Abstract
The structural variability of lateral ventricles is poorly understood notwithstanding that enlarged size has been identified as an unspecific marker for psychiatric illness, including schizophrenia. This paper explores the effects of heritability and genetic risk for schizophrenia reflected in ventricular size and structure. We examined ventricular size and shape in the MRI studies of monozygotic (MZ) twin pairs discordant for schizophrenia (DS), healthy MZ twin pairs, healthy dizygotic twin pairs, and healthy nonrelated subject pairs. Heritability and effect due to disease were analyzed in two tests. First, heritability was examined by ventricle similarity between pairs of co-twins. Results show that co-twin ventricle shape similarity decreases with decreasing genetic identity, an effect not seen in the volume analysis. Co-twin shape similarity of healthy MZ twins did not differ from DS MZ twins. Second, the disease effect was examined through the ventricular differences of DS subjects to a template shape representing healthy subjects. Affected DS twins showed shape differences from healthy subjects on the left and right sides. Interestingly, unaffected DS twins also showed significant shape differences from healthy subjects for both sides. Volume comparisons did not show differences between these groups. Locality of shape difference suggests that the ventricular shape of the anterior and posterior regions is under genetic influence in both healthy controls and schizophrenia patients. Affected and unaffected groups demonstrate main shape differences, compared with healthy controls, only in the posterior region. Our results suggest that genetics have a stronger influence on the shape of lateral ventricles than do the disease-related changes in schizophrenia.
Keywords: brain morphometry, shape analysis, twin study, MRI
Image analysis methods promise to elucidate the influence of genes and disease on brain morphology (1). Extensive prior research has demonstrated that brain morphology, reflected as a measure of size or volume, is altered in schizophrenia. The structural alterations associated with schizophrenia include larger cerebral spinal fluid spaces and smaller cortical and subcortical gray matter structure volumes (2, 3). Contributing factors to volume differences in schizophrenia include genetic inheritance, abnormal genetic or epigenetic events, prenatal or perinatal brain insults, disease pathology, and disease-associated factors.
Published scientific reports suggest that heritability strongly influences brain morphometry. Heritability estimates are high for total brain volume (94%) (4) and for lateral ventricular volume (82-85%) (5). The heritability of brain volume appears to be high for intracranial volume (81%), midline corpus callosum area (79%), and lateral ventricular volume (79%) (6). Evidence for the genetic influence on brain morphology can be obtained by comparing healthy monozygotic (MZ) twin pairs and dizygotic (DZ) twin pairs with nonrelated (NR) normal control pairs. Such studies indicate that there is substantial genetic contribution to corpus callosum size and shape (7), cerebral cortical gyral patterning (4) and morphology (8), and frontal and temporal cortical gray matter density (9). Similarly, a comparison of brain morphometry differences among affected schizophrenia probands, their unaffected MZ twins, and normal subjects provides an opportunity to examine the contributions of genetic inheritance, disease state, and environment, while controlling for anatomical variability in the population.
MRI studies of MZ twins showed that twins with schizophrenia had slightly smaller cerebral hemisphere volumes (10), smaller hippocampal volumes (11), and larger cerebral ventricles (11, 12). However, human raters were not able to distinguish differences in gyral patterns among twins with schizophrenia, unaffected twins, or healthy controls (10).
The results of MRI studies of the unaffected family members of schizophrenia patients suggest that the volume of many brain structures is heritable. Structural abnormalities are more frequently observed in the unaffected relatives and siblings of schizophrenic patients than in normal controls. Subjects with schizophrenia appear to have larger ventricles and smaller hippocampal volume than their unaffected siblings (3). Recently, the contributions of genetics, schizophrenia, and environment have been examined for cortical gray matter deficits (13), hippocampal morphology (14), and corpus callosum morphology (15). The latter appears to be related to lateral ventricular enlargement.
Although measures of brain volume have revealed differences between affected subjects and controls, the magnitude of these differences is moderate, and there is almost always substantial overlap in the distributions of the comparison groups. Shape analysis is an approach to quantify subtle differences in brain structure that could potentially enhance the identification of brain structural abnormalities associated with neurologic and psychiatric illnesses such as schizophrenia. Different shape analysis methods have been suggested in recent years, such as shape analysis through sparse 3D landmarks (16, 17), dense point models (14, 18, 19), parametric boundary models (20, 21), medial models (22-24), or deformable registration (25-28). We developed methods of shape analysis, which have been adequately validated with reliable results (19, 20). In the study presented in this document, these methods of shape analysis were applied to MZ twins with and without schizophrenia, to DZ twins, and to healthy controls. The analysis includes ventricle shape comparison between pairs of co-twins to examine shape similarity in relation to genetic identity. Further, shape deformation due to illness is studied by measuring shape difference from a healthy template. In addition to quantitative analysis, color-coded surface displays illustrate locality and extension of shape deformation. The shape-defining processes are not directly observed because the lateral ventricles are fluid-filled spaces whose shape is governed by the neuroanatomical structures surrounding them, such as the hippocampus or the caudate.
