Abstract
Mycobacterial infections are of primary health concern in NHP colonies in biomedical research. NHP are constantly monitored and screened for Mycobacterium spp. We report 6 Chinese-origin rhesus macaques infected with Mycobacterium kansasii that exhibited positive tuberculin skin tests in the absence of disease. Two of these macaques were being used for research purposes; the remaining 4 macaques were residing at the contract quarantine company. Histopathology and acid-fast staining of fixed tissues from all macaques showed that all were free of disease. Thoracic radiographs were negative for any signs of disease or infection. Samples from bronchial lavage and tissues including lung, spleen, hilar and mesenteric lymph nodes tested negative by PCR assay for Mycobacterium spp. One of the research macaques tested culture-positive for M. kansasii and a poorly characterized M. avium complex organism. One macaque from the contract quarantine facility tested culture positive for M. kansasii. Genomic testing and target gene RNA expression analysis of the 2 M. kansasii isolates were performed to evaluate possible kinship and affected genes that might contribute to susceptibility to mycobacterial infection. Genotyping of the 2 isolates revealed 2 genetically distinct strains (strains 1 and 4). The presence of positive tuberculin skin tests in the absence of disease raises serious concerns regarding diagnostic methods used for infected NHP.
Abbreviations: MOT, mammalian old tuberculin; TST, tuberculin skin test
Mycobacterial infections among NHP is of significant concern, because they can result in high rates of morbidity and mortality.4,14,35 Mycobacterium kansasii infection is one of the most common causes of mycobacterial disease in people36,45 and produces tuberculosis-like disease that is clinically indistinguishable from that of M. tuberculosis in both humans and NHP.12 According to microbiologic criteria, M. kansasii is classified as nontuberculosis Mycobacterium on the basis of in vitro colony appearance, morphology, rate of growth, and color.2
NHP colonies used in biomedical research are carefully screened through intradermal tuberculin tests (TST) using mammalian old tuberculin (MOT). TST have been the mainstay of Mycobacterium spp. screening and antemortem diagnosis in NHP since the 1940s and are currently the only ILAR–CDC-approved method for mycobacterial testing of animals in primary import quarantine.16,20,34 Here we report 6 TST-positive, infected rhesus macaques that did not exhibit any signs of clinical disease. Biologic specimens were collected from these 6 NHP, and Mycobacterium kansasii was cultured from one research macaque and from another at the contract quarantine company. To our knowledge, this clinical report is the first description of TST-positive NHP yielding Mycobacterium-positive cultures in the absence of disease. The infected macaques showed no evidence of disease according to clinical assessment, radiography, gross pathology and histopathology.
Mycobacterial diseases in NHP not only interfere with animal health and research but pose zoonotic health risks to all who have contact with the infected animals. Therefore, positive TST results are taken seriously by veterinarians, researchers, NHP suppliers, and primate colony managers.20 In particular, this situation was an important issue in terms of the health of their NHP colony and the integrity of the affected primates for the upcoming research study. Likewise, these cases posed a dilemma for the contract quarantine company.
A cohort of 85 Chinese-origin rhesus macaques (Macaca mulatta) were imported into the United States by an AAALAC-accredited contract quarantine facility. The animals yielded the final results in a series of 3 negative TST while in quarantine in China within 30 d of being exported. Measles live-attenuated vaccine was administered to the macaques after their first TST test during the 30 d of Chinese export quarantine. After importation to the United States, 3 more TST were administered 2 wk apart, and the entire cohort of animals successfully cleared import quarantine requirements as described in the Federal Quarantine Regulations (42 CFR 71.53), and all animals were subsequently released from quarantine for sale and distribution. Immediately after this release, 24 of these animals were shipped to the USDA-registered, AAALAC-accredited facility at the University of Maryland School of Medicine as part of a research study. The 24 macaques immediately entered the facility's primate quarantine program, which consisted of additional TST once every 2 wk for 6 consecutive negative tests prior to release to the investigator for experimental use.
Three weeks after receipt and quarantine initiation at the research institution, one animal (macaque UMSOM1) exhibited a 4+ response to MOT testing in the left eyelid; this was the 2nd TST at the research institution and the 8th since the initiation of quarantine in China. The animal was isolated immediately and the TST repeated in the contralateral eyelid as well as in the abdominal skin. Both of these tests yielded positive (4+) results. After this finding, the contract quarantine company immediately isolated the 55 remaining animals from this import cohort, and the research institution that had purchased the remaining 6 macaques from this group was notified. Aggressive repeat testing of all remaining animals was initiated to obtain 5 additional consecutive (2 wk apart) TST from the macaques. Of the 55 macaques tested, 4 at the contract quarantine company (HL1 through HL4) and a second macaque at the research facility (UMSOM2) tested positive by TST.
Bronchial lavage samples from the 4 TST-positive animals at the contract quarantine company were collected for PCR testing, and digital radiographs of both lateral and anterior views were obtained. All lavage samples were PCR-negative, and radiographic results were negative for clinical evidence of disease.
