Abstract
The organization of living cells is based on networks of interacting molecules. Systematic analysis of protein interactions of 3-aa loop extension (TALE) homeodomain proteins, fundamental regulators of plant meristem function and leaf development, revealed a highly connected, complex network. The network includes nine members of Arabidopsis thaliana ovate family proteins (AtOFPs), a plant-specific protein family, indicating a close functional connection to TALE homeodomain proteins. Evidence is provided that AtOFP1 is an essential pleiotropic developmental regulator. AtOFP1 and AtOFP5 are shown to associate with the cytoskeleton and to regulate subcellular localization of TALE homeodomain proteins, suggesting a previously unrecognized control mechanism in plant development.
Keywords: cytoskeleton, network modules, protein-protein interactions, BELL proteins, KNOX proteins
Networks of molecular interactions are central to the functional organization of living cells. Graph-theoretical analysis of data sets obtained from large-scale expression profiling, metabolome analysis, and protein interaction mapping reveals universal characteristics in the architecture of complex networks, allowing us to deduce hierarchies and higher-level organizational principles of biological systems (1). Comprehensive protein interaction data of yeast, Caenorhabditis elegans, and Drosophila melanogaster support the idea that protein complexes and/or dynamic functional units are reflected in the network topology in the form of locally highly interconnected modules (2-4). Functional predictions based on modules represent, in turn, testable hypotheses guiding the investigation of complex cellular functions.
Similar to their function in animal development, homeobox genes of the 3-aa loop extension (TALE) superfamily - class I and II Knox and Bell genes - play a central role in plant developmental processes (5, 6). Morphological events are correlated with precisely regulated spatiotemporal patterns of TALE gene expression (7), and corresponding cis-regulatory elements present in promoters and introns of these genes have been identified (8).
In animals, additional cofactors are essential for TALE protein activity and subcellular localization (9, 10). In plants, no such cofactors have been described so far. Similar to PREP/MEINOX proteins in animals, KNOX and BELL proteins form heterodimers that are thought to constitute the functional entities regulating plant development (11-13). KNOX/BELL heterodimerization is implicated in nuclear import of plant TALE proteins and has been shown to increase DNA-binding affinity and specificity (13-15). Further, the capacity of tobacco and potato TALE proteins to bind regulatory sequences of the gibberellin (GA) hormone-synthesizing gene GA20-oxidase1 was interpreted as evidence of KNOX protein-mediated negative regulation of GA biosynthesis in the meristem (15-17).
Despite recent progress, the molecular function of TALE homeodomain proteins is still poorly understood. Here, we present a comprehensive survey of TALE protein interactions that result in a network with the characteristics of a functional module. Members of a previously unrecognized plant protein family [Arabidopsis thaliana ovate family proteins (AtOFPs)] are included in this module. Evidence is provided that these proteins control the intracellular localization of TALE proteins and are important regulators of plant development.
Materials and Methods
Plasmid Construction and Yeast Methods. Full-length cDNAs of TALE genes were obtained by RT-PCR and cloned into pACT-attR and pAS-attR vectors (J.F.U., unpublished data) by using the Gateway system (Invitrogen). Transformation of Saccharomyces cerevisiae AH109 and large-scale yeast two-hybrid screens were performed as described in ref. 18. Accession numbers of TALE and OFP genes are listed in the legend of Fig. 5, which is published as supporting information on the PNAS web site.
Network Analysis. Graphs were drawn by using a modified Fruchterman-Reingold graph layout algorithm as implemented in the pajek 0.97 program package (http://vlado.fmf.uni-lj.si/pub/networks/pajek). Network parameters were calculated as described in refs. 2 and 19.
Plant Transformation and Culture. AtOFP1 cDNA was cloned into pLEELA (M. Jakoby, unpublished data) containing a double 35S promoter. Vectors were electroporated into Agrobacterium tumefaciens GV3101. A. thaliana Col-0 plants were transformed by using the floral-dip method (20). Plants were selected for BASTA resistance and grown under long-day greenhouse conditions. For expression in tobacco, AtOFP1 cDNA was cloned into pLX222-attR (J.H., unpublished data). Nicotiana tabacum cv. SR1 plants were transformed by Agrobacterium-LBA4404-mediated leaf disk transformation (21). Calli were selected on Murashige and Skoog (MS) medium containing 100 mg/liter kanamycin. T-DNA (transferred DNA) insertion lines were obtained from the SALK collection (22).
