Abstract
Identification of effective combination therapies is critical to address the emergence of drug-resistant cancers, but direct screening of all possible drug combinations is infeasible. Here we introduce a CRISPR-based double knockout (CDKO) system that improves the efficiency of combinatorial genetic screening using an effective strategy for cloning and sequencing paired single-guide RNA libraries and a robust statistical scoring method for calculating genetic interactions (GIs) from CRISPR-deleted gene pairs. We applied CDKO to generate a large-scale human GI map, comprising 490,000 double-sgRNAs directed against 21,321 pairs of drug targets in K562 leukemia cells and identified synthetic lethal drug target pairs for which corresponding drugs exhibit synergistic killing. These included the BCL2L1 and MCL1 combination, which was also effective in imatinib-resistant cells. We further validated this system by identifying known and previously unidentified GIs between modifiers of ricin toxicity. This work provides an effective strategy to screen synergistic drug combinations at high-throughput and a CRISPR-based tool to dissect functional GI networks.
Despite progress in the development of targeted cancer therapies, evolution of resistance is common. To counter this, combination therapy is rapidly becoming the standard of care in a range of cancers where single agents are ineffective1. Repurposing existing drugs in combinations could provide new therapeutic possibilities with reduced cost and time for development, while potentially minimizing side effects by lowering the dosage requirement for each drug1–3. Discovering such drug combinations, however, is a major challenge since the number of possible combinations is too large to be empirically validated using traditional assays4.
Genetic interaction (GI) maps have been used successfully to study the coordinated behaviors of genes, and consist of systematic pairwise measures of the extent to which the phenotype of one mutation is modulated by the presence of a second mutation5. The pattern of synergistic and buffering interactions serves as a ‘phenotypic signature’ for each gene, and can be used to cluster genes with similar functions into pathways and complexes. These maps have been useful tools for predicting gene function, allowing dissection of complexes and pathways6–10 in a range of organisms5,7,9,11–15. Notably, a recent study identified conserved synthetic lethal interactions using a yeast GI map that translated into mammalian cells as potential cancer therapies16. We15 and others17 recently demonstrated scalable, rapid strategies to create pooled combinatorial shRNA and miRNA libraries which facilitated GI maps in mammalian cells. Creation of such maps using the CRISPR-Cas9 system, which allows for precise gene disruption with minimal off-target effects18–20, would be a transformative tool for dissection of genetic interaction networks.
Here, we have developed a scalable CRISPR-based double knockout (CDKO) system that enables massively parallel pairwise gene knockout. Although a number of groups have used CRISPR-Cas9 for multiplexed genome engineering20–23, our library design minimizes possible recombination24,25 and positional bias while enabling simple cloning and direct paired-end sequencing of sgRNAs. Furthermore, we develop a robust statistical scoring method for GIs from CRISPR-deleted gene pairs. Using this system in K562 chronic myeloid leukemia (CML) cells, we demonstrate two diverse applications: first, we conduct an ultra-high-throughput search for rare interactions, generating the largest mammalian GI map to date to our knowledge, comprising ~490,000 double-sgRNAs corresponding to 21,321 drug combinations. Based on the genetic data, we identify synergistic drug target combinations and show that the predicted target pairs translate to potent synergistic drug combinations in cell culture. In a second application, we independently validate the method on a dense network of genetic interactions by creating a GI map that uses interaction patterns to correctly classify known and novel regulators of ricin toxicity into functional complexes.
RESULTS
A scalable, efficient CRISPR double knockout (CDKO) system
We first aimed to design a pairwise sgRNA expression system that incorporated several key features (Fig. 1a): (1) efficient double-knockout, (2) limitation of lentiviral vector recombination due to long homologous sequences, (3) compatibility with paired-end deep sequencing, and (4) capacity for easy cloning and multiplexing. We tested two approaches to express pairs of sgRNAs from a lentiviral vector: a dual promoter system and a single promoter Csy4 sgRNA system. For the first, we designed a vector to limit homologous sequences by employing two distinct promoters (human and mouse U6) driving expression of each sgRNA (Fig. 1b). In the second approach, we adapted the Csy4-based multiplex gRNA expression system in which two sgRNAs are transcribed as a single RNA and cleaved into two by Csy4 RNase21. We compared the efficiency of both systems to delete GFP and mCherry in cells stably expressing the corresponding targets and Cas9 (or Cas9 and Csy4). We found that the two-promoter system showed far higher double knockout efficiency (86–88%) than the Csy4-based system (37%) without displaying substantial bias when the orientation of GFP and mCherry sgRNAs was flipped (Fig. 1b) and thus selected this strategy.
Figure 1. Development of a CRISPR double knockout (CDKO) platform to identify novel cancer drug combinations in high throughput.
(a) Drug combinations will be modeled by developing a dual sgRNA system to simultaneously knock out corresponding drug targets. (b) Design of i) a Csy4-based double-sgRNA expression system and ii) a dual promoter-based system; flow cytometry analysis of GFP and mCherry knockout efficiency. (c) Schematic of double-sgRNA library generation. Two sets of pooled oligos were synthesized, amplified, and ligated into separate lentiviral vectors with either human or mouse U6 promoters. sgRNA cassettes were digested out from the hU6 library and ligated into the mU6 library as a pool to create the double-sgRNA CDKO library. (d) Design of drug-targeting sgRNA library. (e) Experimental strategy: Separate mU6-driven and hU6-driven single sgRNA libraries were ligated together into a pooled dual sgRNA library. Cas9-expressing K562 cells were infected with the pooled high-coverage CDKO library, selected with puromycin, and grown in bioreactors at 1000X coverage for ~14 days. The frequency of the dual sgRNA elements in the final population and original plasmid library were quantified by deep sequencing to calculate the growth phenotype and genetic interaction of each DKO gene pair. Potent synergistic pairs were validated in vitro using pairs of drugs.
Oligonucleotide pools were separately cloned into lentiviral vectors with either hU6 or mU6 promoters to generate two single-sgRNA libraries (Fig. 1c). Subsequently, hU6 single-sgRNA cassettes were removed by restriction digest and ligated into the mU6 library to create a CDKO library. This system allows direct paired-end sequencing of double-sgRNA cassettes (Supplementary Fig. 1a,b) without relying on barcodes. This not only simplifies the structure of the oligos and the cloning steps, but should also limit the potential for confounding results from possible vector recombination, which could lead to incomplete sgRNA pairs or mismatched barcodes. Indeed, a vector with two mU6 promoters exhibited substantially reduced activity and increased recombination (Supplementary Fig. 1c–f).
To identify synergistic combinations of drug targets in K562 CML cells, we selected genes that fit a number of criteria (Fig. 1d). First, we chose genes with corresponding drugs in TTD26, DrugBank27, or IUPHAR/BPS28. To enhance the likelihood that targets would have activity in the selected cancer type, we chose genes expressed in K562 cells whose perturbation caused a moderate negative growth phenotype in both CRISPR-Cas929 and shRNA screens30 we previously performed in K562 cells. To enrich for synergistic pairs, genes with lethal single gene deletion phenotypes were removed as their phenotypes would not be further enhanced by additional gene deletions. For the 207 genes that met these criteria (Supplementary Table 1), we hypothesized that drugs targeting these genes would be toxic to K562 cells, and searched for interactions between them.
To generate the GI map, we selected the 3 most effective sgRNAs for each gene based on their phenotypes in our previous K562 genome-wide screens29. We included 79 sgRNAs that target regions of the genome with no annotated function (safe-targeting sgRNAs) as negative controls29. A total of 700 sgRNAs (Supplementary Table 2) directed against 207 drug targets (plus safe-targeting controls) were used to create a CDKO library (DrugTarget-CDKO) comprised of 490,000 double-sgRNAs, corresponding to 21,321 drug target combinations (Fig. 1e). Cas9-expressing K562 cells were infected with this library and maintained in exponential growth phase for 14 days (~14 cell doublings) in duplicate, after which the frequencies of double-sgRNA cassettes were quantified by deep sequencing.
Precise phenotypic measurements of a large CDKO library
To assess whether the diversity of the large DrugTarget-CDKO library was maintained throughout library generation, delivery, and screening, we examined the distribution of library elements in the plasmid library, post-infection and puromycin selection day 0 (T0), and day 14 (T14) (Fig. 2a). 92% of the expected 490,000 double-sgRNAs were detected in both the plasmid and T0 samples, suggesting this large library could be efficiently cloned and delivered to cells. 98.7% of the observed gene pairs had at least 6 double-sgRNAs, with 75.2% having the maximum of 9 double-sgRNA combinations per gene pair (Fig. 2b). Since low sequencing read depth could add noise to the data, we filtered out double-sgRNAs that had read counts below the determined minimum threshold (Supplementary Fig. 2a, Supplementary Table 3).
Figure 2. 490,000-element DrugTarget-CDKO library can be efficiently assembled and delivered into cells and shows no positional bias.
(a) Cumulative distribution of sequencing reads for double-sgRNAs. Read counts were normalized by total reads of each sample and the cumulative sums of double-sgRNAs were plotted as relative percentages of the number of expected double-sgRNAs. (b) Histogram showing the number of double-sgRNAs per gene pair. 98.7% of the 42,319 detected gene pairs have more than 6 double-sgRNA combinations. (c) Growth (γ) phenotypes for two single genes and the corresponding gene pair were calculated from the double-sgRNA frequencies in the T14 sample and plasmid library. For γ phenotypes of single genes, all possible double-sgRNA combinations of the 3 gene-targeting sgRNAs and 79 safe-sgRNAs were measured. For the γ phenotype of a given gene pair, 9 double-sgRNA combinations were measured. Blue dotted lines mark the minimum threshold for read counts (50). (d) Minimal positional bias in the DrugTarget-CDKO library. γ phenotypes of double-sgRNAs were compared between both orientations. (e) High reproducibility of measured γ phenotypes between two experimental replicates.
To quantify GI (deviation of an observed double knockout phenotype from that expected from the corresponding two single knockouts), high-precision phenotypic measurements of both single and double knockouts are required. We defined growth phenotype quantitatively as gamma (γ) adapted from previous shRNA and CRISPRi-based screens (see methods)15,31,32. γ phenotype is expressed as a phenotype Z score (pZ)32 by dividing the phenotype by the standard deviation of all safe-sgRNA pairs (Safe_Safe) (see methods). To calculate γ for a single gene, we used the median of all double-sgRNAs in which the 3 sgRNAs targeting this gene are paired with the 79 safe-sgRNAs (Fig. 2c). To calculate the γ of a gene pair, we used the median of the 9 double-sgRNAs (3 × 3) targeting the gene pair.