Despite the development of multiple testing modalities, including IFNγ assays and antibody assays to several mycobacterial antigens, the TST remains the most common and widely used assay for screening NHP for tuberculosis.6,20,23,24,32 MOT is a poorly defined preparation composed of various mycobacterial antigens that are known to be highly crossreactive regarding the sensitization of hosts by environmental mycobacteria. TST evaluates the presence of a delayed hypersensitivity response to intradermal injection of MOT in the superior eyelid of NHP, which allows for monitoring for response to injection without the need for sedation to the examine injection site.20,24 The injection site is evaluated at 24, 48, and 72 h after injection for the development of erythema, induration, swelling, and eyelid ptosis or necrosis. Tissue response is graded on a scale of 0 to 5; grades 0 through 2 are considered negative; grade 3 is equivocal; and grades 4 and 5 are considered positive for infection.32 Known causes of false-negative results include anergic animals and those with compromised immune systems.9,11 Causes of false-positive results include previous administration of complete Freund adjuvant, traumatic MOT injection, and vaccination with the Bacillus Calmette–Guerin vaccine.8,21,37
Two important issues for both the research facility and the contract quarantine company were the determination of other tuberculosis or mycobacterial disease carriers that might be present in this group of macaques and the safety of the other NHP housed in any facility that received members of the imported group of 85 animals. A summary of the group of macaques and their TST results is shown in Figure 1. To our knowledge, the cases we report are the first naturally occurring cases of NHP with positive TST latently infected with Mycobacterium spp. The presence of positive TST in the absence of disease raises serious concerns regarding the diagnostic methods available to detect mycobacterial infections in NHP. The reported cases underscore the need for improved, reliable diagnostic assays.
Figure 1.
Distribution of a group of 85 macaques obtained from China by a US commercial vendor.
Materials and Methods
Gross pathology, histopathology, and PCR and bacterial culture assays.
Macaques that tested positive on TST were euthanized, and tissues were collected from each animal for histopathologic analysis. Each animal was first sedated with ketamine (10 mg/kg IM; KetaVed, Vedco, St Joseph, MO) and then euthanized by pentobarbital overdose (100 mg/kg IV; Fatal-Plus, Vortech Pharmaceuticals, Dearborn, MI). Organs were observed at necropsy of each animal for gross pathology. Multiple tissues (lung, hilar lymph node, axillary lymph node, mesenteric lymph node, liver, spleen, kidney, heart, pancreas, thymus, parotid salivary gland, duodenum jejunum, ileum and colon) were submitted for histopathology (for both hematoxylin and eosin as well as acid-fast staining). Samples of spleen and hilar lymph nodes were submitted for PCR analysis (Zoologix, Chatsworth, CA) and mycobacterial culture (National Veterinary Services Laboratories, Ames, IA).
Genotypic analysis of Mycobacterium-positive isolates.
Given that 2 isolates for M. kansasii were obtained, they were further characterized to determine whether they were genetically related. Further characterization involved genotyping analysis of the clinical isolates as well as RNA expression of genomic and target genes was also investigated.
Mycobacterial genotyping methods.
The genotyping analysis (National Veterinary Service Lab) of the M. kansasii strains used culture isolates from the clinical samples and followed published protocols.1,42 Briefly, a 288-bp segment of the esat6 gene (a secretory antigen) and a 303-bp segment of cfp10 (culture filtrate protein 10) were amplified from each strain and sequenced. These genes are components of the so-called ‘region of difference’ (RD1 region) of the Mycobacterium genome that is associated with bacterial virulence. After alignment of the nucleotide sequences and their analysis by neighbor-joining tree algorithms, single nucleotide polymorphisms in these sequences were used to differentiate among strains or lineages of M. kansasii. In addition to the 2 clinical isolates of M. kansasii from the animals in this report, 2 other isolates from the reference lab's collection were included in the analysis: isolate TB 10–7992 was recovered from a July 2010 submission to the reference lab of a lymph node from a female spider monkey housed in a zoo in Texas, whereas isolate TB 11–01519 was recovered from the lymph nodes from the head of a Holstein cow (age, older than 30 mo) that had been slaughtered in Pennsylvania and received at the reference lab in November 2010. The analyses of the esat6 and cfp10 sequences included 6 reference strains present in GenBank (M. kansasii strains 1, 2, 3, 3B, 4, and 5) and M. tuberculosis strain H37Rv as an outgroup. The GenBank accession numbers for these reference strains are provided in the phylogenetic trees illustrated in Figures 2 through 4. The analysis of the esat6 and cfp10 sequences and of concatemers of both sequences (that is, 288 bp + 303 bp = 591 bp) involved performing a ClustalW alignment19 of the sequences followed by construction of neighbor-joining trees with 1000 bootstrapping replicates by using MEGA 5.0.40 This process resulted in the generation of phylogenetic trees that depict the relatedness of the various isolates.
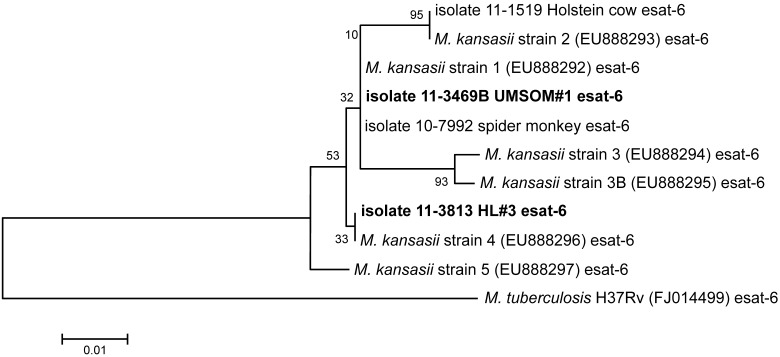
Figure 2.