Methods
Summary. High-resolution MRI scans were acquired from three different subject groups [10 MZ twin pairs discordant for schizophrenia (DS), 9 healthy MZ twin pairs, and 10 healthy DZ twin pairs imaged on the same scanner]. All three groups were matched for age, gender, and handedness. A fourth group consisting of 10 healthy NR subject pairs also matched for age, gender, and handedness was selected from the two healthy groups. Volumetric differences as well as 3D maps representing magnitude and significance of shape differences between the twin pairs and between subject groups were computed and visualized.
Healthy MZ and DZ Subjects. Volunteers were screened for a history of neurological, psychiatric, and other major medical illnesses. All scans were read by a radiologist and deemed qualitatively normal. The mean age of the 10 MZ pairs (4 female pairs) was 31 years (range 19-54), and the mean age for the 10 DZ twins (6 female pairs) was 23 years (range 18-30). Only twins who matched on 19 red blood cell antigens were considered MZ, which predicts monozygosity at a minimum 97% confidence level (29). Information about chorion status was not available. One twin pair was excluded from the analysis because of a history of car accident with closed head injury at age 7.5 years with coma of 6 weeks' duration. We selected 20 of the 38 subjects into 10 NR pairs that were matched for age, gender, and handedness (6 female pairs). The average age was 25 years (range 18-35). All volunteers gave informed consent for the MRI scans after the study protocol was reviewed and approved by the institutional review board.
Patients. The population of DS twins consisted of 10 MZ twin pairs in which one twin had schizophrenia and the co-twin was psychiatrically normal (30). Psychiatric diagnoses of the ill twins were determined according to Diagnostic and Statistical Manual III Revised (DSM-III-R) criteria by using the Structured Clinical Interview for DSM-III-R-Patient Version (31). The mean severity of illness (axis V of DSM-III-R) during the period just preceding the investigation was 39.7 (range 26-55, SD 11.1). Psychiatric normality in the co-twins was defined as the absence of an axis I or II disorder, determined by DSM-III-R criteria by using the Structured Clinical Interview for DSM-III-R Non-Patient Version (31). Monozygosity was determined by examination of 19 red blood cell antigens and physical appearance. The twins were born in the United States and Canada (mean age 32 years, range 28-40, SD 6.6). Five of the pairs were female. In 8 of the 10 pairs, the ill twin was the first-born in the pair. Mean age at time of adult assessment was 30.9 years (range 25-40, SD 6.5). Mean age at time of illness onset was 21.6 years (range 19-30, SD 4.6). The probability that any of the DS pairs would become concordant in the future was presumed to be very low, because an average of 9.5 years (range 6-16, SD 4.3) had elapsed from illness onset of the proband until the time of the investigation. Illness onset in a previously normal co-twin is very infrequent after 4 years of discordance (32). In one case, the well twin was found to have hydrocephaly of unknown origin, so the pair containing this case was excluded from the analyses. All subjects gave informed consent to the MRI scans after the study protocol was reviewed and approved by the institutional review board.
MRI. All 3D MRI datasets were acquired by using the same 1.5-tesla scanner (Signa, General Electric) with a T1-weighted spoiled gradient-recalled acquisition in the steady-state sequence (TR = 24 ms, TE = 5 ms). A single sagittal series of 124 contiguous 1.5-mm-thick slices with an in-plane field of view of 240 mm across a 256 × 256 pixel matrix (0.9375 × 0.9375 × 1.5 mm3) was collected.
Image Processing. Every dataset was processed by using a rater-independent, automatic tissue-segmentation method (33) that generates detailed maps of gray matter, white matter, and cerebrospinal fluid. This processing includes a bias field correction, which adjusts for intensity inhomogeneities. A rater-guided, connectivity-based method was used to separate the lateral ventricles. An automatic morphological closing operation smoothes the boundary and leads to spherical topology of the segmented lateral ventricles. The smoothing maximally amounts in changes along the boundary of a single voxel amplitude and is thus not expected to influence the data analysis. Because of the young age of the subjects, the segmentations of the temporal horn were disconnected from the main body and were excluded from the analysis. All segmentations were finally inspected by an expert and were judged to be adequate for ventricular body, occipital horn, and lateral horn. Fig. 1 illustrates surface displays of segmented and aligned ventricle pairs. Ventricles of co-twins are shown side by side by using the same color code.
Fig. 1.
Graphical view of aligned and size-normalized ventricles. (Left) Superior view of left ventricles of five MZ twin pairs (Upper) and five DZ twin pairs (Lower) displayed from the top. Ventricles of co-twins are shown by using the same color. (Right) Sagittal view of right ventricles of 10 DS pairs, with affected and unaffected shown side by side. The third pair (Upper Right) was excluded because of hydrocephaly in the unaffected twin.
Statistical Analysis. We analyzed the volumetric and shape differences between twin pairs and between groups of normal controls and patients. All analyzed measurements were corrected for gender and age influence by using the jmp statistics software (SAS Institute, Cary, NC).
