evident by the return of the T1D incidence to preSFB levels in offspring of antibiotic- and bedding-treated mice (Figure 5 B). We were unable to save antibiotic-treated mice that were not reconstituted with SFB-negative gut microbiota as a comparison control. The reestablishment of normal T1D prevalence in the treated mouse colonies also suggests that the identical gut microbiota found in NOD colonies contaminated by SFB was not required to reconstitute the immunologic capacity to induce autoimmune diabetes, as demonstrated by the microbial diversity and community structure (Figures 2 and 3).
We have proposed that SFB was the primary source of T1D prevention in the NOD colonies used by the research group in this case. However, other organisms might have been involved in the observed reduction in T1D incidence. For example, several other organisms have been identified as single-species inhibitors of T1D in animal models, including Lactobacillus johnsonii in BioBreeding rats37 and Bacillus cereus20 in NOD mice. Other currently unidentified microbial species may also have this effect on T1D development in animal models. Nonetheless, it is important to note that the NOD mouse colonies in our current study are long-term residents of the room in which they were housed, are free of Lactobacillus johnsonii and Bacillus cereus as determined by regular health monitoring, and did not suffer a loss in the incidence of T1D until after SFB-positive mice were brought into the mouse room. In addition, the principal investigator states that this specific colony of NOD mice has one of the highest incidence rates of T1D in the world. Therefore, although other species of bacteria potentially contributed to the loss of T1D development in these mouse colonies, it remains highly likely that SFB is the primary culprit. In this context, note that the mouse colonies described in this manuscript are free of trichomonads, which have recently been implicated to have a direct effect on immune homeostatis in the gut of mice, including an influence on Th17 cell immunity.8,12
References
- 1.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson MS, Bluestone JA. 2005. The NOD mouse: a model of immune dysregulation. Annu Rev Immunol 23:447–485. [DOI] [PubMed] [Google Scholar]
- 3.Bach JF. 2002. The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med 347:911–920. [DOI] [PubMed] [Google Scholar]
- 4.Brugman S, Klatter FA, Visser JTJ, Wildeboer-Veloo ACM, Harmsen HJM, Rozing J, Bos NA. 2006. Antibiotic treatment partially protects against type 1diabetes in the Bio-Breeding diabetes-prone rat. Is the gut flora involved in the development of type 1 diabetes? Diabetologia 49:2105–2108. [DOI] [PubMed] [Google Scholar]
- 5.Candon S, Perez-Arroyo A, Marquet C, Valette F, Foray AP, Pelletier B, Milani C, Ventura M, Bach JF, Chatenoud L. 2015. Antibiotics in early life alter the gut microbiome and increase disease incidence in a spontaneous mouse model of autoimmune insulin-dependent diabetes. PLoS One 10:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chase DG, Erlandsen SL. 1976. Evidence for a complex life cycle and endspore formation in the attached, segmented filamentous bacterium from murine ileum. J Bacteriol 127:572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chudnovskiy A, Mortha A, Kana V, Kennard A, Ramirez JD, Rahman A, Romain Remark R, Mogno I, Ng R, Gnjatic S, Amir ED, Solovyov A, Greenbaum B, Clemente J, Faith J, Belkaid Y, Grigg ME, Merad M. 2016. Host–protozoan interactions protect from mucosal infections through activation of the inflammasome. Cell 167:444–456.