Neighbor-joining tree analysis of esat6 sequences. Briefly, isolates that cluster with one another in ‘branches’ of the tree are more similar to each other than to isolates located elsewhere in the tree. The numerical values provided at the tree branching points in the figures indicate the percentage of the 1000 bootstrapping replicates that generated that particular branching pattern, with higher values implying greater confidence in that branching pattern. The small scale located adjacent to the phylogenetic tree indicates evolutionary distance in base substitutions per site.
Genomic analysis methods.
DNA samples from all 6 TST-positive or -suspect animals (Figures 2 and 3) and from 6 additional TST-negative animals from the same group were sent for genomic testing (Molecular Anthropology Laboratory, University of California, Davis, CA). This lab has developed an assay method using 96 SNP selected from among 4000 SNP identified by pyrosequencing technology that identify the region of origin of rhesus macaques with a high degree of reliability. We used these SNP and the program STRUCTURE30 to estimate the proportion of Indian-origin rhesus macaque and cynomolgus macaque ancestry of these rhesus macaques that are alleged to be of full Chinese rhesus macaque ancestry.
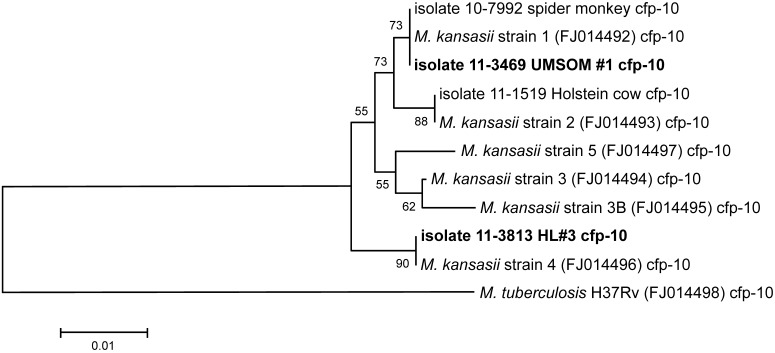
Figure 3.
Neighbor-joining tree analysis of cfp10 sequences. See the legend to Figure 2 for a brief explanation regarding interpreting the phylogenetic tree shown.
Genotyping was performed by using the SNaPshot assay (SNaPshot Mutliplex System, Applied Biosystems, Foster City, CA),27 which is based on fluorescently labeled single-nucleotide base extension. A multiplex reaction to identify 10 to 12 different SNP is set up using dideoxyNTP and primers specific the sequence adjacent to each SNP that differ in length and that end at the nucleotide just before the SNP being assayed. Each dideoxyNTP was labeled with a different fluorescent dye that identifies the nucleotide added to the 3′ end of a primer; 5 or 6 different multiplex reactions are sufficient to genotype all the SNP that have been found to be most informative for genomic analysis. The region of origin (or admixture) test combines SNP unique to Indian and Chinese rhesus macaques with other SNP unique to cynomolgus macaques in each of the following 5 geographic regions: Indochina, Indonesia, Philippines, Mauritius, and Malaysia. All tests were run on a 16-capillary genetic analyzer (model 3130xl, Applied Biosystems, Foster City, CA).
RNA expression analysis.
The goal of this gene expression analysis was to identify peripheral blood lymphocyte expression profiles consistent with tuberculosis-positive animals. RNA from peripheral blood lymphocytes was extracted (RNeasy Mini Kit, Qiagen, Valencia, CA) and quantified prior to cDNA synthesis according to the manufacturer's protocol (High-Capacity cDNA Reverse Transcription Kit, Applied Biosystems, Foster City, CA). The primers and probes for IL17 (Hs00174383 m1), IL6 (Rh02621719_u1), IL12 (Hs00168405_m1), TLR4 (Hs00152939_m1), TLR2 (Rh02787279_s1), and IL2 (Rh02789779_m1) were obtained from Applied Biosystems. Primers for IFNγ (forward, 5′ AAG CTG ACC AAT TAT TCG GTA ACT G 3′; reverse, 5′ AGT TCA GCC ATC ACT TGG ATG A 3′; probe, 5′ TCA AAT GTC CAA CGC AAA GCA GTA CAT GA 3′), TNFα (forward, 5′ AGG CAG TCA GAT CAT CTT CTC GAA 3′; reverse, 5′ GGA GCT GCC CCT CAG CTT 3′; probe, 5′ AGC CTG TAG CCC ATG TTG TAG CAA ACC CT 3′), and prGAPDH (forward, 5′ GCA CCA CCA ACT GCT TAG CAC C 3′; reverse, 5′ TCT TCT GGG TGG CAG TGA TG 3′; probe, 5′ TCG TGG GAA GGA CTC ATG ACC ACA GTC C 3′) were designed, optimized, and validated by the Lucy Whittier Molecular Core Lab at the University of California Davis and have been used in other studies.31
Results
Gross and histopathology.