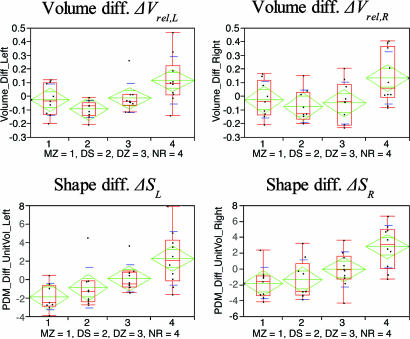
Twin-Pair Volumetric Analysis.We analyzed the volumetric similarity of the left and right lateral ventricles for each subject pair as the relative volume difference: ΔVrel = ABS(VTwinA - VTwinB)/(VTwinA + VTwinB). Because ΔVrel is relative to the overall size, no normalization with the intracranial cavity volume is required. This measurement expresses the percentage difference between ventricle pairs. The group mean difference tests of ΔVrel are shown in Fig. 4.
Fig. 4.
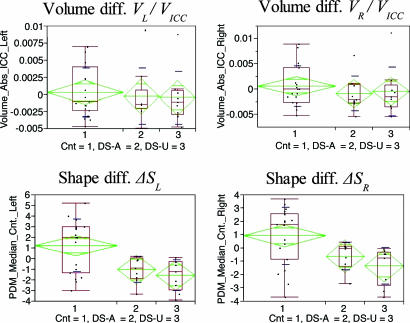
Statistical analysis plots of the relative volume difference (Upper) and the global shape difference (Lower). Parametric statistics is shown in green and blue (mean, confidence interval about mean, and standard deviation). Nonparametric statistics with median and quartiles are shown in red.
Shape Description and Shape Difference. Each lateral ventricle was transformed into the spherical harmonic-based shape description (34), which continuously describes the surface of objects by a set of coefficients weighting the spherical harmonic basis functions. The spherical harmonic-based shape description is then uniformly sampled into a set of surface points. Spherical harmonic correspondence between the surface points of two objects was established by parameter-space rotation based on the first-order expansion of spherical harmonics (20). The lateral ventricles were rigidly aligned by using a rigid-body Procrustes alignment (35). To create a shape measurement that is independent of size, we uniformly scaled all lateral ventricles to the same volume, which was chosen at the average volume (20). The results are illustrated in Fig. 1, with co-twin ventricles displayed side by side by using the same color code.
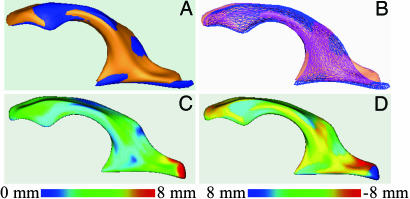
The local ventricle shape difference between aligned subject pairs was computed as the Euclidean distance between corresponding points. These distances create a distance map on the ventricular surface for every subject pair. Distances can be used either as absolute differences for a measurement of deviation or as signed differences with additional information about outward or inward deformations with respect to a reference shape. Fig. 2 illustrates the absolute and signed shape difference absolute distances with the example of a ventricle pair of DZ co-twins. A detailed description of the processing steps is found in refs. 23 and 36.
Fig. 2.
Concept of shape analysis by using distance maps. The example of a right lateral ventricle pair (DZ, twin 1, and twin 2 in blue and orange) is shown. (A) Lateral ventricles after alignment and scaling to unit volume. (B) Same as A, but ventricles are shown as transparent and grid-mesh surfaces. (C) Distance map with color-coded absolute distances between corresponding points. The reference is the surface of twin 2. Distances are coded in blue to red. (D) Distance map with color-coded signed distances between corresponding points. Negative distances are computed outside the reference object and positive distances inside.
Twin-Pair Shape Analysis. In the twin-pair analysis, the absolute distance between corresponding points is used as a metric for shape similarity. This measure is calculated for co-twin pairs of all groups (see Fig. 1). The global shape difference ΔS for each subject pair was computed by averaging the distances across the surface, which results in the mean absolute distance. The statistical analysis of the mean absolute distance between groups is shown in Fig. 4. A visual assessment of the locality and magnitude of differences is obtained by using the color-coded average distance maps representing locally averaged distances mapped onto a template ventricle surface (see Fig. 2). A comparison of the average distance maps is shown in Fig. 5. Local group statistics are then computed on these co-twin distance maps as shown in Fig. 3. The local P values are plotted with a color code, resulting in statistical significance maps (e.g., Fig. 6). These P values were computed by using the nonparametric permutation approach provided by the snpm (Tom Nichols and Andrew Holmes, University of Michigan, Ann Arbor) software (37, 38), which is part of the spm package (39). This permutation approach accounts for the multiple comparisons problem and has strong control over experiment-wise type 1 errors (37).
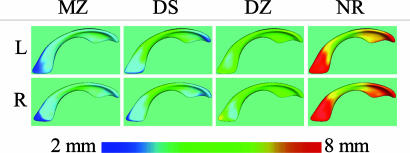
Fig. 5.
Average distance maps visualize the distances between co-twins, averaged over the group. The figure shows absolute distances for each group. The distances are color-coded, as shown in the color map. The displays show increasing distance from MZ and DS over DZ to NR.
Fig. 3.
Concept of statistical significance maps. For two groups of objects, surface distance maps are compared locally, resulting in a map of statistical tests, which displays locations of significant differences between groups.
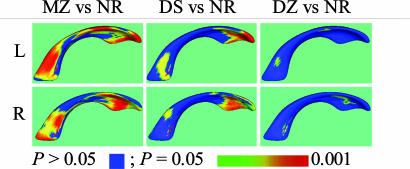
Fig. 6.