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis CP, Savage DC. 1974. Habitat, succession, attachment, and morphology of segmented filamentous microbes indigenous to the murine gastrointestinal tract. Infect Immun 10:948–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis CP, Savage DC. 1976. Effect of penicillin on the succession, attachment, and morphology of segmented filamentous microbes in the murine small bowel. Infect Immun 13:180–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeSantis TZ, Jr, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM, Phan R, Andersen GL. 2006. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res 34 Suppl 2:W394–W399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Escalante NK, Lemire P, Cruz Tleugabulova MC, Prescott D, Mortha A, Streutker CJ, Girardin SE, Philpott DJ, Mallevaey T. 2016. The common mouse protozoa Tritrichomonas muris alters mucosal T cell homeostasis and colitis susceptibility. J Exp Med 13:2841–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Funda DP, Kaas A, Tlaskalova-Hogenova H, Buschard K. 2007. Gluten-free but also gluten-enriched (gluten+) diet prevent diabetes in NOD mice; the gluten enigma in type 1 diabetes. Diabetes Metab Res Rev 24:59–63. [DOI] [PubMed] [Google Scholar]
- 14.Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, Eberl G, Snel J, Kelly D, Cerf-Bensussan N. 2009. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31:677–689. [DOI] [PubMed] [Google Scholar]
- 15.Hansen AK, Ling F, Kaas A, Funda DP, Farlov H, Buschard K. 2006. Diabetes preventive gluten-free diet decreases the number of caecal bacteria in nonobese diabetic mice. Diabetes Metab Res Rev 22:220–225. [DOI] [PubMed] [Google Scholar]
- 16.Hermitte L, Vialettes B, Naquet P, Atlan C, Payan MJ, Vague P. 1990. Paradoxical lessening of autoimmune processes in nonobese diabetic mice after infection with the diabetogenic variant of encephalomyocarditis virus. Eur J Immunol 20:1297–1303. [DOI] [PubMed] [Google Scholar]
- 17.Hu Y, Peng J, Tai N, Hu C, Zhang X, Wong SF, Wen L. 2015. Maternal antibiotic treatment protects offspring from diabetes development in nonobese diabetic mice by generation of tolerogenic APCs. J Immunol 195:4176–4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Institute for Laboratory Animal Research 2011. Guide for the care and use of laboratory animals, 8th ed. Washington (DC): National Academy Press. [Google Scholar]
- 19.Joseph J, Bittner S, Kaiser FMP, Wiendl H, Kissler S. 2011. IL17 silencing does not protect nonobese diabetic mice from autoimmune diabetes. J Immunol 188:216–221. [DOI] [PubMed] [Google Scholar]
- 20.King C, Sarvetnick N. 2011. The incidence of type 1 diabetes in NOD mice is modulated by restricted flora not germ-free conditions. PLoS One 6:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klaasen HL, Kooman JP, Poelma FG, Beyen AC. 1992. Intestinal segmented filamentous bacteria. FEMS Microbiol Rev 8:165–180. [DOI] [PubMed] [Google Scholar]
- 22.Klaasen HL, Van der Heijden PJ, Stok W, Poelma FG, Koopman JP, Van den Brink ME, Bakker MH, Eling WM, Beyen AC. 1993. Apathogenic, intestinal, segmented bacteria stimulate the mucosal immune system of mice. Infect Immun 61:303–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koopman JP, van den Brink ME, Scholten PM, Hectors MPC, Nagengast FM. 1987. Influence of the antibiotics roxithromycin and erythromycin on the gastrointestinal ecology of mice. Z Versuchstierkd 30:79–83. [PubMed] [Google Scholar]