Gross necropsy of all TST-positive NHP revealed no pulmonary lesions. The spleen had a thickened capsule, with a pitted serosal surface and prominent white nodular lesions throughout the parenchyma. All other gross findings were within normal limits. Histopathologic analysis of the lung, hilar lymph node, axillary lymph node, mesenteric lymph node, liver, spleen, kidney, heart, pancreas, thymus, parotid salivary gland, duodenum jejunum, ileum, and colon revealed no abnormalities. No lesions consistent with mycobacterial organisms were found in any of these tissue sections. All acid-fast staining results were negative.
PCR and culture analyses of spleen and lymph nodes.
PCR analysis of spleen and hilar lymph node samples as well as bronchial lavage samples collected from all TST-positive macaques were negative for mycobacteria according to both a generalized nested PCR assay, which detects a wide range of mycobacterial species (Zoologix assay P0007), and an ultrasensitive assay that detects only M. tuberculosis and M. bovis (Zoologix assay B0067). Culture of tissues at the National Veterinary Service Labs recovered 2 isolates from the hilar lymph nodes of macaques UMSOM1 and HL3: M. kansasii and an organism that could only be differentiated to the M. avium complex on the basis of 16S rRNA and rpoB nucleotide sequence data.
Genotyping results.
The analysis of esat6 sequences is provided in Figure 2. The UMSOM isolate clustered with the spider monkey isolate from the zoo in Texas and M. kansasii reference strain 1 (all these isolates have exactly the same esat6 sequence). The HL3 isolate clustered with M. kansasii reference strain 4. The isolate from the Holstein cow clustered with M. kansasii reference strain 2.
The analysis of the cfp10 sequences is provided in Figure 3. Results were similar to those observed with the esat6 sequences; namely, the UMSOM isolate clustered with the spider monkey isolate and M. kansasii strain 1 (all these isolates have exactly the same cfp-10 sequence). The HL3 isolate clustered with M. kansasii strain 4, and the Holstein cow isolate clustered with M. kansasii strain 2.
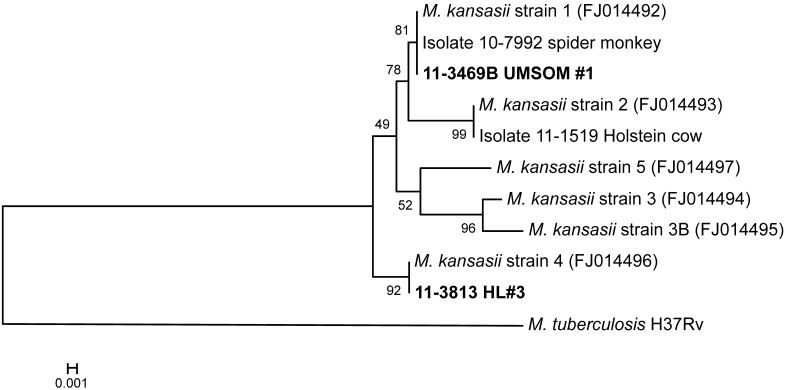
A phylogenetic tree constructed from an alignment of concatemers (a long continuous DNA molecule that contains multiple copies of the same DNA sequences linked in series) of both the esat6 and cfp10 sequences is provided in Figure 4; this tree also placed the UMSOM isolate with strain 1 and the HL3 isolate with strain 4. Thus, results from all 3 phylogenetic analyses were equivalent, indicating that the genetic relationships among the isolates can be interpreted with confidence. Taken together, the results of the sequence analyses indicate that the strains of M. kansasii infecting UMSOM1 and HL3 are different from each other. If both animals originated in the same source colony, it appears that multiple strains of M. kansasii are circulating in these animals.
Figure 4.
Neighbor-joining tree analysis of concatenated esat6 and cfp10 sequences. See the legend to Figure 2 for a brief explanation regarding interpreting the phylogenetic tree shown.
Genomic analysis results.
This analysis revealed that all animals tested were pure Chinese-origin rhesus macaques (no geographic or rhesus–cynomolgus admixture). Thus, variability in TST results between known culture-positive compared with culture-negative macaques and M. kansasii strain differences are unlikely to be attributed to genomic variability due to either geographic origin or rhesus–cynomolgus admixture.
Target gene RNA expression analysis.