Statistical maps displaying the locations of significant differences between groups for the co-twin analysis. The colors indicate the level of significance as shown in the color map. Results for group comparisons not shown in this figure did not have significant regions
Group Tests Between Controls and Schizophrenics. Whereas co-twin similarity analysis reflects shape difference in relation to genetic similarity, it does not show difference between healthy control subjects and schizophrenia patients. We therefore performed group tests to investigate differences between healthy controls and the affected and unaffected groups of the DS twins. The healthy control group was selected by randomly sampling one of the co-twins from each healthy MZ and healthy DZ pair, resulting in a set of 20 nonrelated subjects. All healthy subjects were used to calculate the template representing the average healthy shape model. The schizophrenia patient group was constructed from all schizophrenia subjects in the DS group. Their psychiatrically normal co-twins represented the unaffected group.
Volumetric Analysis Between Groups. We analyzed the volumes of the left and right lateral ventricles for each group of subjects. These measurements were normalized with the intracranial cavity volume: Vabs = Vvent/VICC. Tests for mean differences of volumes are shown in Fig. 7.
Fig. 7.
Statistical analysis plots of the individual volume measurements normalized with the intracranial volume (Upper) and the global shape difference ΔS (Lower) from the average healthy template.
Shape Analysis Between Groups. Similarly to the twin-pair shape analysis, we used surface distance maps to determine differences in shape between groups. We use a shape-difference metric based on the distance to a template shape. The ventricle templates were calculated as the weighted average ventricle shape from all healthy subjects.
The twin-pair shape analysis described previously used absolute surface point distances because there was no preference of a reference object for each pair. Shape analysis based on a template, however, defines the template shape as the reference object. This template allows the computation of signed distances with negative distances inside the reference object and positive distances outside (see Fig. 2 Right). The global shape/distance measurement is computed as the median across the surface to increase robustness in the presence of outliers. Average distance maps and statistical significance maps are computed the same way as in twin-pair shape analysis.
Results
Co-Twin-Pair Ventricle Differences. Results of the comparison of ventricle size and shape between co-twin pairs are plotted in Fig. 4 and illustrated as 3D displays in Fig. 5. These plots and color-coded surfaces illustrate the degree of ventricular similarity of co-twin pairs, separately studied for left and right. This co-twin similarity can be tested among groups to test the hypothesis that size and shape difference is related to genetic inheritance. Results of group tests are listed in Table 1 for a quantitative global measure and in Fig. 6 for depicting local surface regions where groups differ significantly.
Table 1. P values for testing group mean differences between groups.
|
P value
|
||||
|---|---|---|---|---|
| Comparison | ΔVrel, L | ΔVrel, R | ΔSL | ΔSR |
| MZ vs. NR | 0.0537 | 0.0514 | 0.0013 | 0.0006 |
| MZ vs. DZ | 0.82 | 0.71 | 0.0082 | 0.0399 |
| MZ vs. DS | 0.18 | 0.42 | 0.28 | 0.68 |
| DS vs. NR | 0.0033 | 0.011 | 0.018 | 0.0026 |
| DS vs. DZ | 0.069 | 0.70 | 0.25 | 0.24 |
| DZ vs. NR | 0.057 | 0.027 | 0.050 | 0.016 |
Values significant at the 5% level are printed in boldface.
Statistical volumetric analysis. There was a clear trend for the volume difference in MZ pairs to be smaller than in NR pairs (see Table 1). This finding was close to statistical significance in the left (PΔV,L = 0.054) and right (PΔV,R = 0.051) lateral ventricles. Volume differences in DZ pairs were significantly smaller than in NR pairs in the right (PΔV,R = 0.027) but not in the left (PΔV,L = 0.057) lateral ventricles. No test among the MZ, DS, and DZ groups was significant. Ventricle volume difference in DS pairs was significantly smaller than ventricle volume difference in NR pairs. This finding reached statistical significance for both the left (PΔV,L = 0.0033) and right (PΔV,R = 0.011) lateral ventricles. These findings suggest that lateral ventricle volumes of co-twin pairs are more similar than are volumes of NR pairs of subjects.
Pairwise co-twin shape similarity. Relative volume differences for each group are plotted in Fig. 4 Upper. Healthy MZ, DS, and DZ show smaller volume differences between pairs than NR. Qualitatively, the variability is larger for right ventricles than for left ventricles. Average distance maps (see Fig. 5) of the healthy MZ twin pairs suggested similarity in lateral ventricular shape along the entire length of the structure. The same is found for DS MZ twin pairs. Both findings are confirmed by the quantitative analysis of integrated shape differences (Fig. 4), which shows small differences (vertical axis) for the MZ and DS groups. The DZ revealed slightly larger shape differences between co-twin ventricles localized along the entire length of the structure. The quantitative shape difference plotted in Fig. 4 shows larger DZ co-twin shape differences, compared with the MZ and DS groups. Fig. 4 clearly demonstrates the larger pairwise shape differences for NR pairs, compared with all of the other groups, but also shows the larger variability in this group. The plots indicate an increase in co-twin ventricle shape difference in the order MZ ≈ DS < DZ < NR. The average NR distance maps (Fig. 5) suggested large differences in lateral ventricular shape localized to the anterior and posterior aspects of the structure. The NR group appeared to have higher shape similarity localized at the middle aspect, compared with anterior and posterior aspects. However, similarity of shape localized to this middle aspect in the NR group did not appear as similar as in the MZ group. The decrease of shape similarity with decrease of genetic similarity as seen in the quantitative results also is clearly illustrated in Fig. 5.