Single geneγ phenotypes in the CDKO vector showed strong correlation (Pearson coeff.=0.87) with γ from previous single-sgRNA genome-wide screens29 (Supplementary Fig. 2e), suggesting that effectiveness of deletion is preserved in our double-sgRNA system. Furthermore, γ phenotypes of double-sgRNAs (Fig. 2d) and corresponding gene pairs (Supplementary Fig. 2c) were similar in both orientations and γ correlated well between experimental replicates (Fig. 2e, Supplementary Fig. 2d). Thus, the order of sgRNAs within the double-sgRNA cassettes had minimal impact on knockout efficiency, and measured phenotypes were highly reproducible.
Calculation of GI scores from CRISPR knockout phenotypes
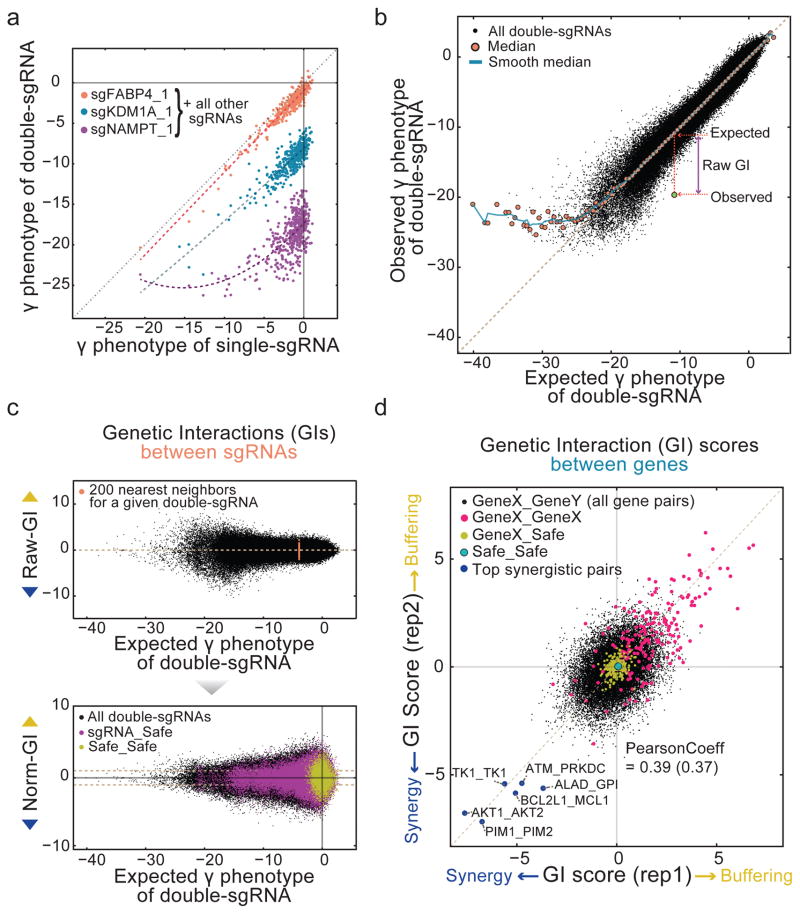
Previously, we calculated GI of double-shRNAs as the deviation from a linear fit of all double-shRNA phenotypes that combine the same single-shRNA with second shRNAs of interest15. To determine whether the same linear relationship could be used to calculate GIs from pairwise CRISPR knockouts, we examined three groups of sgRNAs, with either a phenotypically weak (sgFABP4_1), medium (sgKDM1A_1), or strong (sgNAMPT_1) sgRNA in one position of the double-sgRNA (Fig. 3a). Notably, the NAMPT-containing double-sgRNA phenotypes leveled off as the negative γ phenotype of the second sgRNAs increased, whereas the FABP4- or KDM1A-containing double-sgRNAs showed linear relationships with second sgRNAs. This suggested that the assumed linear relationship would not always hold true as the phenotype of a double-sgRNA increased.
Figure 3. Strategy to calculate quantitative genetic interaction scores.
(a) Single-sgRNA γ phenotypes plotted against corresponding double-sgRNA γ phenotypes in combination with a sgRNA of interest. sgRNAs paired with a weak or moderate γ phenotype sgRNA (FABP4 or KDM1A) showed a linear relationship between single-sgRNA and double-sgRNA γ phenotypes. For sgRNAs paired with a strong γ phenotype sgRNA (NAMPT), γ phenotypes of double-sgRNAs quickly leveled off as γ phenotypes of single-sgRNAs become more negative. Dotted lines mark polynomial fitting (n=2).
(b) Measuring genetic interactions of double-sgRNAs as deviations from the expected double-sgRNA γ phenotype. Observed and expected γ phenotypes of all double-sgRNAs were plotted (black dots). Expected γ phenotypes were calculated by sum of the two single sgRNA γ phenotypes. Medians of binned data were plotted (orange closed circle) and connected by a smooth median line (blue line). Deviations from the median line were defined as genetic interactions (see methods). (c) Raw-GIs were plotted against the expected γ phenotypes of double-sgRNAs (top panel). Positive raw-GIs are buffering while negative Raw-GIs are synergistic. Each Raw-GI was normalized by the standard deviation of the 200 nearest neighbors in terms of the expected γ phenotype. Normalized GIs (Norm-GI) were then plotted (bottom panel). In this plot, double-sgRNAs comprised of only safe-sgRNA pairs (yellow dots) and all sgRNAs paired with safe-sgRNAs (purple dots) showed symmetric distributions centering around 0 Norm-GI, confirming that safe-sgRNAs generally do not interact with other sgRNAs. (d) T-value based GIT scores (see methods) plotted between two experimental replicates. GI scores of control pairs - genes paired with safe-sgRNAs (yellow dots) and Safe_Safe pairs (cyan closed circle) - showed negligible genetic interactions as expected. Gene pairs comprised of same genes are marked as pink dots. The five most synergistic pairs by rank-sum of GIT scores of two replicates are marked as dark blue dots. The Pearson correlation after same-gene targeting pairs were removed is reported in parentheses.
Therefore, to systematically analyze how this relationship changed with the γ phenotype of double-sgRNAs, we plotted the observed phenotypes against the expected phenotypes for all double-sgRNAs. Expected double phenotypes were calculated as the phenotypic sum of the corresponding single-sgRNAs (Fig. 3b). For most sgRNA pairs, we observed a strong linear relationship with slope close to 1, indicating that the observed γ phenotypes of most double-sgRNAs could be reliably predicted by the simple sums of individual sgRNA phenotypes. However, as the expected γ phenotypes dropped below ~−20 pZ, the observed γ phenotypes abruptly leveled off. While limited sequencing depth can contribute to the plateau (Supplementary Fig. 2f), as discussed below this can also occur due to phenotypic saturation.
To account for this phenotypic plateau, we calculated GIs between sgRNAs (Raw-GIs) as the deviation from the smoothed line connecting the medians of the observed phenotypes of double-sgRNAs binned over the expected phenotypes (see methods) as similarly used for saturable GI phenotypes8. We defined GIs as buffering (positive sign) if the double knockout phenotype shifted toward that of the Safe_Safe controls, while deviations away from Safe_Safe controls were defined as synergistic (negative sign). Buffering GIs have also been referred to as alleviating7 or positive interactions6, whereas synergistic GIs are sometimes referred to as aggravating7 or negative6 interactions.
We also found that the variation of Raw-GIs increased as the negative γ phenotypes of double-sgRNAs increased (Fig. 3c, top panel). This systematic bias would thus over-estimate the GI for double-sgRNAs with large γ phenotypes. To account for the increased variation associated with phenotype size, the Raw-GI for a given double-sgRNA was divided by the standard deviation of Raw-GIs from the 200 nearest neighbors. This normalized GI (termed Norm-GI) should allow fair comparison of GIs across the range of γ phenotypic strengths and experimental replicates (Fig. 3c, bottom panel, see methods).
To quantify GIs at the gene level (Fig. 3, Supplementary Fig. 3), we took advantage of the fact that each pair of genes has 18 different double-sgRNAs (9 for each of two orientations). Furthermore, the library contains many non-interacting double-sgRNA controls: Safe_Safe (6,241) and sgRNAs paired with a safe-sgRNA (49,059) (Fig. 3c, bottom panel). This allowed us to compare the double-sgRNAs for each gene pair to these non-interacting sgRNA pairs to calculate scores that reflect both the consistency and strength of GIs. We then tested two scoring systems - GIT score (Fig. 3d, Supplementary Fig. 3a) and GIM score (Supplementary Fig. 3b) - which are based on a modified t-value33 and a signed logarithmic p value determined by Mann-Whitney U test15, respectively (Supplementary Table 4, see methods). Nearly identical synergistic and buffering gene pairs were identified using each method, suggesting that both scoring systems robustly measure GIs in the DrugTarget-CDKO map.
We evaluated gene-level interactions between drug targets in the library as GIT scores (Fig. 3d). As expected, genes showed minimal interactions with safe-targeting controls. Furthermore, strong buffering GIs were observed between a gene and itself (Supplementary Fig. 3c), suggesting that overall the CDKO system achieved efficient double knockout since incomplete knockout would lead to more additive or synergistic interactions between two sgRNAs targeting the same genes. A few genes appeared self-synergistic (e.g. TK1_TK1), likely from a disproportionate increase in gene deletion efficiency when two different ineffective sgRNAs are used to target the same gene (Supplementary Fig. 4, see Supplementary Text).
As drug targets were chosen from unrelated pathways, most gene pairs did not interact and had small GIT scores, consistent with low interaction frequencies in previous GI maps for unrelated genes6,34,35 (see Supplementary Text). Note that the low density and overall small effect of interactions result in lower correlation between GIs than the primary gene effects (Fig. 2d,e), which are highly reproducible. However, when we looked at the rare synergistic gene pairs that were well-separated from the cloud of weakly interacting pairs, a number of strong interactions were observed. These consisted of several pairs with no annotated interaction, as well as many gene pairs for which synergistic interactions are either supported by previous studies or readily explained by the known biological functions of the corresponding genes. For example, the AKT1_AKT2 and PIM1_PIM2 pairs are highly homologous kinase isoforms with known functional redundancy36,37 and the BCL2L1_MCL1 and ATM_PRKDC pairs have similar biological functions (apoptosis and DNA damage repair, respectively). Notably, it has been shown that when both PIM1 and PIM2 are deleted, oncogenic ABL-mediated transformation in hematopoietic cells cannot occur38. Because K562 growth is driven by BCR-ABL, the results suggest that cancer-specific synthetic lethalities can be identified with the CDKO system. Overall these results suggest that even when interactions are rare, the CDKO system can efficiently identify biologically meaningful synergistic relationships.