RT-PCR Analysis. Five micrograms of total RNA from leaves were used for cDNA synthesis with an oligo(dT) primer and Stratascript RT (Stratagene). Thirty PCR cycles were performed by using primers specific for AtGA20ox1 (AtGA20ox1-F, GCCGTAAGTTTCGTAACAACATCTCC; AtGA 20ox1-R, GAGAGAGGCATATCAAAGAA GCGG) and R AN3 (RAN3-F, ACCAGCAAACCGTGGATTACCCTAGC; R-AN3-R, ATTCCACAAAGTGAAGATTAGCGTC). Experiments were conducted at least three times with three independent RNA preparations.
Transient Expression of Proteins. TALE, AtOFP1, and AtOFP5 cDNAs were cloned into pBatTL vectors (K.R., unpublished work) containing a double 35S promoter, a translation enhancer, and a GFP or red fluorescent protein (RFP) tag, respectively. Leaves of 3- to 5-week-old Nicotiana benthamiana plants were coinfiltrated with A. tumefaciens LBA4404 strains containing the pBatTL constructs and a viral silencing suppressor gene, respectively, according to ref. 23. Localization of fluorescent proteins was monitored 3-7 days after infiltration, the period when RFP fluorescence was optimal, by using a confocal laser scanning microscope (Zeiss LSM510/ConfoCor 2).
Results and Discussion
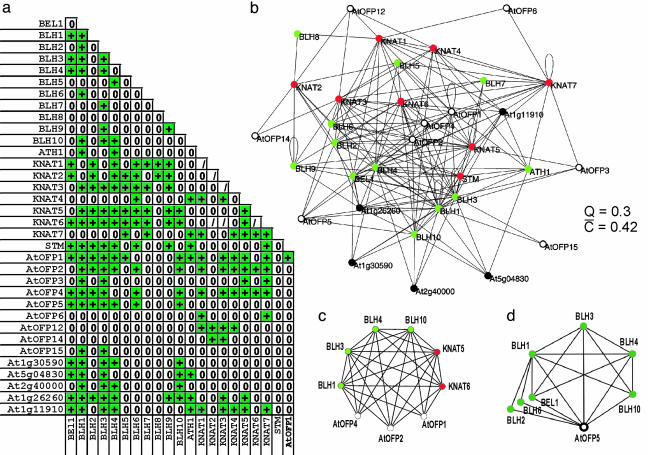
An Interaction Network of TALE Homeodomain Proteins. We have used a large-scale yeast two-hybrid technology to systematically analyze the interactions of A. thaliana TALE homeodomain proteins. A combination of cDNA-library screenings and an all-against-all pairwise interaction test revealed a densely connected network of interactions between and within the two TALE families (Fig. 1 a and b). Every member of the KNOX family has at least one interaction partner amongst the BELL proteins and vice versa, supporting the notion that BELL/KNOX heterodimerization plays a general role in TALE protein function. In addition, several homo- and heterodimerizations within each TALE family, including dimerizations of BELL proteins not previously described, have been detected (Fig. 1 a and b), bearing implications for further functional TALE complexes. Our experimental setup, however, failed to detect one interaction published previously (BEL1/KNAT5) (12) and revealed interactions between TALE proteins previously suggested to not interact (BLH9/KNAT3 and BLH9/KNAT5) (14). These discrepancies presumably are due to the differences in the yeast two-hybrid systems used (LexA- or Gal4-based) and/or the stringency of selection. The number of shared interactions might be taken as a crude measure of the overlap in two proteins' functions. Therefore, the dense TALE network indicates potential functional redundancy amongst TALE proteins, which might account for the fact that null mutations in TALE genes frequently do not exhibit obvious aberrant phenotypes (unpublished data). Apart from TALE/TALE interactions, we identified a number of additional proteins forming a homogenous, highly interconnected network (Fig. 1 b, c, and d). Significantly, this network involves nine members of a previously unrecognized plant-specific protein family denominated AtOFPs.
Fig. 1.