Shape of MZ vs. NR and DS vs. NR. The shape of lateral ventricles in MZ pairs was significantly more similar than in NR subjects (see Fig. 4) for both the left (PΔS,L = 0.0013) and right (PΔS,R = 0.0006) lateral ventricles. Lateral ventricular shape in DS pairs also was significantly more similar than in NR subjects for both the left (PΔS,L = 0.018) and right (PΔS,R = 0.0026) lateral ventricles. Statistical maps showing the location of statistically significant differences in shape between the MZ and NR groups confirmed the findings suggested by visual inspection of the average distance maps (see Fig 6). Statistically significant shape differences are located along the entire length of the lateral ventricles. Smaller P values were localized to the anterior and posterior aspects, compared with the middle aspects of the left and right lateral ventricles. The statistical maps confirmed the presence of statistically significant differences between the DS and NR groups localized to the anterior and posterior aspects of the left and right lateral ventricles.
MZ vs. DZ and DS vs. DZ. The shape of lateral ventricles was significantly more similar for MZ pairs than for the DZ pairs for both the left (PΔS,L = 0.0082) and the right (PΔS,R = 0.0399) lateral ventricles. There were no significant differences between the DS and DZ groups for shape. Statistical maps (Fig. 6) did not yield any significant locality for shape differences between the MZ and the DZ groups in any regions (Fig. 6). Similarly, no significant regions were found between the DS and DZ groups.
DZ vs. NR. Significant shape differences between the DZ and NR groups were observed in the left (PΔS,L = 0.050) and the right (PΔS,R = 0.016) lateral ventricles. Statistical maps (Fig. 6) showed a few small regions of local differences between the DZ and NR groups. MZ vs. DS. No significant results were observed for shape differences between the MZ and DS groups in the left (PΔS,L = 0.28 or right PΔS,R = 0.68) lateral ventricles (Fig. 4). Average distance maps did not suggest that significant shape differences between the MZ and DS twin pairs would be observed, and the statistical maps (Fig. 6) show no local significant differences for the left or right lateral ventricles.
Control for Schizophrenic Difference.
Volume comparison. Lateral ventricular volume of neither the affected group (PVL/VICC = 0.71, PVR/VICC = 0.35) nor the unaffected group (PVL/VICC = 0.60, PVR/VICC = 0.56) differed significantly from that of the healthy control group (Fig. 7 Upper). Also, lateral ventricular volume of the affected group did not differ significantly from that of the unaffected group. The plots and group statistics illustrate that volumetric measurements do not discriminate healthy controls from either the affected or unaffected groups. Further, the affected group in this small sample does not show an increase of ventricular volume, an effect often found in many, but not all, studies of schizophrenia patients.
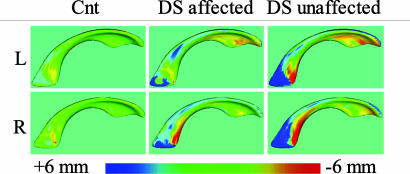
Shape comparison. In contrast to lateral ventricular volume, lateral ventricular shape of the affected group differed from the healthy group, with left being significant and right showing a trend (PΔS,L = 0.039, PΔS,R = 0.058), as listed in Table 2. Interestingly, the unaffected group showed a similarly significant shape distance to the healthy group (PΔS,L = 0.0042, PΔS,R = 0.0089). The average distance maps suggested that differences among the three groups were localized to the anterior and posterior aspects of the left and right lateral ventricles, relative to the middle (see Fig. 8). Lateral ventricular shape distance to healthy subjects between affected and unaffected groups did not differ significantly, although the average distance maps (Fig. 8) showed more pronounced shape difference from normal in the unaffected group, compared with the affected group. These findings lead to the conclusion that both the affected and unaffected groups show a significant shape change, compared with a healthy control template.
Table 2. P values for testing mean difference between the groups.
|
P value
|
||||
|---|---|---|---|---|
| Comparison | ΔVrel, L | ΔVrel, R | ΔSL | ΔSR |
| Cnt vs. DS-A | 0.71 | 0.35 | 0.039 | 0.058 |
| Cnt vs. DS-U | 0.60 | 0.56 | 0.0042 | 0.0089 |
| DS-A vs. DS-U | 0.91 | 0.82 | 0.39 | 0.22 |
Values significant at the 5% level are printed in boldface.
Fig. 8.
Signed average distance maps to the healthy template. Distances are color-coded as shown in the color map. Surface locations with outward deformations and shown in blue, whereas surface locations with inward deformations are shown in red. The figure clearly demonstrates small average distances around zero for controls and indicates locations of large differences in the affected and unaffected groups, mostly located in the atrium and occipital horn regions.