Validation of the CDKO system on a dense ricin GI map
Although the DrugTarget-CDKO map revealed rare potent synergistic targets, another useful feature of GI maps is the ability to cluster functionally related genes by the similarity of their genetic interaction patterns7,33. Since genes for the DrugTarget-CDKO library were selected across a wide variety of pathways, genetic interactions were sparse6,7,34,35 (see Supplementary Text), and consequently did not result in meaningful clustering (with a few exceptions) (Supplementary Fig. 5). Therefore, we sought to rigorously evaluate our CRISPR-based GI map system in a different context, where frequent genetic interactions among functionally related genes yield interaction patterns that can be used to cluster genes into pathways and complexes. For this purpose, we generated a GI map focused on the biology of the AB-type protein toxin ricin, for which we have extensive data on the dense network of known physical, genetic, and functional interactions between regulators of toxicity15,39,40. We chose a set of 79 non-essential genes that modulated the cellular susceptibility to ricin (Supplementary Table 5,6) in a genome-wide CRISPR-Cas9 ricin screen29 (Supplementary Fig. 6a), and included both genes identified as ricin regulators in our previous shRNA-based interaction map15, as well as novel modifiers identified only in the CRISPR screen (discussed below). We generated a Ricin-CDKO library (80,656 double-sgRNAs for 3,081 gene pairs), and measured the ricin-resistance phenotype (ρ) of double-sgRNAs as previously described15 (Supplementary Fig. 6b–f, see methods).
As expected, we observed more frequent and larger genetic interactions between ricin pathway regulators compared to the DrugTarget-CDKO library (Supplementary Fig. 6g). GIs were highly reproducible between experimental replicates (Supplementary Fig. 6g, Supplementary Table 7), as were the correlations of GI patterns between sgRNA pairs (Fig. 4a), though we also observed phenotypic saturation for highly ricin-resistant gene pairs (Supplementary Fig. 6f, top right). Furthermore, GI patterns were more well correlated between two sgRNAs targeting the same gene, as expected for on-target sgRNAs acting through the same mechanism (Fig. 4a,b).
Figure 4. A CRISPR-based GI map of ricin pathway regulators validates the CDKO platform.
(a) Correlations of GI profiles between two sgRNAs were compared between two experimental replicates. sgRNAs targeting the same gene are marked in pink. The pearson correlation after same-gene targeting pairs were removed is reported in parentheses. (b) The distributions of correlations of GI profiles for all sgRNA pairs (blue) and for sgRNAs pairs targeting the same gene (orange). Medians of the distributions are marked with the dotted lines. (c) GI map of ricin modulators. GIM scores of all gene pairs were calculated and color-coded by a yellow-cyan heatmap. Genes were hierarchically clustered by their correlation of GI profiles. ρ phenotypes of individual genes were marked in sidebars with a red-blue heatmap. Previously reported protein complexes are labeled. (d) Plot showing the percentage of the top N gene pairs that have corresponding protein interactions in the STRING database, sorted by correlation of GI profiles (orange), buffering GIs (blue), or synergistic GIs (brown) for known protein interactions. Randomly sorted gene pairs were marked in pink. Data were plotted for N>10. (e) Relationship between the correlation of GI profiles and the predictive power for known protein interactions. Percentage of reported PPIs from STRING were averaged over a moving window of 20 data points against the correlation of GI profiles.
We next used hierarchical clustering to generate an interaction map for the 79 × 79 matrix of ricin interactors (Fig. 4c). The resulting map clearly identified a number of functional complexes with known involvement in ricin biology, including the vesicle trafficking TRAPP, COPII and GARP complexes. Our previous shRNA-based interaction map identified two distinct mammalian TRAPP complexes, mTRAPPII and mTRAPPIII, which contain unique subunits (TRAPPC9/10 or TRAPPC12/13, respectively) and associate with either COPI or COPII complexes, respectively15. Not only did the Ricin-CDKO map separate the TRAPPC9/10 cluster clearly from the TRAPPC12/13 cluster, but these clusters were also grouped with their corresponding COPI or COPII modules.
When we compared 91 common genetic interactions between the Ricin-CDKO map and our previous shRNA-based Ricin GI map15, they showed relatively high correlation (Pearson coeff.=0.606, Supplementary Fig. 7a), suggesting that the new CRISPR-based map recapitulated previously reported interactions. We do note some differences between two ricin maps, which is not unexpected given that we and others have observed significant differences between shRNA-based knockdown and CRISPR KO studies30,41. However, both maps identified common protein complexes such as TRAPPII, TRAPPIII, COPII, and GARP (Supplementary Fig. 7b,c). Overall, these data suggest that functional clustering of genes is robust regardless of gene perturbation methods.
The Ricin-CDKO map identified a number of known and previously unidentified protein complexes whose roles in ricin biology remain unexplored. For example, a number of genes involved in protein N-glycosylation, the mTOR pathway or chromatin remodeling/transcription control complexes were identified as protective factors against ricin toxicity when deleted. (Fig. 4c, see legend). In each case, the corresponding genes were correctly clustered together in complexes according to previous annotations42–44.
Next, we systematically tested the ability of GIs to predict the 246 known PPIs present in the STRING45 database among the 79 genes in the ricin map (Supplementary Table 8). Similar approaches have been successful for systematic validation of GI maps in yeast10,33. We sorted the 3,081 gene pairs present in the Ricin-CDKO library by the correlation of their GI patterns, their buffering GIs, or their synergistic GIs and quantified the percentage of the known protein interactions captured by the top N pairs (Fig. 4d, Supplementary Fig. 8a). The correlation of GI patterns had a high predictive power for known protein interactions (Fig. 4d,e): among the 66 gene pairs with GI correlation > 0.5, 32% of the pairs had previously validated interactions, significantly higher than the 4.8% of 66 randomly sorted gene pairs (Supplementary Fig. 8b, Supplementary Table 9). Additionally, we observed that buffering GIs had strong predictive power for PPIs. This is consistent with previous work showing that gene pairs with highly correlated GIs and buffering interactions often exist in the same protein complexes or pathways7,33. Together these results show that the Ricin-CDKO map could correctly classify complex functional relationships among ricin modulators.
Validation of top interacting drug-target gene pairs
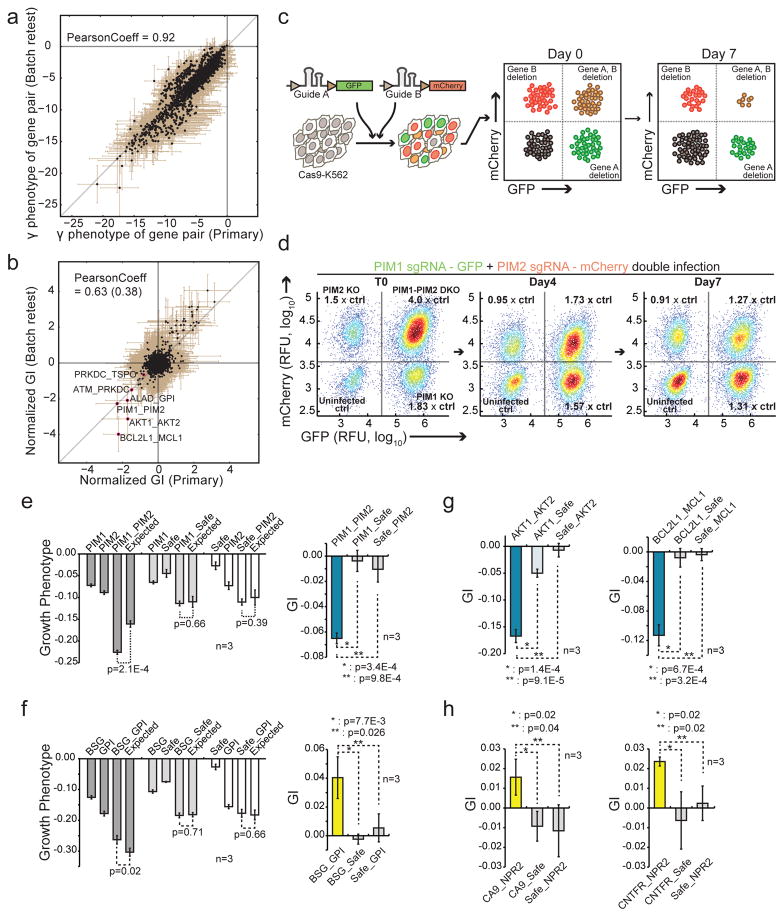
We next sought to verify drug target pairs from the initial DrugTarget-CDKO map in high throughput using a ‘batch retest’ mini-map. In previous studies, we15 and others46,47 have seen that measurement of smaller libraries at higher coverage reduced false positives/false negatives from larger primary screens. We selected the top synergistic and buffering pairs from the DrugTarget-CDKO map and generated a mini-map (Supplementary Table 10,11) consisting of 82,369 (287 × 287) double-sgRNAs targeting 3,081 drug combinations (79 X 79 genes). Compared to the DrugTarget-CDKO map, the batch retest was performed at 3X higher cell coverage and ~3X higher read-coverage (see methods). Both γ phenotypes (Fig. 5a) and measured GIs (Fig. 5b) in the primary DrugTarget-CDKO and batch retest screens correlated well and identified the same highly synergistic pairs, indicating that hits observed in the large interaction map were robust.
Figure 5. Validation of predicted genetic interactions using individual sgRNAs.
(a) Reproducibility between γ phenotypes in the primary and batch retest DrugTarget-CDKO screens. Data represent mean ± SEM. (b) Reproducibility between Norm-GIs in the primary and batch retest screens. Data represent mean ± SEM. The Pearson correlation after same-gene targeting pairs were removed is reported in parentheses. (c) Schematic for dual sgRNA validation assay. Cas9-expressing K562 cells were infected with lentiviruses expressing one sgRNA in a GFP vector and another sgRNA in an mCherry vector. The abundances of the four resulting fluorescent populations (uninfected, GFP, mCherry, and GFP+mCherry) were measured after 7 days. (d) K562 cells expressing a PIM1-targeting sgRNA (GFP) and a PIM2-targeting sgRNA (mCherry). The growth phenotype is calculated by measuring the relative depletion of the single-infected and double-infected cells at day 7 vs. day 0. (e,f) Quantification of growth phenotypes and genetic interaction scores for indicated double sgRNA infections compared to single sgRNA_safe-targeting controls (see methods). The PIM1_PIM2 sgRNA pair is synergistic as predicted while the BSG_GPI pair is buffering as predicted. Data represent mean ± SD from 3 replicate cultures. (g,h) Additional high-confidence synergistic and buffering gene pairs validate with predicted genetic interactions. Data represent mean ± SD (n=3) from replicate cultures.