TALE-protein interaction network. (a) Protein interactions in the TALE cluster. +, interaction; 0, no interaction;/, not determined. (b) Network of protein interactions among TALE proteins and cofactors. Green, BELL proteins; red, KNOX proteins; black, nonhomeodomain cofactors; open circles, AtOFPs. The clustering coefficients Q and C are given as a measure of the degree of local clustering. (c) Quasi-clique formed by a fully connected hexagon motif of TALE proteins (BLH1, BLH3, BLH4, BLH10, KNAT5, and KNAT6) and three AtOFPs. (d) A highly connected quasi-clique formed by seven BELL proteins and AtOFP5.
TALE Homeodomain Proteins and AtOFPs Interact in Vivo. To confirm protein-protein interactions in vivo, we applied the bimolecular fluorescence complementation technique (24, 25). Selected TALE proteins (BLH1 and BLH7) and AtOFP1 were fused to an N-terminal (YFP-N) or a C-terminal (YFP-C) fragment of yellow fluorescent protein (YFP), respectively. Transient coexpression of these fusion constructs in agroinfiltrated N. benthamiana leaves revealed homodimerization of BLH1, homodimerization of AtOFP1, and heterodimerization of BLH1 with AtOFP1 in vivo by reconstitution of fluorescence of the split YFP. However, no fluorescence was detected when either of the constructs was coexpressed with negative controls or when AtOFP1-YFP-C was coexpressed with BLH7-YFP-N, a BELL protein shown in yeast to not interact with AtOFP1 (Fig. 6, which is published as supporting information on the PNAS web site).
The very close interconnectedness of AtOFPs in the network hint at a central role of these proteins as regulators or cofactors of TALE homeodomain proteins in plants. AtOFPs are characterized by a conserved C-terminal domain shared with the tomato OVATE protein (Fig. 5a), and most members of this family contain a predicted nuclear localization signal. In tomatoes, a premature stop codon abolishing most of the conserved C-terminal domain of the OVATE protein has been shown to cause pear-shaped fruit development. Tomato lines ectopically expressing the ovate gene exhibit pleiotropic developmental aberrations (26).
Network Topologies Indicate a Close Functional Connection of TALE Homeodomain Proteins and AtOFPs. Functional modules emerging from a network representation of protein interactions are locally highly interconnected clusters on the mesoscale level; i.e., they comprise between 5 and 50 elements (2). The TALE cluster, consisting of 34 proteins (nodes in the network) with an average node degree of 9.7, has characteristics similar to functional modules in large-scale protein interaction networks. The mean clustering coefficient, the number of observed interactions between the neighbors of a node divided by the number of possible interactions (19), indicates a high degree of local clustering (Fig. 1b). The density of the entire “TALE module,” measured by the parameter Q = 2m/n*(n - 1), where m is the number of edges and n is the number of nodes in the module, is exceptionally high and, according to the statistical analyses by Spirin and Mirny (2), can be considered significant, indicating a close functional association of the components of the TALE module. The core of the cluster is formed by four BELL and two KNOX proteins (BLH1, BLH3, BLH4, BLH10, KNAT5, and KNAT6), exhibiting the highest mutual clustering coefficients and forming a fully connected hexagon motif (Q = 1; Fig. 1c). Ranking of the clustering coefficients places AtOFPs in high positions of the hierarchy in the TALE module. AtOFP1, AtOFP2, and AtOFP4 form a quasi-clique (Q = 0.86) with the six TALE proteins indicated (Fig. 1c). AtOFP5 is integrated in a highly connected quasi-clique with seven BELL proteins (Fig. 1d), but it does not interact with any of the KNOX proteins. Taken together, the data indicate that the TALE network is an evolutionary conserved module, providing strong evidence for a close functional link between TALE proteins and the AtOFP family.
To identify the interaction domains of A. thaliana TALE proteins and AtOFPs, nested deletion constructs were analyzed by using the yeast two-hybrid system. The single conserved domain of the AtOFPs, the C-terminal 60-70 aa, was found to mediate the interaction with the homeodomains of both BELL and KNOX proteins (data not shown). Involvement of homeodomains in both DNA-binding and protein-protein interactions has previously been observed in metazoan proteins, indicating a potential general role of this domain in cofactor binding (27, 28).
The BELL and KNOX families of the TALE proteins have split early in plant evolution (12, 29). The finding that four AtOFPs interact with members of both TALE families supports an ancient functional connection that is conserved despite the fast turnover of interactions in proteome evolution (30). Evolutionary conservation of BELL and KNOX interactions with AtOFPs is corroborated by our observation that a number of TALE proteins from barley, a monocot plant species, can interact with AtOFPs (data not shown).