Discussion
This study examined the contribution of genetics, disease, and environment to lateral ventricle morphometry. The study design controlled for genetics, disease, family environment, and anatomical variability in the population by using schizophrenia patients, their unaffected MZ twins, healthy MZ and DZ twin pairs, and NR pairs of healthy controls. The NR healthy control population can be considered biased because it was composed of a random selection of one subject of each healthy MZ and DZ twin pair. The bias arises only when comparing differences between healthy twins with differences between nonrelated subjects. Because we assume that the selected unrelated population is a true sample of the general healthy population, we do not anticipate any significant effects on the observed results.
In this shape study, all lateral ventricles are scaled to the same volume to create shape measurements that are independent of size. We compared the presented results with the results of the shape analysis without scaling normalization. The latter are omitted for reasons of brevity. Because of the large volume variability, the shape analysis without scaling normalization showed a decreased statistical significance in the pairwise analysis and no statistical significance in the groupwise analysis. This scaling normalization would not be used in longitudinal studies, because the volume variability is not expected to be larger than the shape variability.
Pairwise Shape Differences Between Co-Twins. There was a trend for lateral ventricular volumes to be more similar in pairs of highly genetically related individuals, compared with NR pairs. Specifically, the trend for greater similarity in lateral ventricular volume between MZ twins, compared with NR pairs, as well as the significantly greater similarity in right lateral ventricular volume for DZ twins, compared with NR pairs, observed in the pairwise co-twin analysis, suggests that a relationship may exist between lateral ventricular size and degree of genetic relatedness. This finding is consistent with the results of previous studies (3), which show that the unaffected relatives of schizophrenia patients also tend to have enlarged lateral ventricular volumes. The results of the co-twin shape analysis imply that a relationship exists between lateral ventricular shape and genetic relatedness. Lateral ventricular shape was significantly more similar in healthy MZ twin pairs, DS MZ twin pairs, and healthy DZ twin pairs, compared with NR twin pairs. This conclusion also is supported by the finding that lateral ventricular shape was more similar in the highly genetically similar MZ twin pairs, compared with the less genetically similar DZ twin pairs.
Differences in lateral ventricular volume or shape similarity between pairs of DS MZ twins and healthy MZ twins are attributable to disease, factors related to disease (i.e., treatment with antipsychotic medication and differences in body weight, nutrition, or hydration) or to environmental factors not controlled for in the study (i.e., prenatal or perinatal injury). Because no statistically significant differences in volume or shape were observed between the DS MZ twin pairs and the healthy MZ twin pairs (see Table 2) and because the results of the average distance maps (Fig. 5) and the statistical maps (Fig. 6) are consistent with these findings, the results of the co-twin analysis show that genetic relatedness exerts a stronger influence than disease or disease vulnerability on lateral ventricular shape and volume.
Shape Difference Between Groups. In the co-twin analysis, similarity of volume and shape between different groups of subject pairs is examined. In contrast, the group analysis measures differences in mean ventricular volume between, and deviation of individual subjects from a healthy ventricle template in, three groups. The three groups examined are normal controls, schizophrenic patients, and their unaffected MZ twins. The group analysis addresses the following scientific question: does disease or vulnerability for disease alter ventricular size or shape? The results of the group analysis are shown in Figs. 7 and 8 and are discussed below.
Statistically significant differences in lateral ventricular volume could not be detected among groups of healthy controls, schizophrenics, or their unaffected co-twins. Volume measurements were not adequately sensitive to differentiate these groups and showed similar means and variances for all three groups. Volumes were corrected with intracranial cavity size to account for variations in head size. Statistically significant differences in lateral ventricular shape, normalized for unit size, were observed between the control group and the affected group (Table 2). Similarly, a significant difference in lateral ventricular shape was observed between the control group and the unaffected subjects. This finding demonstrates that ventricles of not only schizophrenics show alterations from healthy subjects but also their unaffected co-twins. There was no significant difference between this shape-deformation measure for the affected and unaffected groups.
A possible interpretation of the findings observed in the co-twin and group analysis is that lateral ventricle shape is strongly influenced by genetics. MZ twins DS have very similarly shaped lateral ventricles, suggesting that the influence of disease or disease-related factors on lateral ventricular shape is relatively small, compared with genetic factors. The significant ventricular shape differences between the healthy control population and the MZ twins discordant for schizophrenia, observed in the group analysis, could be explained by a combination of disease, disease-related, and genetic influences.
The analysis of the volume measurements was not adequately sensitive to show ventricular volume enlargement in schizophrenics, compared with healthy controls, an abnormality observed in many schizophrenia studies. There are several possible explanations for this finding. Most importantly, the sample size of the study is too low in comparison with the large ventricular volume variability. The combination of both female and male subjects in the same analysis further lowers the power of the study, because lateral ventricles are smaller in female subjects, compared with male subjects (40). Furthermore, enlargement of lateral ventricle size in schizophrenia also has been correlated with age, and thus the relatively young age of our subjects could be an additional factor.
Because differences in lateral ventricular shape between pairs and also across groups consistently reached statistical significance, whereas differences in lateral ventricular volume did not, our shape measure appeared superior to volume measurements for the purposes of differentiating groups with varying genetic inheritance and discordant for disease.