To test individual gene pairs, their respective single sgRNAs (Supplementary Table 12) were cloned into vectors tagged with either GFP or mCherry. To measure their GIs, we simultaneously infected Cas9-expressing K562 cells with lentiviruses containing each sgRNA to generate four populations of cells: cells that contain both sgRNAs (GFP-mCherry double-positive), one sgRNA (GFP or mCherry single-positive), and uninfected cells (Fig. 5c). Tracking the change in abundance of each population relative to the uninfected control over time allowed us to measure both the single and double knockout phenotypes of the sgRNAs, which were subsequently used to calculate the GI (see methods). For PIM1_PIM2, a top synergistic pair (Fig. 5d,e), the double knockout phenotype was significantly larger (p=2.1E-4) than the sum of the single knockout phenotypes, resulting in a synergistic GI, whereas the corresponding negative control pairs did not interact.
GIs for the 6 most synergistic and 3 most buffering pairs (ordered by rank-sum of GIT and GIM scores) were individually validated (Fig. 5e–h, Supplementary Fig. 9a). Testing several pairs with a second pair of sgRNAs yielded similar results (Supplementary Fig. 9b). A subset of the same pairs were also validated as synergistic with the double-sgRNA vector used in the DrugTarget-CDKO library, whereas the sgRNAs shuffled into predicted non-interacting pairs were confirmed to be non-synergistic (Supplementary Fig. 9c,d). A summary of the sgRNA validations is presented in Supplementary Table 13. Efficient knockout by the sgRNAs was confirmed by analysis of indel frequency using TIDE48 (Supplementary Fig. 9e).
Identification of synergistic drug pairs using a GI map
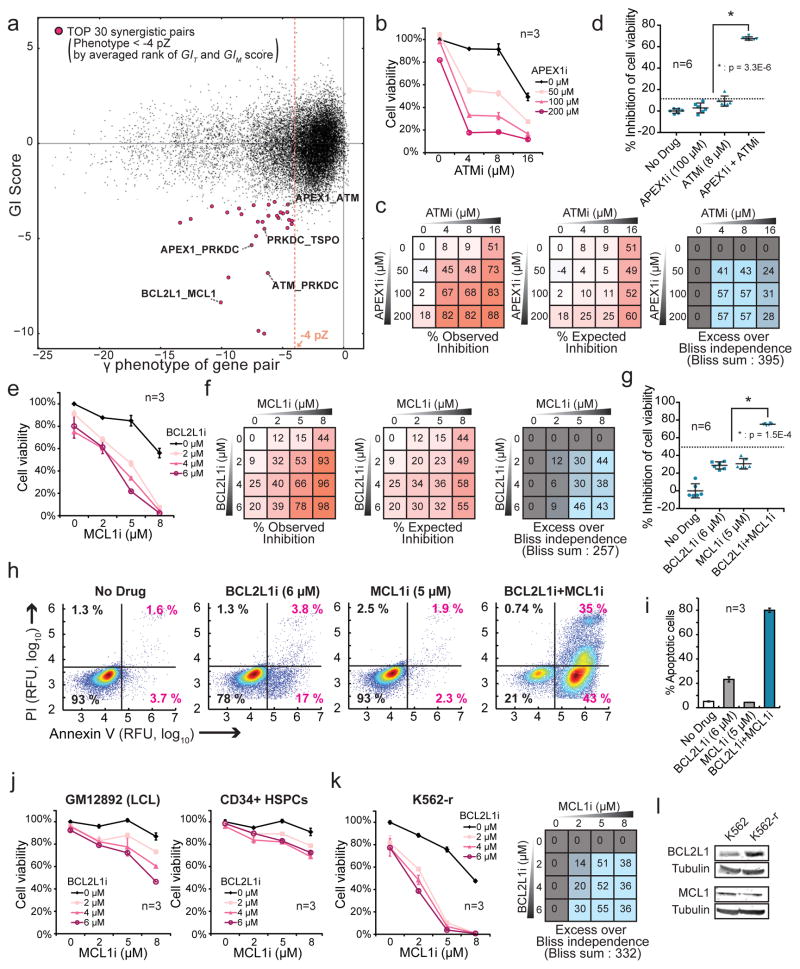
As cancer therapeutic targets, gene pairs that exhibit both strong synergistic GIs and a severe growth phenotype when targeted in combination would be ideal. To identify these pairs, we exploited the ability of the DrugTarget-CDKO map to simultaneously measure two critical features of gene pairs - γ phenotypes and GIs. The 30 most synergistic gene pairs with severe γ phenotypes (< −4 pZ) (Fig. 6a) included pairs for which no annotated synergy had been observed, as well as gene pairs in related pathways, such as apoptosis modulators, epigenetic modulators, and DNA repair genes (Supplementary Table 14). We selected several candidates based on the availability of highly selective inhibitors and tested whether the pairs of corresponding drugs synergistically inhibited the growth of K562 cells as predicted in the GI map. Overall, we tested 11 drug pairs which resulted in 5 true positive synergistic interactions, 4 true non-interactions, 1 false positive, and 1 false negative (Supplementary Table 13).
Figure 6. Predicted synergistic gene pairs translate to synergistic drug combinations.
(a) GIT scores and γ phenotypes of gene pairs were plotted together. We set a γ phenotype cutoff of −4 pZ for strongly toxic drug combinations and highlight the 30 most synergistic gene pairs by rank-sum of GIT and GIM scores (marked in pink). (b,e) K562 cells were treated with (b) APEX1 (CRT0044876) and ATM (KU-60019) inhibitors or (e) BCL2L1 (A-1155463) and MCL1(A-1210477) inhibitors alone and in combination at the indicated concentrations for 72 h. Cell viability relative to the no drug control was determined by measuring live cell number using flow cytometry (FSC/SSC). (c,f) Drug synergy represented by excess over Bliss independence is calculated by subtracting the % expected inhibition from the % observed inhibition at each combination of drug doses. (d,g) % inhibition of cell viability upon treatment with the indicated inhibitors in a separate experiment with 6 replicate cultures. P-values represent significant differences by one-tailed t-test between the observed effect of the drug combination and the expected effect (dotted lines) based on the Bliss independence model. (h,i) K562 cells were treated with indicated drugs for 48 h and assessed by flow cytometry for Annexin V-FITC and propidium iodide (PI) staining. Plots in h are representative of three independent experiments and the percentages of Annexin V positive cells are quantified in i. (j,k) Cell viability plots for GM12892 (LCL) and CD34+ (bone marrow-derived hematopoietic stem-progenitor) cells in j or imatinib-resistant K562 cells in m treated with BCL2L1 (A-1155463) and MCL1(A-1210477) inhibitors alone and in combination at the indicated doses for 72 h. (l) BCL2L1 and MCL1 protein levels were compared by immunoblot analysis between lysates from parental and imatinib-resistant K562 cells. Data represent mean ± SD from 3 replicate cultures unless otherwise noted.
We first examined APEX1_ATM because of its mechanistic similarity to the well-known synthetic lethality between BRCA and PARP. Synthetic lethality between base excision repair (BER) and double-strand break (DSB) repair has been successfully exploited to treat BRCA-deficient ovarian cancer patients with PARP inhibitors49, and APEX1 (BER) and ATM (DSB repair) are involved in the same pathways. We observed potent synergistic inhibition of cell viability using a combination of inhibitors against APEX1 and ATM as calculated by the Bliss independence model (Fig. 6b–d). Furthermore, the drug pair synergistically induced DNA damage γ (Supplementary Fig. 10a–c) and apoptosis (Supplementary Fig. 10d,e). Drug synergy was also observed between inhibitors of APEX1 and PRKDC and of ATM and PRKDC (Supplementary Fig. 11a,b). These results are consistent with recent studies demonstrating synthetic lethality between APEX1 shRNA knockdown and inhibitors of DSB repair, including ATM and PRKDC inhibitors50 and with the observed behavior of PRKDC inhibitors used in ATM-deficient cancers51. Additionally, a synergistic interaction was identified between inhibitors of PRKDC and TSPO (Supplementary Fig. 11c), which encodes a mitochondrial translocator protein that is thought to play a role in cell death regulation52. As expected, TXN_XPO1, MCL1_PRKDC, TSPO_XPO1, and CARM1_XPO1, predicted to have little interaction, did not show substantial synergy with small molecule inhibitors (Supplementary Fig. 11d–g). However, NAMPT_XPO1, predicted to be synergistic did not show drug synergy (false positive) (Supplementary Fig. 11h), and BCL2L1_XPO1 demonstrated drug synergy despite having little GI (false negative) (Supplementary Fig. 11i). Overall, these data show that the use of targeted inhibitors could generally recapitulate genetic interactions observed in the DrugTarget-CDKO map.
Because of its strong synergy in the DrugTarget-CDKO map, we also examined combination treatment using BCL2L1 (BCL-XL) (A-1155463) and MCL1 (A-1210477) inhibitors. Although synergy might be expected between BCL2L1 and MCL1, which encode anti-apoptotic proteins of the Bcl-2 family, cancers differ markedly in their dependence on particular pro- and anti- apoptotic proteins53, and a direct test of combined drug inhibition of BCL2L1 and MCL1 has not been tested in BCR-ABL-driven cancers to our knowledge. We observed robust synergy (Fig. 6e–g) and induction of apoptosis with these drugs (Fig. 6h,i). In addition, strong synergy could be seen when the BCL2L1 inhibitor was coupled with another MCL1 inhibitor (UMI-77) (Supplementary Fig. 11j), indicating the observed synergy is likely due to on-target effects. Furthermore, combined BCL2L1 and MCL1 treatment was synergistic in MV4;11 AML cells (Supplementary Fig. 11k). Because effective cancer therapeutics require a therapeutic window for cancer-selective toxicity, we additionally tested the BCL2L1 and MCL1 combination treatment in two non-cancerous cell lines - immortalized GM12892 lymphoblastoid cell line (LCL) and primary CD34+ hematopoietic stem-progenitor cells (HSPCs) isolated from human cord blood. The treatment was less effective and synergistic in both cell types (Fig. 6j), suggesting this combination may be selective for certain cancer cells over normal cell types.