AtOFP1 Is Essential for Pollen Function. To address a possible functional involvement of AtOFP1 in TALE protein-dependent developmental processes, we analyzed three independent A. thaliana lines with T-DNA insertions in the single exon and in the 5′ upstream region (Fig. 5b). Among >200 individuals analyzed, no plants homozygous for the T-DNA insertion were obtained upon selfing of heterozygous plants, strongly supporting the conclusion that the AtOFP1 gene is required for essential processes in gametophyte or sporophyte development. Because neither pollen nor ovules of heterozygous plants displayed any apparent morphological aberrations, male and female transmission of the T-DNA insertion was investigated by reciprocal crosses with wild-type A. thaliana Col-0 plants and the analysis of progeny by PCR for presence of the insertion. A total of 75 plants were analyzed, and in the case of the heterozygous mutant lines as pollen donators, not a single plant containing the insertion in the AtOFP1 gene was among them. The female transmission efficiency, however, was not affected. These data suggest an essential role of AtOFP1 in male transmission and pollen function.
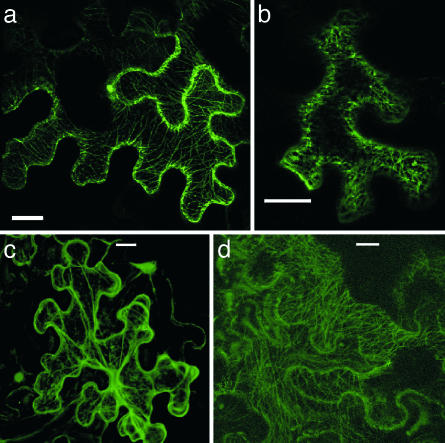
Ectopic Expression of AtOFP1 Causes Pleiotropic Developmental Abnormalities. Transgenic A. thaliana plants expressing AtOFP1 under control of the CaMV35S promoter exhibited dominant pleiotropic phenotypes (Fig. 2). Plants were stunted and characterized by thickened stems, delayed development, and general shortening and thickening of all aerial parts. Leaves were irregularly heart-shaped and lobed and displayed curved surfaces (Fig. 2 a, b, and e). Anthers were short with a thick filament, and style and stigma protruded from the flower (Fig. 2 c and d); siliques were short and uneven and produced few seeds (Fig. 2f). Identity, number, and order of floral organs, however, were unchanged. Tobacco plants expressing AtOFP1 from the CaMV35S promoter displayed similar phenotypical alterations (Fig. 2g). We conclude that the coordinated growth of plant organs or their parts, known as allometry, is hindered in AtOFP1-overexpressing plants, supporting a hierarchically high-ranking role of this gene in plant growth and shape regulation.
Fig. 2.
Ectopic expression of AtOFP1. Overexpression of AtOFP1 from the CaMV 35S promoter causes dominant pleiotropic developmental aberrations in A. thaliana (a-f) and tobacco (g). (a and b) Stunted growth of 35S::AtOFP1 plants. (c) Flowers exhibit oval petals and sepals and protruding styles. (d) Stamens have shorter filaments than do wild-type plants. (e) Leaves are lobed and asymmetric and have broader midveins than wild-type leaves. (f) Siliques are shortened and contain only a few fertile seeds. (g) Flowers, leaves, and siliques of tobacco plants overexpressing AtOFP1 are altered in a similar way as in A. thaliana. (h) RT-PCR analysis reveals decreased levels of AtGA20ox1 mRNA in vegetative leaf tissue of AtOFP1-overexpressing plants. Control amplification of RAN3 transcript indicates equal amounts of cDNA. [Scale bars: c, d, and f, 1 mm; e, 20 mm (enlargement, 4 mm); g, 20 mm.]
Aberrant leaf phenotypes of Knox-overexpressing plants have been shown previously to be at least partially caused by decreased levels of bioactive gibberellins due to a direct repression of the GA20-oxidase1 gene (15-17). To examine whether ectopically expressed AtOFP1 acts in this functional context, expression of all A. thaliana Bell and Knox genes and of the GA20-ox1 gene in wild-type A. thaliana and in plants overexpressing AtOFP1 were analyzed. Whereas the transcript accumulation of TALE genes was unaffected, expression of the GA20-ox1 gene was decreased by 80% compared with the wild-type level (Fig. 2h). This result agrees with the view that a hormonal imbalance in AtOFP1-overexpressing plants contributes to their particular phenotype and suggests that AtOFP1 regulates TALE protein-mediated processes at the posttranslational level rather than the transcriptional level.