Along with prior shape studies of different brain structures (1, 19, 41), our results support the finding that shape differences confirm volume differences and show higher statistical significance, even when the volumetric differences do not reach significance. These results further emphasize the importance of shape-analysis studies when studying populations of naturally low sample sizes, such as in discordant twin studies.
Acknowledgments
This work was supported by National Cancer Institute Grant CA47982, a University of North Carolina Intel grant, National Institutes of Health Grant 156001393A1, and the Foundation of Hope (Raleigh, NC).
Author contributions: M.S. and G.G. designed research; M.S. performed research; M.S., D.R.W., D.W.J., and G.G. contributed new reagents/analytic tools; M.S. analyzed data; and M.S., J.A.L., R.K.M., and G.G. wrote the paper.
Abbreviations: MZ, monozygotic; DZ, dizygotic; DS, discordant for schizophrenia; NR, nonrelated; DSM-III-R, Diagnostic and Statistical Manual III Revised.
References
- 1.Gerig, G., Styner, M., Shenton, M. E. & Lieberman, J. A. (2001) in Medical Image Computing and Computer-Assisted Intervention, Lecture Notes in Computer Science, eds. Niessen, W. & Viergever, M. (Springer, Berlin), Vol. 2208, pp. 24-32. [Google Scholar]
- 2.Powchik, P., Davidson, M., Haroutunian, V., Gabriel, S. M., Purohit, D. P., Perl, D. P., Harvey, P. D. & Davis, K. L. (1998) Schizophr. Bull. 3, 325-341. [DOI] [PubMed] [Google Scholar]
- 3.McClure, R. K. & Weinberger, D. R. (2001) in Current Issues in the Psychopharmacology of Schizophrenia, eds. Breier, A., Tran, P. V., Herrera, J. M., Tollefson, G. D. & Bymaster, F. P. (Lippincott William & Wilkins, Philadelphia), pp. 27-56.
- 4.Bartley, A., Jones, D. & Weinberger, D. (1997) Brain 120, 257-269. [DOI] [PubMed] [Google Scholar]
- 5.Reveley, A. M., Chitkara, B. & Clifford, C. (1984) Psychiatry Res. 13, 261-266. [DOI] [PubMed] [Google Scholar]
- 6.Pfefferbaum, A. S., Swan, G. E. & Carmellia, D. (2000) Neurobiol. Aging 21, 63-74. [DOI] [PubMed] [Google Scholar]
- 7.Oppenheim, J. S., Tramo, M. J. & Gazzaniga, M. S. (1989) Ann. Neurol. 26, 100-104. [DOI] [PubMed] [Google Scholar]
- 8.Biondi, A., Nogueira, H., Dormont, D., Duyme, M., Hasboun, D., Zouaoui, A., Chantome, M. & Marsault, C. (1998) Am. J. Neuroradiol. 19, 1361-1367. [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson, P., Cannon, T. D., Narr, K. L., van Erp, T., Poutanen, V. P., Huttunen, M., Lonnqvist, J., Standertskjold-Nordenstam, C. G., Kaprio, J., Khaledy, M., et al. (2001) Nat. Neurosci. 4, 1-6. [DOI] [PubMed] [Google Scholar]
- 10.Noga, T. J., Bartley, A., Jones, D. W., Torrey, E. F. & Weinberger, D. R. (1996) Schizophr. Res. 22, 27-40. [DOI] [PubMed] [Google Scholar]
- 11.McNeil, T. F., Cantor-Graae, E. & Weinberger, D. R. (2000) Am. J. Psychiatr. 157, 203-212. [DOI] [PubMed] [Google Scholar]
- 12.Reveley, A. M., Clifford, C. A. & Murray, R. M. (1982) Lancet i, 540-541. [DOI] [PubMed] [Google Scholar]
- 13.Cannon, T. D., Thompson, P. M., van Erp, T. G., Toga, A. W., Poutanen, V., Huttunen, M., Lonnqvist, J., Standerskjold-Nordenstam, C. G., Narr, K. L., Khaledy, M., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 3228-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Narr, K. L., van Erp, T. G., Cannon, T. D., Woods, R. P., Thompson, P. M., Jang, S., Blanton, R., Poutanen, V., Huttunen, M., Lonnqvist, J., et al. (2002) Neurobiol. Dis. 11, 83-95. [DOI] [PubMed] [Google Scholar]
- 15.Narr, K. L., Cannon, T. D., Woods, R. P., Thompson, P. M., Kim, S., Asunction, D., van Erp, T. G., Poutanen, V., Huttunen, M., Lonnqvist, J., et al. (2002) J. Neurosci. 22, 3720-3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bookstein, F. L. (1997) Comput. Vis. Image Und. 66, 97-118. [Google Scholar]
- 17.Dryden, I. L. & Mardia, K. V. (1993) Sankhya 55, 460-480. [Google Scholar]
- 18.Cootes, T., Taylor, C. J., Cooper, D. H. & Graham, J. (1995) Comput. Vis. Image Und. 61, 38-59. [Google Scholar]
- 19.Styner, M., Lieberman, J. A. & Gerig, G. (2003) in Medical Image Computing and Computer-Assisted Intervention, Lecture Notes in Computer Science, eds. Ellis, R. & Peters, T. (Springer, Berlin), Vol. 2879, pp. 464-471. [Google Scholar]
- 20.Gerig, G., Styner, M., Jones, D., Weinberger, D. & Lieberman, J. (2001) in Mathematical Methods in Biomedical Image Analysis, eds. Staib, L. & Rangarajan, A. (IEEE Computer Society, Washington, DC), pp. 171-178.