Although tyrosine kinase inhibitors such as imatinib that target BCR-ABL have been effective in treating CML, around 33% of CML patients treated with imatinib do not achieve complete cytogenetic response, largely due to resistance54. Notably, when applied to imatinib-resistant K562 (K562-r) cells, the BCL2L1_MCL1 drug combination was even more synergistic compared to parental K562 cells (Fig. 6k). We found that K562-r had markedly increased levels of BCL2L1 expression, suggesting a greater dependence upon anti-apoptotic machinery (Fig. 6l). Given that overexpression of BCL2L1 and MCL1 has been associated with cancer survival and drug resistance55 and that K562 cells actively regulate anti-apoptotic machinery in response to imatinib (Fig. 6l)56,57, co-treatment with BCL2L1 and MCL1 inhibitors in addition to imatinib may be an effective strategy to prevent resistance in CML.
DISCUSSION
High-throughput interrogation of cancer dependencies have successfully revealed oncogenes58,59, drug resistance mechanisms60,61, and synthetic lethal interactions62,63, and have led to the development of new therapies that are now being tested in clinical trials62,63. However, these studies have primarily used single-gene targeting libraries, limiting their capacity to interrogate combinations of targets for effective therapies.
To address this, we developed and optimized a scalable pairwise CRISPR-Cas9 gene deletion platform that allows for easy cloning, minimized recombination, and paired-end sequencing. We also establish robust scoring methods for calculating genetic interactions between CRISPR-deleted gene pairs. This system enabled a rapid search through combinations of drug targets at an unprecedented scale - 490,000 sgRNA pairs directed against 21,321 drug target combinations. From this large space of potential candidates, we identified a small number of highly synergistic drug combinations in K562 CML cells. In particular, the BCL2L1_MCL1 drug combination demonstrated a strong synergy. Given the importance of apoptosis evasion in cancer55, there has been enormous interest in targeting the anti-apoptotic BCL2 family for cancer therapy64. BH3 mimetics such as the BCL2L1/BCL2-targeting navitoclax and BCL2-specific venetoclax have demonstrated efficacy in patients in a variety of cancers. Additionally, these molecules can sensitize cancers to other drugs; numerous clinical trials are underway to evaluate these in combination with standard of care therapies55.
The anti-apoptotic Bcl-2 family members act in parallel to block apoptosis, and MCL1 upregulation has been reported as a mechanism of resistance to Bcl-2 family inhibitors in certain cancers55,64 and co-targeting of anti-apoptotics has been explored in certain contexts65,66. Our CRISPR-based drug target interaction map identified BCL2L1_MCL1 as an especially potent synergistic pair in BCR-ABL-driven CML (K562). Furthermore, we showed that imatinib-resistant K562 cells have markedly increased sensitivity to combination BCL2L1 and MCL1 drug treatment. Given that previous studies have shown synergy between imatinib and BCL2 family inhibitors67, combining imatinib with both BCL2L1 and MCL1 inhibition may represent an effective strategy to combat resistance in CML.
The results of genetic screens can suggest promising candidates for drug development when potent, selective drugs are unavailable. For example, the GI map identified many promising synergistic drug target pairs such as CDC25A_RELA and CIT_STMN1 that currently lack specific drugs, but represent promising synergistic targets based on their known biological roles in NF-kB signaling68 and microtubule regulation69 respectively (Supplementary Table 14). The map also identified a number of drug targets which show weak synergy or additivity that might have therapeutic value in combination (for example, TXN_XPO1, for which potent drugs in clinical trials exist, Supplementary Fig. 11d).
Beyond the ability to search for rare synthetic lethal pairs, we show here that systematic, quantitative phenotypic measurements of interactions between CRISPR-deleted genes can be effectively used to define functional relationships and pathways. Focusing on the well-validated physical and genetic interactions of the ricin pathway, we created a dense map of interactions between regulators that correctly classified genes - including previously unidentified ricin modifiers from a genome-wide CRISPR screen - into known physical complexes and pathways. Furthermore, the similarity of interaction patterns accurately predicted membership in protein complexes independently reported in the STRING PPI database (Fig. 4d).
In summary, our work demonstrates a highly robust, efficient method that can be used to search through the expansive space of potential synergistic drug target pairs for use in cancer therapies, and might even facilitate personalized targeted cancer therapies when applied in diverse cancer types or patient-derived samples. More generally, we expect that this system will enable systematic dissection of genetic networks in mammalian cells.
METHODS
Cell culture
K562 cells (ATCC) were cultured in RPMI 1640 (Gibco) supplemented with 10% FBS (HyClone), penicillin/streptomycin, and L-glutamine. GM12892 cells (gift from S. B. Montgomery) were cultured in RPMI, supplemented with 10% FBS, penicillin/streptomycin, and GlutaMAX (Gibco). The imatinib-resistant K562-r cell line was generated by culturing K562 cells in the presence of increasing dosages of imatinib for 6 weeks. Dosages of imatinib were doubled weekly from 50 nM to 1.6 uM. CD34+ HSPCs isolated from primary bone marrow (gift from S. Mantri and M. Porteus) were cultured in StemSpan SFEM II media (StemCell Technologies) supplemented with 100 ng/mL recombinant human SCF, TPO, Flt3-L, and IL-6 (PeproTech), 0.75 μM StemRegenin 1 (Cellagen Technology), and penicillin/streptomycin. HEK293T cells were cultured in DMEM (Gibco) supplemented with 10% FBS, penicillin/streptomycin, and L-glutamine. Cells were maintained in logarithmic growth in a humidified incubator (37°C, 5% CO2) for all biological assays.
Lentivirus production and infection
Lentivirus production and infection were performed as previously described21. Briefly, HEK293T cells were transfected with third generation packaging plasmids and individual sgRNA vectors or CDKO libraries. Lentivirus was harvested after 48 h and 72 h and filtered through a 0.45 μm PVDF filter (Millipore). Spin infections were used to deliver sgRNA vectors and libraries into K562 cells.
Construction of CDKO library
The mouse U6 (mU6) single sgRNA vector was derived from a pSico lentiviral vector which expresses GFP and a puromycin-resistance cassette separated by a T2A sequence from EF-1 alpha promoter as previously described32,70. We then replaced the mU6 promoter with a human U6 promoter and moved a XhoI restriction site to the 5′ end of the hU6 promoter to generate the hU6 single-sgRNA vector. This allowed us to cut out the hU6-sgRNA-tracrRNA cassette and ligate it into the mU6 vector following the mU6-sgRNA-tracrRNA cassette to generate the CDKO library (Fig. 1c). To clone the mU6 and hU6 single sgRNA libraries with 700 sgRNAs targeting 207 druggable genes, two sets of DNAs that have identical sgRNA sequences but with different adaptors were PCR-amplified from single pooled-oligo chip (Agilent) and separately ligated into the mU6 and hU6 vectors using T4 ligase-mediated ligation and Gibson assembly, respectively. hU6-sgRNA-tracrRNA cassettes were cut out by XhoI and BamHI and ligated into the mU6 library that was also cut with the same restriction enzyme pair. This generated the DrugTarget-CDKO library which has 490,000 double-sgRNA combinations. To prevent cutting inside sgRNA sequences by restriction enzymes, we excluded sgRNA that have target sequences of XhoI, BamHI, BstXI or BlpI. (The last two enzymes were used to cut the mU6 vector and amplified sgRNA cassettes during cloning of the mU6 single sgRNA library) The batch retest and Ricin-CDKO libraries were constructed in the same way.
Selection of 207 drug target genes for the DrugTarget-CDKO library and 79 genes for the Ricin-CDKO library
For the DrugTarget-CDKO library, genes with moderate negative growth phenotypes in K562 cells were first selected from genome-wide CRISPR screens previously performed in our lab; these were genes for which the estimated effect size calculated by the casTLE algorithm41 was between −4 and −0.2. We then filtered out genes which do not have corresponding drugs in any of the following database: Therapeutic Target Database, DrugBank, and the IUPHAR/BPS Guide to Pharmacology (http://guidetopharmacology.org/). We further removed genes if their knockdown positively affected K562 cell growth in the genome-wide shRNA screen previously performed in our lab30,41 or they were not expressed in K562 cells (NCBI GEO Database) (Supplementary Table 1). For the Ricin-CDKO library, we first measured the ricin resistance phenotype (ρ) of genes in a genome-wide CRISPR screen29. From the hits of this screen, we first chose genes whose roles in ricin biology are known, and which are parts of well-characterized protein complexes15. In addition, we included genes whose roles in ricin biology are unknown. Essential genes (casTLE41 effect size < −3.6) were removed from the final list (Supplementary Table 5).
CRISPR-Cas9 screens with DrugTarget-CDKO, batch retest, and Ricin-CDKO libraries
For the CRISPR-Cas9 screens, we used a K562 cell line stably expressing the Cas9 endonuclease as previously described35. In short, a SFFV-Cas9-BFP lentiviral vector was transduced into K562 cells and cells were sorted for BFP. We infected the CDKO libraries into the Cas9-expressing K562 cells, waited for three days and applied puromycin selection for another three days until over 90% of the population contained the library (measured by GFP signal). At this point, aliquots of the library were frozen in liquid nitrogen as T0 samples, while the rest were used for the screen. For the DrugTarget-CDKO and batch retest screens, cells were grown for 14 days (~14 doublings), after which genomic DNA from the plasmid library and the T14 sample were isolated for PCR amplification of double-sgRNA cassettes and deep sequencing. The DrugTarget-CDKO screen was performed at ~1000X coverage (maintained above 500 × 106 cells in 1 liter culture) while the batch retest was performed at ~3000X coverage (maintained above 250 × 106 cells in 500 ml culture). For the Ricin-CDKO screen, cells were split into two conditions, untreated and ricin-treated, each maintained at ~3000X coverage. For the untreated sample, cells were grown for 14 days (~14 doublings). For the ricin-treated sample, cells were treated with four pulses of ricin at LD50 (first two pulses: 0.25 ng/ml ricin, third pulse: 0.3 ng/ml, fourth pulse: 0.35 ng/ml) so that there were about 6 doubling differences between the untreated and the ricin-treated samples. Genomic DNA from both samples were isolated and frequencies of double-sgRNAs were compared between the untreated and the ricin-treated sample through deep sequencing. All screens were carried out in two experimental replicates starting from the same T0 population and phenotypes of double-sgRNAs were normalized and pooled together before the medians of phenotypes and genetic interactions were measured.