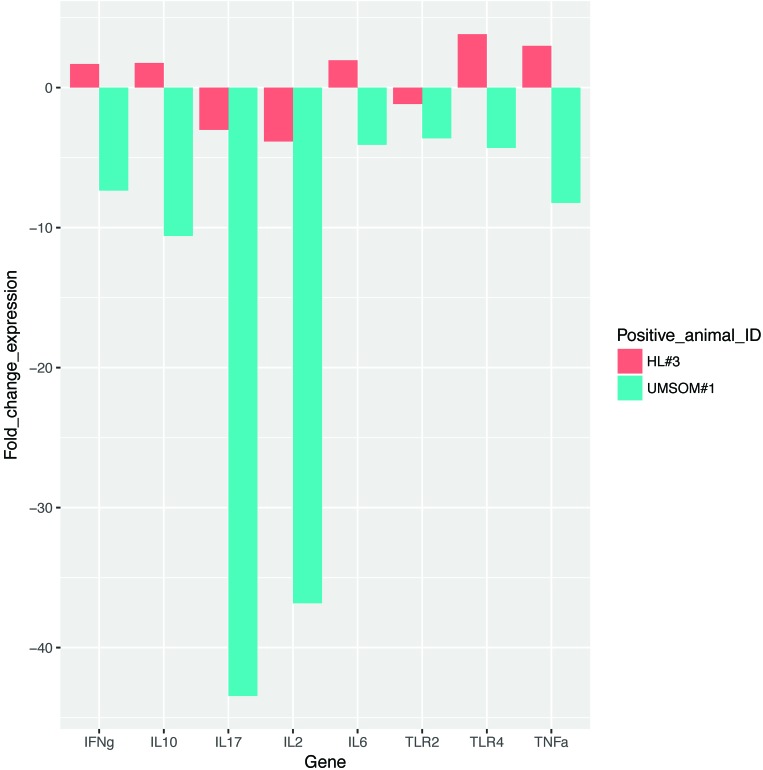
The expression levels of several host innate immune-response genes in peripheral blood lymphocytes from all 6 TST-positive animals indicated that 3 of the candidate genes (IL2, IL17, and TLR2) were downregulated in M. kansasii-positive macaques compared with negative controls. These genes have been reported to be altered in human tuberculosis patients.15 Downregulation of these genes might correlate with either previous measles vaccination or M. kansasii infection. This observation can be evaluated by using a larger sample size. The fold changes in the expression levels of these candidate genes in the 2 M. kansasii-positive macaques compared with 4 M. kansasii-negative animals are summarized in Figure 5.
Figure 5.
Fold change in expression levels of candidate genes in 2 M. kansasii-positive macaques compared with 4 M. kansasii-negative animals.
Discussion
Mycobacterial infection in NHP is usually a rapidly spreading disease, leading to high morbidity and mortality. Here we report tuberculin-positive test results in Chinese rhesus macaques latently infected with M. kansasii. Mycobacteriosis is usually acquired through human, animal, or environmental contact.13,43 Infection can produce a variety of clinical signs, including cough, dyspnea, weight loss, diarrhea, and lymphadenopathy, or infection may be asymptomatic. Gross lesions of the disease include caseous nodules in hilar lymph nodes and lung, coalescing lesions within the lung, and tubercles extending into the thoracic pleura. In advanced disease demonstrates secondary spread of tubercles to other organs including the spleen and various lymph nodes, resulting in multifocal miliary disease. Acid-fast bacilli can be identified within lesions by using an acid-fast stain.44
Mycobacteriosis due to M. kansasii infection has been reported in Indian rhesus macaques.18,41 M. kansasii is a recognized pathogen in NHP and human populations.43 The organism has been detected in wild animals and in water, which may be the natural reservoir. The presence of M. kansasii has been reported infrequently in asymptomatic wild or domestic animals such as birds, wild deer, pigs, dogs, and squirrel monkeys (Saimiri sciureus sciureus).3,28 Infection with M. kansasii accompanied by inflamed lymph nodes or pneumonic lesions has been described in rhesus monkeys, squirrel monkeys, cattle, llamas, goats, camels, and both domestic and feral pigs. In addition, M. kansasii has been isolated occasionally from unpasteurized cow milk.3
A clinical case of M. kansasii affected an Indian rhesus macaque, which demonstrated a positive TST, cavitation of the left and right apical pulmonary nodes, and enlarged bronchial lymph node. Histopathology identified granulomatous abscesses with caseous centers and cavitation of both apical lobes. The lesions of interest contained macrophages, lymphocytes, Langerhans giant cells, and acid-fast bacilli. M. kansasii was cultured. Gross and histologic lesions were characteristic of tuberculosis.18 A colony outbreak of M. kansasii infection in a breeding colony of Indian rhesus macaques led to 74 TST-positive cases; cultures from 60 of the animals yielded M. kansasii. In addition, gross lesions and histopathology were characteristic of tuberculosis in all animals. This colony outbreak underscores the seriousness of M. kansasii as a mycobacterial pathogen in NHP.41 In squirrel monkeys, M. kansasii infected 4 of 5 tuberculin-positive animals, and mycobacterial DNA PCR-amplified from bronchial lymph nodes ruled out M. tuberculosis, M. avium, and M. intracellularae. In these cases, M. kansasii was confirmed by culture from bronchial lymph nodes causing multiple granulomas in the mesenteric lymph nodes, lungs, liver, kidneys and spleen.5 M. kansasii and M. avium, along with recently discovered atypical mycobacteria, are associated with clinical tubercular disease in humans.42
The clinical cases we report represent the first description of TST-positive rhesus macaques infected with a Mycobacterium species in the absence of disease signs. Experimental studies of low-dose M. tuberculosis in cynomolgus monkeys led to latent infections in some animals.7
Latent clinical infection with M. kansasii without disease has been reported previously, in a rhesus monkey that was positive to QuantiFERON-TB Gold+ (Quest Diagnostics, Madison, NH) testing yet negative on TST.26 There were no gross pathologic or histopathologic lesions in the macaque, and M. kansasii was cultured from a tracheobronchial lymph node. In contrast to the comparative medicine perspective, TST positivity in the absence of disease occurs in approximately 30% of the world's human population.46 In these macaques, the lack of gross and histopathologic lesions despite TST positivity and positive culture results of M. kansasii from lymph nodes supports the presence of latent infection in the absence of overt mycobacteriosis. The negative PCR results from the macaques might be explained by low numbers of organisms, their sporadic location in tissues relative to sampling sites, or mutations in the organism.