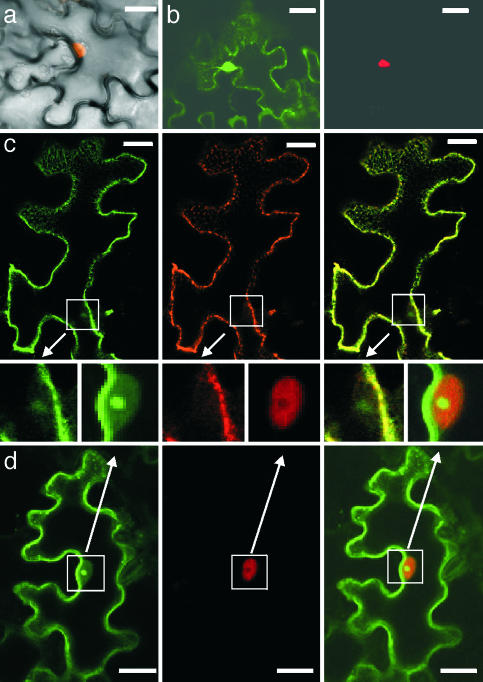
AtOFPs Regulate Subcellular Localization of TALE Proteins. To get insights into the mechanism of the functional interaction between TALE proteins and AtOFPs, their subcellular localization was analyzed by means of transient expression of fluorescently tagged proteins in N. benthamiana leaves. Both AtOFP1-GFP and AtOFP5-GFP accumulated in subnuclear structures, presumably nucleoli, and associated with structures reminiscent of the cortical cytoskeleton (Fig. 3a; see also Fig. 7d, which is published as supporting information on the PNAS web site). Visualization of the actin and tubulin cytoskeleton by using GFP-tagged MAP4 or talin, respectively, and the application of the actin depolymerizing drug cytochalasin D suggest that these structures correspond to microtubules (Fig. 3 b, c, and d). In contrast, BELL and KNOX proteins such as BLH1 and KNAT1 were located exclusively in the nucleus but not in the nucleolus (Figs. 4 a and b and 7 a and b). Coexpression of RFP-tagged BLH1 or KNAT1 with AtOFP1-GFP resulted in a relocalization of the TALE proteins from the nucleus to the cytoplasmic space. Colocalization with AtOFP1 occurred in discrete punctate structures apparently aligned with the cytoskeleton and in peripheral regions of the cytoplasm (Figs. 4c and 7c). A similar alteration of the subcellular distribution of BLH1 was observed upon coexpression with AtOFP5 (Fig. 7e). Recruitment of BLH1-RFP to the cytoplasmic space was consistently observed in four independent infiltration experiments with >60 cells coexpressing BLH1 and AtOFP1 analyzed in each experiment. The relocalization depends on the interaction of AtOFP1 with specific BELL or KNOX proteins; AtOFP1-GFP had no effect on the localization of BLH7-RFP, a BELL protein unable to interact with AtOFP1 (Fig. 4d). Similarly, coexpression of BLH1 with a truncated AtOFP1 protein lacking the conserved C-terminal domain did not alter the intracellular localization of BLH1, confirming the significance of the protein interaction for the observed effects (Fig. 7a).
Fig. 3.
AtOFP1 is associated with the cytoskeleton. Localization of transiently expressed GFP-fusion proteins in N. benthamiana leaves was analyzed by confocal laser scanning microscopy. (a) AtOFP1-GFP accumulates in the nucleolus and is located at the cytoskeleton. (b) Localization of AtOFP1-GFP is not altered after 20-min treatment with the actin depolymerizing drug cytochalasin D. (c) GFP fusion of the I/LWEQ domain of talin localizes to the actin filaments. (d) MAP3-GFP associates with microtubules. (Scale bars: 20 μm.)
Fig. 4.