Paired-end deep-sequencing of double-sgRNA cassettes
For each screen, cells at ~1,000X coverage (i.e. 500 × 106 cells for the DrugTarget-CDKO screen) were lysed and genomic DNA was purified using QIAGEN Blood Maxi kits. Two rounds of PCR were carried out to amplify double-sgRNA cassettes from the genomic DNA using Herculase II Fusion Polymerase (Agilent) as previously described97. For every ~5,000 unique double-sgRNAs, 10 ug of genomic DNA was used as template in each 100 ul reaction: about 100 ul × 100 reactions were used to amplify double-sgRNA cassettes from the genomic DNA of the DrugTarget-CDKO library which has 490,0000 double-sgRNAs. In the first PCR, the forward PCR primer that binds to 3′ end of the mU6 promoter (5′-ggcttggatttctataacttcgtatagc-3′) and the reverse PCR primer that binds to the 3′ end of a typical double-sgRNA cassette (5′-ccgcctaatgg-atcccctaggaaa-3′) were used. Adapters that have Illumina P5, P7, a 6bp index, and sequencing primers binding sites were added in the second round of PCR using the following forward and reverse PCR primers: 5′-aatgatacggcgaccaccgagatctacactgtgtgttttgagactataagtatcccttggag-3′ and 5′-caagcagaagacggcatacgagatagacagcagtcccgtgttccggttcattctatcaNNNNNNggatcccctaggaaaaaaa-gcaccg-3′, respectively, where Ns indicate Illumina index barcodes. From these two rounds of PCR, a PCR fragment around 640bp that contains both sgRNAs was amplified and gel-purified. The amplicons were then sequenced on a NextSeq 550 (Illumina) using its paired-end sequencing protocol. The paired-end sequencing protocol was slightly modified to read two sgRNAs and a sample index barcode (Supplementary Fig. 1a,b) with three custom sequencing primers listed below. The typical paired-end sequencing protocol proceeds with Read1, Index Read, and Read2 steps, but the modified protocol uses the Index Read step to read the second sgRNA and the Read2 step to read the illumina index barcode.
Custom Read1 primer : 5′-gtgtgttttgagactataagtatcccttggagaaccaccttgttgg-3′
Custom Read2 primer : 5′-agacagcagtcccgtgttccggttcattctatca-3′
Custom Index Read primer : 5′-ttgaaagtatttcgatttcttggctttatatatcttgtggaaaggacgaaacaccg-3′
In short, the custom Read1 primer that binds to the 3′ end of the mU6 promoter reads 20bp of the front sgRNA. The custom Read2 primer that binds to the 3′ end of hU6 promoter reads 20bp of the rear sgRNA in the same direction as the first read. Clusters in the flow cells are then stripped off and flipped over through cluster regeneration. Finally, the custom Index Read primer reads 6bp of the Illumina index barcode. From one NextSeq run (NextSeq 500/550 High Output Kit, 75 cycles), we can acquire ~700–800M reads. Sequencing was performed at ~200X read-coverage for the DrugTarget-CDKO screen and ~600X coverage for the batch retest screen. The ricin-CDKO screen was sequenced at about 500X read-coverage. The two sgRNA sequences were then aligned to the known library sequences using Bowtie71 with one mismatch allowed and we typically acquire about 400M aligned reads of double-sgRNAs.
Minimum count threshold
We determined a minimum threshold for read counts (Supplementary Fig. 2a) and masked out double-sgRNAs that have read counts below the threshold. To do this, we first measured frequencies of the single-sgRNAs in both the hU6 and mU6 libraries, calculated expected frequencies of the double-sgRNAs, and compared them to observed frequencies in the CDKO library (Supplementary Fig. 2a). Ratios of the two frequencies showed that under ~50 read counts, the observed frequencies markedly fell below the expected; therefore, we removed double-sgRNAs with less than 50 read counts at T=0 from further analyses. Zero count in T14 samples were replaced with count=1.
Measurement of phenotypes and genetic interactions
To calculate the phenotype of double-sgRNAs, we compared frequencies of double-sgRNAs between the plasmid library and T14 sample to calculate the growth phenotype (γ) and between the ricin-treated T14 and untreated T14 samples to calculate the ricin-resistance phenotype (ρ) as previously described31. In short, we measured the frequency of each double-sgRNA between two samples and calculated the log2 enrichment ratio (log2e) between them, which was then normalized to the median log2e of the negative control double-sgRNAs, comprised of 79 × 79 Safe_Safe sgRNA pairs. Equations used here are as follows:
Where:
N14: the read counts of a double-sgRNA in the T14 sample
Nctrl-14: the read counts of a Safe_Safe double-sgRNA in the T14 sample
Np: the read counts of a double-sgRNA in the plasmid library
Nctrl-p: the read counts of a Safe_Safe double-sgRNA in the plasmid library
Cp: the total read counts of the plasmid library
C14: the total read counts of the T14 sample
D: the number of doublings of the infected cells between infection and T14
Ricin-resistance phenotype (ρ) is calculated in the same way except that read counts are compared between the ricin-treated T14 sample and the ricin-untreated T14 sample instead of the T14 sample and the plasmid library in γ Phenotype.
Lastly, we calculated a phenotype Z score (pZ)32 by dividing the phenotype of each double-sgRNA by the standard deviation of Safe_Safe pairs. To calculate log fold enrichment of double-sgRNAs, we compared counts in T14 to counts in the plasmid library instead of those in the T0, since we observed that genes with large negative growth phenotypes dropped out quickly even between infection and T0 (6 days after infection, puromycin selection for lentiviral integration, and expansion) (Supplementary Fig. 2b). To calculate the expected phenotype of double-sgRNAs, we first calculated the phenotype of individual sgRNAs: a phenotype of an individual sgRNA was measured as the median phenotype of all double-sgRNAs comprised of this sgRNA and one safe-sgRNA. For example, to calculate the phenotype of AKT1 sgRNA-1, we measured phenotypes of all AKT1 sgRNA-1s paired with any of the 79 Safe sgRNAs and calculated the median of the phenotype distribution. The expected double-sgRNA phenotype was then calculated by summing the phenotypes of the two individual sgRNAs. To measure the genetic interaction between two sgRNAs, the expected phenotype of double-sgRNAs were plotted against their observed phenotypes (Fig. 3b). Data were then binned along the expected phenotype so that each bin contains 200 data points. The median of each bin were measured and the line connecting the median values was then smoothed by a three-point moving averaging filter. The smoothed line represents the typical behavior of double-sgRNAs and deviations from this line were measured to calculate the GIs between sgRNA pairs. Deviations that moved the double-sgRNA phenotypes closer to the phenotype of the Safe_Safe controls were given a positive sign and defined as buffering GIs, while those that moved away from the Safe_Safe phenotype were given a negative sign and defined as synergistic GIs. A signed GI (Raw-GI) was then divided by the standard deviation of the Raw-GIs of the 200 (arbitrary number) nearest neighbors along the expected phenotype. This normalized GI (Norm-GI) measures GI between sgRNA pairs and we found that the normalization stabilizes variance of genetic interactions, improving reproducibility of genetic interactions between experimental replicates (Supplementary Fig. 12) To calculate a quantitative genetic interaction score for a given gene pair, we pooled all double-sgRNAs targeting the same gene pair in both orientations from two experimental replicates. This would give us 36 data points at most for any given gene pair (9 combinations × 2 orientations × 2 replicates). We filtered out gene pairs if they have less than 3 data points. Then, genetic interaction scores between gene pairs were defined with two scoring methods. GIT score for a given gene pair was calculated based on the modified t-value score using the following equation (A similarly modified t-value score was used to calculate a quantitative genetic interaction score, S score, in budding yeast33):
Where:
Uexp: the median of Norm-GIs of all double-sgRNAs targeting the gene pair
Nexp: the number of all double-sgRNAs targeting the gene pair
Vexp: the variance of Norm-GIs of all double-sgRNAs targeting the gene pair
Uctrl: the median of Norm-GIs of genetically non-interacting double-sgRNAs (all double-sgRNAs containing at least one safe-sgRNA)
Nctrl: the median number of double-sgRNAs per gene pair for all gene pairs
Vctrl: the variance of Norm-GIs of all genetically non-interacting double-sgRNAs
To calculate the GIM score, the probability that the distribution of Norm-GIs of all double-sgRNAs targeting a given gene pair was significantly different from that of non-interacting double-sgRNAs was measured using the Mann-Whitney U test. A similar concept has been used to score phenotypes of genes from ultracomplex pooled shRNA and CRISPRi screens15,32. Log10 of p values were then given a positive sign if gene pairs were buffering according to the median of Norm-GIs and a negative sign if gene pairs were synergistic. The two GI scores generally identified similar top buffering and synergistic hits (Fig. 3e, Supplementary Fig. 3b) and the rank-sum of the two GI scores was used to prioritize synergistic, toxic gene pairs for subsequent validation with corresponding drugs.
Systematic comparison between GI and PPI
To systematically compare GI and PPI, we examined the previously reported interactions among 79 ricin modulators from STRING45. Among all the validated and predicted PPIs for the 79 ricin modulators, we used only the experimentally validated interactions for the comparison (minimum required interaction score was set to 0.15 in STRING). Gene pairs were then sorted by one of the three features of genetic interactions - buffering GI, synergistic GI, or correlation of GI profiles (Pearson correlation coefficient, uncentered). Here, correlation of GI profiles were calculated as previously described15,31,72. Next, for a given top N pairs of genes, we measured how many of the pairs have the reported interactions in STRING and plotted the measured percent coverage by the STRING interactions on the y axis against the N number on the × axis (Fig. 4d). Functional interactomes of the 66 most correlated gene pairs in terms of GI profiles were drawn using Cytoscape software73 in Supplementary Fig. 8b. Edges were drawn between genes whose correlation of GI profiles were greater than 0.5 and the width of the edges were determined by their GI correlation values. If an interaction was already reported in STRING for a given edge, it was colored in red. Comparison of GIs to PPIs in BioGrid101 also showed similar results (data not shown).
Constructing genetic interaction maps
To construct the GI maps, GI profiles of genes were generated based on GI scores. Genes were then hierarchically clustered with Cluster 3.0 software74 based on the uncentered Pearson correlations of their GI profiles as a similarity metric and average linkage as the clustering method. Generated clusters were displayed using Java TreeView75. Both GIM score and GIT score identified similar clusters.