The essential role of MHC class II complex and antigen-presenting cells (for example, macrophages) through which mycobacterial antigens are presented to CD4+ T cells might be related to the variation in response to TST among the rhesus macaques in our report. We measured the relative expression of several host innate immune-response genes in the peripheral blood lymphocytes of all 6 TST-positive animals. Previous studies indicate the important predictive role of these host candidate genes in mycobacterial infection.10 Although we observed downregulation of IL2, IL17, and TLR2 gene expression in the 2 M. kansasii-positive macaques compared with the 4 M. kansasii-negative animals tested, we were unable to make a statistically significant conclusion regarding the association between M. kansasii infection and changes in the expression levels of these host candidate genes because of the small sample size. The statistical significance of these findings can be evaluated in a larger sample of animals naturally infected with M. kansasii. The TST is based on the type IV delayed hypersensitivity reaction,11,38 which might be related to the variation in the response to TST among rhesus macaques. Additional studies to clarify the variation in MHC class II and response to TST in rhesus macaques are warranted.
Of prime importance was the delay of time in detecting the cases until 6 wk after release from USA-CDC importation quarantine. Intensive review of the macaques’ medical records revealed that the monkeys were vaccinated for measles after their first of 3 tuberculin tests prior to exportation from China. This vaccination may have interfered with the delayed hypersensitivity reaction, leading to the lack of positive TST results during the balance of the export quarantine period in China as well as during the United States importation quarantine. Measles vaccine is recognized to immunosuppress animals sufficiently to lead to interference with the tuberculin test.21,22,29,37,39 This immunosuppression is similar to the situation with measles virus, where infectious virus is shed for as long as 14 d, with virion RNA shed for at least 50 d.33 Measles virus RNA is released by persistently infected cells in the form of exosomes or virions that are not detected as infectious units. Immune factors involved include neutralizing antibodies for humoral responses, T-cell responses related to IFNγ-secreting cells in PBMC, and the immunosuppressive influence of regulatory T cells22 and signaling lymphocyte activation molecule.17
This prolonged release of measles virus RNA may account for persistent low-level immunosuppression.29,33 Clinical cases of measles in children yielded viral RNA at multiple sites more than 3 mo after acute disease, indicating that the clearance of cells infected with measles virus can require many months.15 Immune interference due to measles virus most likely continued during the 30-d importation period in the United States, when 3 additional tuberculin tests were conducted. Had measles vaccine not been administered during the Chinese export quarantine, macaques with positive TST might have been detected either prior to China exportation or during the United States importation quarantine.
The contract quarantine company that had imported these macaques conducted an in-depth review of the Chinese primate supplier and discovered that swine were bred and raised on the same site. Swine are recognized as a reservoir of M. kansasii and might have been the source of infection. Although no epizoologic investigations of the swine herd or primate colony at this Chinese supplier were performed, no further purchases have been made from this source. There is growing evidence of animal-to-human infectivity of tuberculosis, with swine being an important zoonotic species.25
The presence of only 6 cases in the shipment of 85 macaques strongly suggests a lack of primate-to-primate infectivity. These cumulative features led to the decision to release the remaining primates from the contract quarantine company's extended quarantine. All of these animals as well as the rest of the macaques from the same cohort remained healthy, with negative TST tests and absence of disease. They were assigned successfully to studies with no interference in research. These cases of tuberculin-positive NHP with absence of disease underscore the need for improved and specific diagnostic testing.
Acknowledgments
We thank Theresa Alexander, Kayla Yi, Michelle Izuka, and Dawn Hunt for their excellent veterinary technical support. This research was supported by departmental funds obtained from the Program of Comparative Medicine at the University of Maryland School of Medicine. We thank Dr Victoria Baxter for her valuable contribution clarifying Figures 1 through 3.We thank Theresa Nolan, Dawn Engel, and Michelle Izuka for technical support.