- 24.Kriegel MA, Sefik E, Hill JA, Wu HJ, Benoist C, Mathis D. 2011. Naturally transmitted segmented filamentous bacterial segregate with diabetes protection in nonobese diabetic mice. Proc Natl Acad Sci USA 108:11548–11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuwahara T, Ogura Y, Oshima K, Kurokawa K, Ooka T, Hirakawa H, Itoh T, Nakayama-Imaohji H, Ichimura M, Itoh K, Ishifune C, Maekawa Y, Yasutomo K, Hattori M, Hayashi T. 2011. The lifestyle of the segmented filamentous bacterium: a nonculturable gut-associated immunostimulating microbe inferred by whole-genome sequencing. DNA Res 18:291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livanos AE, Greiner TU, Vangay P, Pathmasiri W, Stewart D, McRitchie S, Li H, Chung J, Sohn J, Kim S, Gao Z, Barber C, Kim J, Ng S, Rogers AB, Sumner S, Zhang X-S, Cadwell K, Knights D, Alekseyenko A, Bäckhed F, Blaser MJ, [Internet] 2016. Antibiotic-mediated gut microbiome perturbation accelerates development of type 1 diabetes in mice. Nature Microbiol [Cited 30 August 2016]. Available at: https://www.nature.com/articles/nmicrobiol2016140 [DOI] [PMC free article] [PubMed]
- 27.Mathis D, Benoist C. 2012. The influence of the microbiota on type 1 diabetes: on the threshold of a leap forward in our understanding. Immunol Rev 245:239–249. [DOI] [PubMed] [Google Scholar]
- 28.Maurano F, Mazzarella G, Luongo D, Stephanile R, D'Arienzo R, Rossi M, Auricchio S, Troncone R. 2005. Small intestinal enteropathy in nonobese diabetic mice fed a diet containing wheat. Diabetologia 48:931–937. [DOI] [PubMed] [Google Scholar]
- 29.Oldstone MB. 1988. Prevention of type I diabetes in nonobese diabetic mice by virus infection. Science 239:500–502. [DOI] [PubMed] [Google Scholar]
- 30.Pamp SJ, Harrington ED, Quake SR, Relman DA, Blainey PC. 2012. Single-cell sequencing provides clues about the host interactions of segmented filamentous bacteria (SFB). Genome Res 22:1107–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pozzilli P, Signore A, Williams AJ, Beale PE. 1993. NOD mouse colonies around the world—recent facts and figures. Immunol Today 14:193–196. [DOI] [PubMed] [Google Scholar]
- 32.Schmid S, Koczwara K, Schwinghammer S, Lampasona V, Ziegler AG, Bonafacio E. 2004. Delayed exposure to wheat and barley proteins reduces diabetes incidence in nonobese diabetic mice. Clin Immunol 111:108–118. [DOI] [PubMed] [Google Scholar]
- 33.Schnupf P, Gaboriau-Routhiau V, Gros M, Friedman R, Moya-Nilges M, Nigro G, Cerf-Bensussan N, Sansonetti PJ. 2015. Growth and host interaction of mouse segmented filamentous bacteria in vitro. Nature 520:99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwartz RF, Neu J, Schatz D, Atkinson MA, Wasserfall D. 2007. Comment on: Brugman S. et al. (2006) Antibiotic treatment partially protects against type 1 diabetes in the Bio-Breeding diabetes-prone rat. Is the gut flora involved in the development of type 1 diabetes? Diabetelogia 49:2105–2108. Diabetologia 50:220–221. [DOI] [PubMed] [Google Scholar]
- 35.Serreze DV, Leiter EH. 2001. Genes and cellular requirements for autoimmune diabetes susceptibility in nonobese diabetic mice. p 31–67. In: von Herrath MG, editor. Molecular pathology of type 1 diabetes mellitus. Basel (Switzerland): Karger. [DOI] [PubMed] [Google Scholar]
- 36.Snel J, Heinen PP, Blok HJ, Carman RJ, Duncan AJ, Allen PC, Collins MD. 1995. Comparison of 16S rRNA sequences of segmented filamentous bacteria isolated from mice, rats, and chickens and proposal of “Candidatus arthromitus.” Int J Syst Bacteriol 45:780–782. [DOI] [PubMed] [Google Scholar]
- 37.Valladares R, Sankar D, Li N, Williams E, Lai K-K, Abdelgeliel AS, Gonzalez CF, Wasserfall CH, Larkin J, 3rd, Schatz D, Atkinson MA, Triplett EW, Neu J, Lorca GL. 2010. Lactobacillus johnsonii N6.2 mitigates the development of type 1 diabetes in BB-DP rats. PLoS One 5:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilberz S, Partke HJ, Dagnaes-Hansen F, Herberg L. 1991. Persistent MHV (mouse hepatitis virus) infection reduces the incidence of diabetes mellitus in nonobese diabetic mice. Diabetologia 34:2–5. [DOI] [PubMed] [Google Scholar]
- 39.Wolf KJ, Daft JG, Tanner SM, Hartmann R, Kafipour E, Lorenz R. 2014. Consumption and acidic water alters the gut microbiome and decreases the risk of diabetes in NOD mice. J Histochem Cytochem 62:237–250. [DOI] [PMC free article] [PubMed] [Google Scholar]