Validation of identified GIs using individual sgRNAs
For each gene pair, we selected the pair of sgRNAs that showed the greatest GI in the DrugTarget-CDKO screen. The first sgRNA in the pair was cloned into a sgRNA expression vector (pMCB306) which expresses GFP-T2A-puromycin from an EF-1 alpha promoter. The second sgRNA was cloned into a vector (pMCB320) that expresses mCherry-T2A-puromycin. Two lentiviruses were produced separately and pooled together to infect Cas9-expressing K562 cells. We adjusted virus titers so that the infection generated four populations of cells: uninfected, GFP-positive, mCherry-positive, and GFP-mCherry double positive. The single knockout phenotype for each sgRNA was measured by comparing the abundance of either GFP-positive or mCherry-positive cells relative to the uninfected cells between T0 (Day 5 post-infection) and T7 (Day 12 post-infection). Similarly, the double knockout phenotype was measured by comparing the relative abundance of the double positive cells between T0 and T7. Log2 enrichment of each population divided by 7 (the estimated number of doublings) was used to calculate the growth phenotype. GI was calculated as the difference between the expected phenotype (sum of two single knockout phenotypes) and the observed phenotype of double knockout. The GI between two genes-targeting sgRNAs were compared to GIs from each individual sgRNA paired with a safe-sgRNA as non-interacting controls. The statistical significance (p value) used to compare the average phenotype or GI of two samples was determined by unpaired, two-tailed Student’s t-test implemented in Microsoft Excel.
Sequencing analysis for indel detection
Genomic DNA was extracted from cells using a QIAamp DNA mini kit (Qiagen). A ~600–700 bp region containing the sgRNA cut site was amplified by PCR. The PCR product was run on a 0.8% TAE-agarose gel, bands containing the amplicons were excised, and DNA was purified from the gel slice using a QIAquick Gel Extraction kit (Qiagen). Samples were sent for Sanger sequencing. To assess indel frequency, TIDE was used to compare the sequence trace files from the test sample containing the targeting sgRNA to a control sample containing safe harbor sgRNAs and quantify the distribution of indels in the test sample48.
Cell viability assay for drug synergy
Cells were seeded in 24-well plates and treated with no drug, single drug or in combination at the indicated concentrations for 72 hours. For the drug synergy tests, drug pairs were tested in wild-type K562 cells without Cas9, and thus represent effects independent of possible Cas9-induced DNA damage. Following drugs were used for the assay: A-1155463 (Chemietek CT-A115), A-1210477 (Chemietek CT-A121), CRT0044876 (Selleckchem S7449), KU-60019 (Selleckchem S1570), UMI-77 (Sigma SML1492), NU 7441 (Selleckchem S2638), PK-11195 (Enzo BML-CM118), PX-12 (Cayman 14192), KPT-330 (Selleckchem S7252), 1-benzyl-3,5-bis-(3-bromo-4-hydroxybenzylidene)piperidin-4-one (Millipore 217531), FK866 (Selleckchem S2799). K562 cells were seeded at 100,000 cells/ml, MV4;11 cells were seeded at 200,000 cells/ml, and GM12892 cells and CD34+ HSPCs were seeded at 300,000 cells/ml. After 72 hour incubation, the number of viable cells was counted by flow cytometry (FSC/SSC) using a BD Accuri C6 flow cytometer. Drug synergy was evaluated using the Bliss independence model (Bliss CI, 1939). Briefly, the predicted fractional growth inhibition of the drug combination is calculated using the equation FA + FB − (FA × FB), where FA and FB are the fractional growth inhibitions of the individual drugs A and B at a given dose. Bliss excess is the difference between the expected growth inhibition and the observed inhibition from the drug combination. Bliss scores greater than zero, close to zero, and less than zero denote synergy, additivity, and antagonism, respectively. Bliss sum is the sum of individual bliss scores in the 3 by 3 matrix of drug doses. P values where indicated were determined by one-tailed Student’s t-test between the observed effect of the drug combination and the expected effect calculated from the individual drug effects.
Apoptosis assay
K562 cells were seeded in 24-well plates at 100,000 cells/ml and treated with no drug, single drug, or in combination at the indicated concentrations for 48 hours. After 48 hour treatment, apoptosis was measured using an Annexin V-FITC Apoptosis Kit (BioVision). Cells were collected and incubated with Annexin V-FITC and propidium iodide (PI) before quantification using a BD Accuri C6 flow cytometer.
Immunoblot analysis
Whole-cell protein extracts were prepared using lysis buffer containing 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA and 1% Triton X-100 supplemented with Protease Inhibitor Cocktail (Sigma-Aldrich). Samples (100 ug of protein) were separated by SDS-PAGE and transferred to PVDF membranes. Immunoblot analysis was performed using Rabbit anti-Bcl-xL (CST 2762S), Rabbit anti-Mcl-1 (CST 4572S), Mouse anti-tubulin (Sigma T6199), Goat anti-rabbit IRDye 800CW (LI-COR 926-32211), and Donkey anti-mouse IRDye 680LT (LI-COR 926-68022) antibodies, and imaged on a Li-Cor Odyssey Infrared Imaging System.
γH2AX flow cytometric analysis and immunofluorescence microscopy
K562 cells were seeded in 6-well plates at 250,000 cells/ml and treated with no drug, single drug, or in combination at the indicated concentrations for 48 hours. Cells were washed in PBS, fixed in 4% paraformaldehyde for 10 min, washed twice in PBS, permeabilized/blocked in 0.1% Triton/2% BSA/PBS for 30 min, washed twice in PBS, probed with anti-γH2AX monoclonal antibody (Millipore 05–636) for 1 h, washed twice in PBS, incubated with Alexa Fluor 488 goat anti-mouse IgG secondary antibody (Invitrogen A-11029) for 45 min, and washed twice in PBS before quantification using a BD Accuri C6 flow cytometer. Remaining cells were stained with Hoescht 33342 (ThermoFisher Scientific) for 10 min, washed twice with PBS, and mounted on coverslips in ProLong Gold (Thermo Fisher). Cells were imaged with a Nikon Ti-E spinning disk microscope at 100X magnification.
Data availability
Sequencing data are available at Sequence Read Archive accession number SRP099805 under Bioproject accession number PRJNA374750. Plasmids and their sequences are deposited at Addgene. Scripts used for analysis of sequencing data are available upon request.
Supplementary Material
Acknowledgments
We thank S. Collins and members of the Bassik lab for helpful discussions and critical reading of the manuscript, A. Sockell and L. Tonkin for technical assistance in deep sequencing, and M. Porteus and S. Mantri for CD34+ HSPCs. We thank M. Cleary, J. Duque-Afonso, and P. Jackson for helpful discussions regarding drug combination assays. We thank M. Kampmann, M. Horlbeck, L. Gilbert, and J. Weissman for helpful discussions, and Laurakay Bruhn, Carsten Carstens, Peter Sheffield, and Ben Borgo of Agilent technologies for oligonucleotide synthesis and helpful discussions. The work was funded by the NIH Director’s New Innovator Award Program (project no. 1DP2HD084069-01) NIH/NHGRI (training grant T32 HG000044 to D.W.M.), NIH/NCI 1U01CA199261-02, and a seed grant from Stanford ChEM-H. K.H. is supported by the Walter V. and Idun Berry award. E.E.J. was supported by the NIH (2T32CA009302) and a Hubert Shaw and Sandra Lui Stanford Graduate Fellowship.
Footnotes
CONTRIBUTIONS
K.H. and M.C.B. conceived and designed the study. K.H. designed the CDKO system and the scoring systems for GI map. K.H. analyzed the screen data and performed the GI and PPI analyses. K.H., A.L. and E.E.J. performed the CDKO screens. G.T.H., A.L., and D.W.M. performed the genome-wide screens for ricin modulators. D.W.M. selected the best-working sgRNAs to design the CDKO libraries. K.H. and E.E.J. validated the hits from the CDKO screens. E.E.J. performed the drug validations and related experiments. K.H., E.E.J. and M.C.B. wrote the manuscript. All authors discussed the results and the manuscript. M.C.B. supervised the study.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interest.