References
- 1.Arend SM, deHaas P, Leyten E, Rosenkrands I, Rigouts L, Andersen P, Wouter M, vanDissel JT, van Soolinger D. 2005. ESAT6 and CFP10 in clinical compared with environmental isolates of Mycobacterium kansasii. J Infect Dis 191:1301–1310. [DOI] [PubMed] [Google Scholar]
- 2.Attorri S, Dunbar S, Clarridge JE., 3rd 2000. Assessment of morphology for rapid presumptive identification of Mycobacterium tuberculosis and Mycobacterium kansasii. J Clin Microbiol 38:1426–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bercovier H, Vincent V. 2001. Mycobacterial infections in domestic and wild animals due to Mycobacterium marinum, M. fortuitum, M. chelonae, M. porcinum, M. farcinogenes, M. smegmatis, M. scrofulaceum, M. xenopi, M. kansasii, M. simiae, and M. genavense. Rev Sci Tech 20: 265–290. [DOI] [PubMed] [Google Scholar]
- 4.Brack M. 1987. Mycobacteriaceae—tuberculosis type. p 214–235. In: Agents transmissible from simians to man. Berlin (Germany): Springer-Verlag. [Google Scholar]
- 5.Brammer DW, O'Rourke CM, Heath LA, Chrisp CE, Peter GK, Hofing GL. 1995. Mycobacterium kansasii infection in squirrel monkeys (Saimiri sciureus sciureus). J Med Primatol 24:231–235. [DOI] [PubMed] [Google Scholar]
- 6.Brusasca PN, Peters RL, Motzel SL, Klein HJ, Gennaro ML. 2003. Antigen recognition by serum antibodies in nonhuman primates experimentally infected with Mycobacterium tuberculosis. Comp Med 53:165–172. [PubMed] [Google Scholar]
- 7.Capuano SV, 3rd, Croix DA, Pawar S, Zinovik A, Myers A, Lin PL, Bissel S, Fuhrman C, Klein E, Flynn JL. 2003. Experimental Mycobacterium tuberculosis infection of cynomolgus macaques closely resembles the various manifestations of human M. tuberculosis infection. Infect Immun 71:5831–5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaparas SD, Good RC, Janicki BW. 1975. Tuberculin-induced lymphocyte transformation and skin reactivity in monkeys vaccinated or not vaccinated with Bacille Calmette–Guerin, then challenged with virulent Mycobacterium tuberculosis. Am Rev Respir Dis 112:43–47. [DOI] [PubMed] [Google Scholar]
- 9.Corcoran KD, Jaax GP. 1991. An attempt to predict anergy in tuberculosis suspect cynomolgus monkeys. Lab Anim Sci 41:57–62. [PubMed] [Google Scholar]
- 10.de Oliveira LRC, Peresi E, de Assis Golim M, Gatto M, Araujo Jr JP, de Costa EAPN, Ayres JA, Parise Fortes MR, Calvi SA. 2014. Analysis of toll-like receptors, iNOS, and cytokine profiles in patients with pulmonary tuberculosis during antituberculosis treatment. PLoS One. 9:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elferink BG, Geluk A, Otting N, Slierendregt BL. 1993. MHC class II complex. Hum Immunol 38:396–409. [DOI] [PubMed] [Google Scholar]
- 12.Evans SA, Colville A, Evans AJ, Crisp AJ, Johnston IDA. 1996. Pulmonary Mycobacterium kansasii infection: comparison of the clinical features, treatment, and outcome with pulmonary tuberculosis. Thorax 51:1248–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frost PA, Calle PP, Klein HJ, Thoen CO. 2014. Zoonotic tuberculosis in nonhuman primates. In: Thoen CO, Steele JH, Kaneen JB, editors. Zoonotic tuberculosis: Mycobacterium bovis and other pathogenic mycobacteria, 3rd ed. Ames (IA): Wiley and Sons. [Google Scholar]
- 14.Gibson SV. 1998. Bacterial and mycotic diseases, p 59–110. In: Bennett BT, Abee CR, Henrickson R. Nonhuman primates in biomedical research: diseases, vol 2. San Diego (CA): Academic Press. [Google Scholar]
- 15.Gourgouillon N, de Lauzanne A, Cottart CH, Curis E, Debord C, Guérin-El Khourouj V, Pédron B, Faye A, Sterkers G. 2012. TNFα/IL2 ratio discriminates latent from active tuberculosis in immunocompetent children: a pilot study. Pediatr Res 72:370–374. [DOI] [PubMed] [Google Scholar]
- 16.Kennard MA, Schroeder CR, Trask JD, Paul JR. 1939. A cutaneous test for tuberculosis in primates. Science 89:442–443. [DOI] [PubMed] [Google Scholar]
- 17.Koga R, Ohno S, Ikegame S, Yanagi Y. 2010. Measles virus-induced immunosuppression in SLAM knock-in Mice. J Virol 84:5360–5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson RK, Juras RA, Stiefel S, Hall JE. 1989. Mycobacterium kansasii in a rhesus monkey. Lab Anim Sci 39:425–428. [PubMed] [Google Scholar]
- 19.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. [DOI] [PubMed] [Google Scholar]
- 20.Lerche NW, Yee JL, Capuano SV, Flynn JL. 2008. New approaches to tuberculosis surveillance of nonhuman primates. ILAR J 49:170–178. [DOI] [PubMed] [Google Scholar]
- 21.Lin PL, Yee J, Klein E, Lerche NW. 2008. Immunologic concepts in tuberculosis diagnostics for nonhuman primates: a review. J Med Primatol 37 Suppl 1:44– 51. [DOI] [PubMed] [Google Scholar]