AtOFP1 controls subcellular localization of TALE protein BLH1. Localization of transiently expressed GFP- and RFP-fusion proteins in N. benthamiana leaves was analyzed by confocal laser scanning microscopy. (a) Overlay of transmitted and fluorescent images of a BLH1-RFP-expressing cell. BLH1-RFP is localized exclusively in the nucleus. (b) Coexpression of BLH1-RFP with GFP alone does not affect the nuclear localization of BLH1. Shown are the GFP (Left) and RFP (Right) channels. (c) Coexpression of AtOFP1-GFP and BLH1-RFP in N. benthamiana leaves. BLH1-RFP is relocated from the nucleus to the cytoplasmic space and colocalizes with AtOFP1 in punctate structures at the cytoskeleton and in the cell periphery. GFP channel (Left), RFP channel (Center), and merged image (Right) are shown. (d) Coexpression of AtOFP1-GFP does not alter nuclear localization of the noninteracting BLH7-RFP. GFP channel (Left), RFP channel (Center), and merged image (Right) are shown. (Scale bars: 20 μm.)
Microtubule-dependent intracellular localization is an important means to control the activity of TALE proteins and other transcription factors in animals (10, 31). Our results strongly suggest a similar regulatory mechanism to modulate plant TALE protein action. The AtOFPs can be considered previously un-identified cofactors linking TALE protein function and cytoskeleton-dependent subcellular localization. In addition to implications for the regulation of cell-autonomous functions of TALE proteins, the identification of AtOFPs will be instrumental in elucidating the molecular machinery underlying the frequently reported capacity of plant KNOX proteins to rapidly move from cell to cell (32-34).
In conclusion, our study demonstrates that a network approach that revealed a functional module linking TALE proteins with AtOFPs is suitable to identify previously unrecognized regulatory components of complex biological processes. The central and highly crosslinked position of AtOFPs in the network corresponds to a fundamental role of the exemplarily investigated AtOFP1 in plant development. At the phenotypic level, the TALE-AtOFP1 interactions seem to be central in the maintenance of intraorgan and interorgan allometric relationships. AtOFP1 is an essential gene that therefore might be overlooked in systematic forward mutant screenings. The intriguing finding is that AtOFPs, as shown for AtOFP1 and AtOFP5, provide a link to microtubule-dependent regulation of subcellular localization of homeodomain proteins. Our findings open the possibility of investigating the integration of TALE homeodomain protein function in the multilayer intracellular and intercellular networks on a molecular level.
Supplementary Material
Acknowledgments
We thank W. Werr, C. Pozzi, and P. Schreier for critical reading of the manuscript and for helpful discussions and C. Koncz (Max-Planck-Institut für Züchtungsforschung) and H. Sommer (Max-Planck-Institut für Züchtungsforschung) for providing yeast two-hybrid libraries. This work was supported by grants from the Max Planck Society.
Author contributions: J.F.U. designed research; J.H., K.R., and J.M. performed research; J.H., K.R., J.M., F.S., and J.F.U. analyzed data; and J.F.U. wrote the paper.
Abbreviations: AtOFP, Arabidopsis thaliana ovate family protein; TALE, 3-aa loop extension; GA, gibberellin; T-DNA; transferred DNA; YFP, yellow fluorescent protein; RFP, red fluorescent protein.
References
- 1.Barabasi, A. L. & Oltvai, Z. N. (2004) Nat. Rev. Genet. 5, 101-113. [DOI] [PubMed] [Google Scholar]
- 2.Spirin, V. & Mirny, L. A. (2003) Proc. Natl. Acad. Sci. USA 100, 12123-12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li, S., Armstrong, C. M., Bertin, N., Ge, H., Milstein, S., Boxem, M., Vidalain, P. O., Han, J. D., Chesneau, A., Hao, T., et al. (2004) Science 303, 540-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giot, L., Bader, J. S., Brouwer, C., Chaudhuri, A., Kuang, B., Li, Y., Hao, Y. L., Ooi, C. E., Godwin, B., Vitols, E., et al. (2003) Science 302, 1727-1736. [DOI] [PubMed] [Google Scholar]
- 5.Pozzi, C., Müller, K. J., Rohde, W. & Salamini, F. (1999) Development: Genetics, Epigenetics, and Environmental Regulation Leaf Development (Springer, Berlin).