- 21.Shen, L., Ford, J., Makedon, F. & Saykin, A. (2003) in Proc. SPIE Medical Imaging: Image Processing, eds. Sonka, M. & Fitzpatrick, M. N. (International Society for Optical Engineering, Bellingham, WA), Vol. 5032, pp. 253-264. [Google Scholar]
- 22.Pizer, S., Fritsch, D., Yushkevich, P., Johnson, V. & Chaney, E. (1999) IEEE Trans. Med. Imaging, 18, 851-865. [DOI] [PubMed] [Google Scholar]
- 23.Styner, M., Gerig, G., Lieberman, J., Jones, D. & Weinberger, D. (2003) Med. Image Anal. 7, 207-220. [DOI] [PubMed] [Google Scholar]
- 24.Golland, P., Grimson, W. E. L. & Kikinis, R. (1999) in Information Processing in Medical Imaging, Lecture Notes in Computer Science, eds. Kuba, A., Samal, M. & Todd-Pokropek, A. (Springer, Berlin), Vol. 1613, pp. 382-388. [Google Scholar]
- 25.Davatzikos, C., Vaillant, M., Resnick, S., Prince, J. L., Letovsky, S. & Bryan, R. N. (1996) J. Comput. Assist. Tomogr. 20, 88-97. [DOI] [PubMed] [Google Scholar]
- 26.Joshi, S., Miller, M. & Grenander, U. (1997) J. Patt. Recogn. Art. Intell. 11, 1317-1343. [Google Scholar]
- 27.Csernansky, J. G., Joshi, S., Wang, L. E., Haller, J., Gado, M., Miller, J. P., Grenander, U. & Miller, M. I. (1998) Proc. Natl. Acad. Sci. USA 95, 11406-11411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Csernansky, J. G., Wang, L., Jones, D. J., Rastogi-Cruz, D., Posener, J. A., Heydebrand, G., Miller, J. P., & Miller, M. I. (2002) Am. J. Psychiatry, 159, 2000-2006. [DOI] [PubMed] [Google Scholar]
- 29.Vogel, F. & Motulsky, A. G. (1986) Human Genetics: Problems and Approaches (Springer, Berlin), 2nd Ed.
- 30.McNeil, T. F., Cantor-Graae, E., Torrey, E. F., Sjostrom, K., Bowler, A., Taylor, E., Rawlings, R. & Higgins, E. S. (1994) Acta Psychiatr. Scand. 89, 196-204. [DOI] [PubMed] [Google Scholar]
- 31.Spitzer, R. L. & Williams, J. B. W. (1986) Structured Clinical Interview for DSM-III-R-Patient Version (New York State Psychiatric Institute, Biometrics Research, New York).
- 32.Belmaker, R., Pollin, W., Wyatt, R. J. & Cohen, S. (1974) Arch. Gen. Psychiatry, 30, 219-222. [DOI] [PubMed] [Google Scholar]
- 33.Van Leemput, K., Maes, F., Vandermeulen, D. & Suetens, P. (1999) IEEE Trans. Med. Imaging 18, 897-908. [DOI] [PubMed] [Google Scholar]
- 34.Brechbühler, C., Gerig, G. & Kübler, O. (1995) Comput. Vision Image Und. 61, 154-170. [Google Scholar]
- 35.Bookstein, F. L. (1991) Morphometric Tools for Landmark Data: Geometry and Biology (Cambridge Univ. Press, Cambridge, U.K.).
- 36.Styner, M., Gerig, G., Pizer, S. M. & Joshi, S. (2003) Int. J. Comput. Vision 55, 107-122. [Google Scholar]
- 37.Nichols, T. E. & Holmes, A. P. (2001) Hum. Brain Mapp. 1, 1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pantazis, D., Leahy, R. M., Nichols, T. E. & Styner, M. (2004) in IEEE Symposium on Biomedical Imaging, eds. Leahy, R. M. & Roux, C. (IEEE Press, Piscataway, NJ), pp. 1283-1286.
- 39.Friston, K. J., Holmes, A. P., Worsley, K. J., Poline, J. P., Frith, C. D. & Frackowiak, R. S. J. (1995) Hum. Brain Mapp. 2, 189-210. [DOI] [PubMed] [Google Scholar]
- 40.Yotsutsuji, T., Saitoh, O., Suzuki, M., Hagino, H., Mori, K., Takahashi, T., Kurokawa, K., Matsui, M., Seto, H. & Kurachi, M. (2003) Psychiatry Res. 122, 1-12. [DOI] [PubMed] [Google Scholar]
- 41.Shenton, M. E., Gerig, G., McCarley, R. W., Szekely, G. & Kikinis, R. (2002) Psychiatry Res. Neuroimag. 20, 15-35. [DOI] [PMC free article] [PubMed] [Google Scholar]