- 22.Lin WH, Kouyos RD, Adams RJ, Grenfell BT, Griffin DE. 2012. Prolonged persistence of measles virus RNA is characteristic of primary infection dynamics. Proc Natl Acad Sci USA 109: 14989–14994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Min F, Wang J, Zhang J. 2015. Performance of ESAT6 for serodiagnosis of nonhuman primate tuberculosis: a metaanalysis. J Adv Vet Anim Res 2:107–114. [Google Scholar]
- 24.Motzel SL, Schachner R, Kornegay R, Fletcher M, Kanaya B, Gomez J, Ngai D, Handt L, Wagner J, Klein H. 1999. Assessment of methods for the diagnosis of tuberculosis in 3 species of nonhuman primates. Abstract presented at the American Association for Laboratory Animal Science 50th Annual Meeting, Indianapolis, Indiana, October 1999. [Google Scholar]
- 25.Muwonge A. 2012. Nontuberculous mycobacteria in swine: is it a public health concern? Mycobact Dis 2:1–2. [Google Scholar]
- 26.Parsons SDC, deVilliers C, Gey van Pittius NC, Warren RM, van Helden PD. 2010. Detection of Mycobacterium kansasii infection in a rhesus macaque (Macaca mulatta) using a modified QuantiFERON-TB Gold assay. Vet Immunol Immunopathol 136:330–334. [DOI] [PubMed] [Google Scholar]
- 27.Pati N, Schowinsky V, Kokanovic O, Magnuson V, Ghosh S. 2004. A comparison between SNaPshot, pyrosequencing, and biplex invader SNP genotyping methods: accuracy, cost, and throughput.J Biochem Biophys Methods 60:1–12. [DOI] [PubMed] [Google Scholar]
- 28.Pierce DL, Dukelow WR. 1988. Misleading positive tuberculin reactions in a squirrel monkey colony. Lab Anim Sci 38:729–730. [PubMed] [Google Scholar]
- 29.Pierson TC, Yewdell JW. 2012. Measles immunometrics. Proc Natl Acad Sci USA 109:14724–14725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pritchard JK, Wen X, Falush D. 2010. Documentation for structure software: version 2.3. Chicago (IL): University of Chicago. [Google Scholar]
- 31.Roodgar M, Lackner A, Kaushal K, Sankaran S, Dandekar S, Satkoski Trask J, Drake C, Smith DG. 2013. Expression levels of 10 candidate genes in lung tissue of vaccinated and TB-infected cynomolgus macaques. J Med Primatol 42: 161–164. [DOI] [PubMed] [Google Scholar]
- 32.Richter CB, Lehner NDM, Henrickson RV. 1984. Primates, p 298–383. In: Fox JG, Cohen BJ, Loew FM, editors. Laboratory animal medicine. Orlando (FL): Academic Press. [Google Scholar]
- 33.Riddell MA, Moss WJ, Hauer D, Monze M, Griffine DE. 2007. Slow clearance of measles virus RNA after acute infection. J Clin Virol 39:312–317. [DOI] [PubMed] [Google Scholar]
- 34.Roberts JA, Andrews K. 2008. Nonhuman primate quarantine: its evolution and practice. ILAR J 49:145–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schrenzel MD. 2012. Molecular epidemiology of mycobacteriosis in wildlife and pet animals. Vet Clin North Am Exot Anim Pract 15:1–23. [DOI] [PubMed] [Google Scholar]
- 36.Shitrit D, Baum GL, Priess R, Lavy A, Shitrit AB, Raz M, Shlomi D, Daniele B, Kramer MR. 2006. Pulmonary Mycobacterium kansasii infection in Israel, 1999 to 2004: clinical features, drug susceptibility, and outcome. Chest 129:771–776. [DOI] [PubMed] [Google Scholar]
- 37.Simmons J, Gibson S. 2012. Bacterial and mycotic diseases. p112–119. In: Abee CR, Mansfield K, Tardif S, Morris T, editors. Nonhuman primates in biomedical research: diseases, 2nd ed. San Diego (CA): Academic Press. [Google Scholar]
- 38.Snyder SB, Fox JG. 1973. Tuberculin testing in rhesus monkeys (Macaca mulatta): a comparative study using experimentally sensitized animals. Lab Anim Sci 23:515–521. [PubMed] [Google Scholar]
- 39.Starr S, Berkovich S. 1964. The effects of measles, γ- globulin–modified measles, and vaccine measles on the tuberculin test. N Engl J Med 270:386–391. [DOI] [PubMed] [Google Scholar]
- 40.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valerio DA, Dalgard DW, Voelker RW, McCarrol NE, Good RC. 1979. Mycobacterium kansasii infection in rhesus monkeys. p 145–150. In: Montali RJ, editor. Mycobacterial infection in zoo animals. Washington (DC): Smithsonian Institute Press. [Google Scholar]
- 42.van Ingen J, de Zwaan R, Dekhuijzen R, Boeree M, van Soolingen D. 2009. Region of difference 1 in nontuberculous Mycobacterium species adds a phylogenetic and taxonomical character. J Bacteriol 191:5865–5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wayne LG, Sramek HA. 1992. Agents of newly recognized or infrequently encountered mycobacterial diseases. Clin Microbiol Rev 5:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whary M. 2015. Nonhuman primates p 855–859. In: Fox JG, Anderson LC, Otto G, Pritchett-Corning K, editors. Laboratory Animal Medicine, 3rd ed. Oxford (UK): Elsevier. [Google Scholar]
- 45.Wolinsky E. 1979. Nontuberculous mycobacteria and associated disease. Am Rev Respir Dis 119:107–159. [DOI] [PubMed] [Google Scholar]
- 46.World Health Organization. [Internet] 2017. Fact Sheet Number 104 March 2015. [Cited 28 April 2017]. Available at: www.who.int/tb/areas-of-work/preventive-care/ltbi/en/