References
- 1.Jia J, et al. Mechanisms of drug combinations: interaction and network perspectives. Nat Rev Drug Discov. 2009;8:111. doi: 10.1038/nrd2683. [DOI] [PubMed] [Google Scholar]
- 2.Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov. 2004;3:673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 3.Al-Lazikani B, Banerji U, Workman P. Combinatorial drug therapy for cancer in the post-genomic era. Nat Biotechnol. 2012;30:679–692. doi: 10.1038/nbt.2284. [DOI] [PubMed] [Google Scholar]
- 4.Sun X, Vilar S, Tatonetti NP. High-Throughput Methods for Combinatorial Drug Discovery. Sci Transl Med. 2013;5:205rv1–205rv1. doi: 10.1126/scitranslmed.3006667. [DOI] [PubMed] [Google Scholar]
- 5.Collins SR, Weissman JS, Krogan NJ. From information to knowledge: new technologies for defining gene function. Nat Methods. 2009;6:721–723. doi: 10.1038/nmeth1009-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costanzo M, et al. A global genetic interaction network maps a wiring diagram of cellular function. Science. 2016:353. doi: 10.1126/science.aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schuldiner M, et al. Exploration of the Function and Organization of the Yeast Early Secretory Pathway through an Epistatic Miniarray Profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 8.Jonikas MC, et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323:1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bandyopadhyay S, et al. Rewiring of Genetic Networks in Response to DNA Damage. Science. 2010;330:1385–1389. doi: 10.1126/science.1195618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins SR, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 11.Dixon SJ, Costanzo M, Baryshnikova A, Andrews B, Boone C. Systematic Mapping of Genetic Interaction Networks. Annu Rev Genet. 2009;43:601–625. doi: 10.1146/annurev.genet.39.073003.114751. [DOI] [PubMed] [Google Scholar]
- 12.Frost A, et al. Functional repurposing revealed by comparing S. pombe and S. cerevisiae genetic interactions. Cell. 2012;149:1339–1352. doi: 10.1016/j.cell.2012.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horn T, et al. Mapping of signaling networks through synthetic genetic interaction analysis by RNAi. Nat Methods. 2011;8:341–346. doi: 10.1038/nmeth.1581. [DOI] [PubMed] [Google Scholar]
- 14.Roguev, et al. Quantitative genetic-interaction mapping in mammalian cells. Nat Methods. 2013;10:432–437. doi: 10.1038/nmeth.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bassik MC, et al. A Systematic Mammalian Genetic Interaction Map Reveals Pathways Underlying Ricin Susceptibility. Cell. 2013;152:909–922. doi: 10.1016/j.cell.2013.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srivas R, et al. A Network of Conserved Synthetic Lethal Interactions for Exploration of Precision Cancer Therapy. Mol Cell. 2016;63:514–525. doi: 10.1016/j.molcel.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong ASL, Choi GCG, Cheng AA, Purcell O, Lu TK. Massively parallel high-order combinatorial genetics in human cells. Nat Biotechnol. 2015;33:952–961. doi: 10.1038/nbt.3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jinek M, et al. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mali P, et al. RNA-Guided Human Genome Engineering via Cas9. Science. 2013;339:823–926. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cong L, et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsai SQ, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie K, Minkenberg B, Yang Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci. 2015;112:3570–3575. doi: 10.1073/pnas.1420294112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong ASL, et al. Multiplexed barcoded CRISPR-Cas9 screening enabled by CombiGEM. Proc Natl Acad Sci. 2016;113:2544–2549. doi: 10.1073/pnas.1517883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vidigal JA, Ventura A. Rapid and efficient one-step generation of paired gRNA CRISPR-Cas9 libraries. Nat Commun. 2015:6. doi: 10.1038/ncomms9083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sack LM, Davoli T, Xu Q, Li M, Elledge SJ. Sources of Error in Mammalian Genetic Screens. G3 (Bethesda) 2016;6:2781–2790. doi: 10.1534/g3.116.030973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu F, et al. Therapeutic target database update 2012: a resource for facilitating target-oriented drug discovery. Nucleic Acids Res. 2012;40:D1128–D1136. doi: 10.1093/nar/gkr797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wishart DS, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pawson AJ, et al. The IUPHAR/BPS Guide to PHARMACOLOGY: An expert-driven knowledgebase of drug targets and their ligands. Nucleic Acids Res. 2014;42:1098–1106. doi: 10.1093/nar/gkt1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morgens DW, et al. Genome-scale measurement of off-target activity using Cas9 toxicity in high-throughput screens. Nat Commun. 2017 doi: 10.1038/ncomms15178. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deans RM, et al. Parallel shRNA and CRISPR-Cas9 screens enable antiviral drug target identification. Nat Chem Biol. 2016;12:361–366. doi: 10.1038/nchembio.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kampmann M, Bassik MC, Weissman JS. Integrated platform for genome-wide screening and construction of high-density genetic interaction maps in mammalian cells. Proc Natl Acad Sci. 2013;110:E2317–E2326. doi: 10.1073/pnas.1307002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilbert LA, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Collins SR, Schuldiner M, Krogan NJ, Weissman JS. A strategy for extracting and analyzing large-scale quantitative epistatic interaction data. Genome Biol. 2006;7:723–733. doi: 10.1186/gb-2006-7-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fischer B, et al. A map of directional genetic interactions in a metazoan cell. Elife. 2015;4:1–21. doi: 10.7554/eLife.05464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blomen VA, et al. Gene essentiality and synthetic lethality in haploid human cells. Science. 2015;350:1092–6. doi: 10.1126/science.aac7557. [DOI] [PubMed] [Google Scholar]
- 36.Brazil DP, Yang ZZ, Hemmings BA. Advances in protein kinase B signalling: AKTion on multiple fronts. Trends Biochem Sci. 2004;29:233–242. doi: 10.1016/j.tibs.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 37.Saurabh K, et al. The PIM family of oncoproteins: small kinases with huge implications in myeloid leukemogenesis and as therapeutic targets. Oncotarget. 2014;5:8503–14. doi: 10.18632/oncotarget.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen JL, Limnander A, Rothman PB. Pim-1 and Pim-2 kinases are required for efficient pre-B-cell transformation by v-Abl oncogene. Blood. 2008;111:1677–1685. doi: 10.1182/blood-2007-04-083808. [DOI] [PubMed] [Google Scholar]
- 39.Johannes L, et al. Tracing the Retrograde Route in Protein Trafficking. Cell. 2005;135:1175–1187. doi: 10.1016/j.cell.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 40.Moreau D, et al. Genome-Wide RNAi Screens Identify Genes Required for Ricin and PE Intoxications. Dev Cell. 2011;21:231–244. doi: 10.1016/j.devcel.2011.06.014. [DOI] [PubMed] [Google Scholar]
- 41.Morgens DW, Deans RM, Li A, Bassik MC. Systematic comparison of CRISPR / Cas9 and RNAi screens for essential genes. Nat Biotechnol. 2016;34:1–4. doi: 10.1038/nbt.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang W, et al. Mannosidase 2, alpha 1 Deficiency Is Associated with Ricin Resistance in Embryonic Stem (ES) Cells. PLoS One. 2011;6:333–9. doi: 10.1371/journal.pone.0022993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bar-Peled L, et al. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340:1100–6. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–9. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Szklarczyk D, et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2014;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parnas O, et al. A Genome-wide CRISPR Screen in Primary Immune Cells to Dissect Regulatory Networks. Cell. 2015;162:675–686. doi: 10.1016/j.cell.2015.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen S, et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell. 2015;160:1246–1260. doi: 10.1016/j.cell.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168–e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lord CJ, Tutt ANJ, Ashworth A. Synthetic Lethality and Cancer Therapy: Lessons Learned from the Development of PARP Inhibitors. Annu Rev Med. 2015;66:455–470. doi: 10.1146/annurev-med-050913-022545. [DOI] [PubMed] [Google Scholar]
- 50.Sultana R, et al. Synthetic lethal targeting of DNA double-strand break repair deficient cells by human apurinic/apyrimidinic endonuclease inhibitors. Int J Cancer. 2012;131:2433–2444. doi: 10.1002/ijc.27512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Riabinska A, et al. Therapeutic Targeting of a Robust Non-Oncogene Addiction to PRKDC in ATM-Defective Tumors. Sci Transl Med. 2013:5. doi: 10.1126/scitranslmed.3005814. [DOI] [PubMed] [Google Scholar]
- 52.Austin CJD, Kahlert J, Kassiou M, Rendina LM. The translocator protein (TSPO): A novel target for cancer chemotherapy. Int J Biochem Cell Biol. 2013;45:1212–1216. doi: 10.1016/j.biocel.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 53.Placzek WJ, et al. A survey of the anti-apoptotic Bcl-2 subfamily expression in cancer types provides a platform to predict the efficacy of Bcl-2 antagonists in cancer therapy. Cell Death Dis. 2010;1:e40. doi: 10.1038/cddis.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bixby D, Talpaz M. Seeking the causes and solutions to imatinib-resistance in chronic myeloid leukemia. Leukemia. 2011;25:7–22. doi: 10.1038/leu.2010.238. [DOI] [PubMed] [Google Scholar]
- 55.Delbridge ARD, Grabow S, Strasser A, Vaux DL. Thirty years of BCL-2: translating cell death discoveries into novel cancer therapies. Nat Rev Cancer. 2016;16:99–109. doi: 10.1038/nrc.2015.17. [DOI] [PubMed] [Google Scholar]
- 56.Alchberger KJ, et al. Identification of mcl-1 as a BCR/ABL-dependent target in chronic myeloid leukemia (CML): Evidence for cooperative antileukemic effects of imatinib and mcl-1 antisense oligonucleotides. Blood. 2005;105:3303–3311. doi: 10.1182/blood-2004-02-0749. [DOI] [PubMed] [Google Scholar]
- 57.Dai Y, Rahmani M, Corey SJ, Dent P, Grant S. A Bcr/Abl-independent, Lyn-dependent Form of Imatinib Mesylate (STI-571) Resistance Is Associated with Altered Expression of Bcl-2. J Biol Chem. 2004;279:34227–34239. doi: 10.1074/jbc.M402290200. [DOI] [PubMed] [Google Scholar]
- 58.KB, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- 59.Friedman A, Perrimon N. A functional RNAi screen for regulators of receptor tyrosine kinase and ERK signalling. Nature. 2006;444:230–234. doi: 10.1038/nature05280. [DOI] [PubMed] [Google Scholar]
- 60.Mendes-Pereira AM, et al. Genome-wide functional screen identifies a compendium of genes affecting sensitivity to tamoxifen. Proc Natl Acad Sci. 2012;109:2730–2735. doi: 10.1073/pnas.1018872108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eichhorn PJA, et al. Phosphatidylinositol 3-kinase hyperactivation results in lapatinib resistance that is reversed by the mTOR/phosphatidylinositol 3-kinase inhibitor NVP-BEZ235. Cancer Res. 2008;68:9221–9230. doi: 10.1158/0008-5472.CAN-08-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Corcoran RB, et al. Synthetic Lethal Interaction of Combined BCL-XL and MEK Inhibition Promotes Tumor Regressions in KRAS Mutant Cancer Models. Cancer Cell. 2013;23:121–128. doi: 10.1016/j.ccr.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Prahallad A, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–3. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 64.Opferman JT. Attacking cancer’s Achilles heel: antagonism of anti-apoptotic BCL-2 family members. FEBS J. 2016;283:2661–2675. doi: 10.1111/febs.13472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leverson JD, et al. Potent and selective small-molecule MCL-1 inhibitors demonstrate on-target cancer cell killing activity as single agents and in combination with ABT-263 (navitoclax) Cell Death Dis. 2015;6:e1590. doi: 10.1038/cddis.2014.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chonghaile TN, et al. Maturation Stage of T-cell Acute Lymphoblastic Leukemia Determines BCL-2 versus BCL-XL Dependence and Sensitivity to ABT-199. Cancer Discov. 2014;4:1074–1087. doi: 10.1158/2159-8290.CD-14-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ko TK, Chuah CTH, Huang JWJ, Ng KP, Ong ST. The BCL2 inhibitor ABT-199 significantly enhances imatinib-induced cell death in chronic myeloid leukemia progenitors. Oncotarget. 2014;5:9033–9038. doi: 10.18632/oncotarget.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hong HY, Choi J, Cho YW, Kim BC. Cdc25A promotes cell survival by stimulating NF-κB activity through IκB-α phosphorylation and destabilization. Biochem Biophys Res Commun. 2012;420:293–296. doi: 10.1016/j.bbrc.2012.02.152. [DOI] [PubMed] [Google Scholar]
- 69.Bassi ZI, Audusseau M, Riparbelli MG, Callaini G, D’Avino PP. Citron kinase controls a molecular network required for midbody formation in cytokinesis. Proc Natl Acad Sci. 2013;110:9782–9787. doi: 10.1073/pnas.1301328110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen B, et al. Dynamic Imaging of Genomic Loci in Living Human Cells by an Optimized CRISPR/Cas System. Cell. 2011;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Langmead B, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009 103. 2009;10:779–785. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kampmann M, Bassik MC, Weissman JS. Functional genomics platform for pooled screening and generation of mammalian genetic interaction maps. Nat Protoc. 2014;9:1825–1847. doi: 10.1038/nprot.2014.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shannon P, et al. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saldanha AJ. Java Treeview - Extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data are available at Sequence Read Archive accession number SRP099805 under Bioproject accession number PRJNA374750. Plasmids and their sequences are deposited at Addgene. Scripts used for analysis of sequencing data are available upon request.