- 6.Chan, R. L., Gago, G. M., Palena, C. M. & Gonzalez, D. H. (1998) Biochim. Biophys. Acta 1442, 1-19. [DOI] [PubMed] [Google Scholar]
- 7.Bharathan, G., Goliber, T. E., Moore, C., Kessler, S., Pham, T. & Sinha, N. R. (2002) Science 296, 1858-1860. [DOI] [PubMed] [Google Scholar]
- 8.Santi, L., Wang, Y., Stile, M. R., Berendzen, K., Wanke, D., Roig, C., Pozzi, C., Muller, K., Muller, J., Rohde, W. & Salamini, F. (2003) Plant J. 34, 813-826. [DOI] [PubMed] [Google Scholar]
- 9.Chariot, A., Gielen, J., Merville, M. P. & Bours, V. (1999) Biochem. Pharmacol. 58, 1851-1857. [DOI] [PubMed] [Google Scholar]
- 10.Huang, H., Paliouras, M., Rambaldi, I., Lasko, P. & Featherstone, M. (2003) Mol. Cell. Biol. 23, 3636-3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muller, J., Wang, Y., Franzen, R., Santi, L., Salamini, F. & Rohde, W. (2001) Plant J. 27, 13-23. [DOI] [PubMed] [Google Scholar]
- 12.Bellaoui, M., Pidkowich, M. S., Samach, A., Kushalappa, K., Kohalmi, S. E., Modrusan, Z., Crosby, W. L. & Haughn, G. W. (2001) Plant Cell 13, 2455-2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith, H. M., Boschke, I. & Hake, S. (2002) Proc. Natl. Acad. Sci. USA 99, 9579-9584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhatt, A. M., Etchells, J. P., Canales, C., Lagodienko, A. & Dickinson, H. (2004) Gene 328, 103-111. [DOI] [PubMed] [Google Scholar]
- 15.Chen, H., Banerjee, A. K. & Hannapel, D. J. (2004) Plant J. 38, 276-284. [DOI] [PubMed] [Google Scholar]
- 16.Sakamoto, T., Kamiya, N., Ueguchi-Tanaka, M., Iwahori, S. & Matsuoka, M. (2001) Genes Dev. 15, 581-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hay, A., Kaur, H., Phillips, A., Hedden, P., Hake, S. & Tsiantis, M. (2002) Curr. Biol. 12, 1557-1565. [DOI] [PubMed] [Google Scholar]
- 18.Soellick, T. R. & Uhrig, J. F. (2001) Genome Biol. 2, RESEARCH0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watts, D. J. & Strogatz, S. H. (1998) Nature 393, 440-442. [DOI] [PubMed] [Google Scholar]
- 20.Clough, S. J. & Bent, A. F. (1998) Plant J. 16, 735-743. [DOI] [PubMed] [Google Scholar]
- 21.Horsch, R. B., Fry, J. E., Hoffman, N. L., Eicholtz, D., Rogers, S. G. & Fraley, R. T. (1985) Science 227, 1229-1231. [Google Scholar]
- 22.Alonso, J. M., Stepanova, A. N., Leisse, T. J., Kim, C. J., Chen, H., Shinn, P., Stevenson, D. K., Zimmerman, J., Barajas, P., Cheuk, R., et al. (2003) Science 301, 653-657. [DOI] [PubMed] [Google Scholar]
- 23.Voinnet, O., Rivas, S., Mestre, P. & Baulcombe, D. (2003) Plant J. 33, 949-956. [DOI] [PubMed] [Google Scholar]
- 24.Walter, M., Chaban, C., Schutze, K., Batistic, O., Weckermann, K., Nake, C., Blazevic, D., Grefen, C., Schumacher, K., Oecking, C., et al. (2004) Plant J. 40, 428-438. [DOI] [PubMed] [Google Scholar]
- 25.Hu, C. D., Chinenov Y. & Kerppola T. K. (2002) Mol. Cell 9, 789-798. [DOI] [PubMed] [Google Scholar]
- 26.Liu, J., Van Eck, J., Cong, B. & Tanksley, S. D. (2002) Proc. Natl. Acad. Sci. USA 99, 13302-13306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piper, D. E., Batchelor, A. H., Chang, C. P., Cleary, M. L. & Wolberger C. (1999) Cell 96, 587-597. [DOI] [PubMed] [Google Scholar]
- 28.Passner, J. M., Ryoo, H. D., Shen, L., Mann, R. S. & Aggarwal, A. K. (1999) Nature 397, 714-719. [DOI] [PubMed] [Google Scholar]
- 29.Becker, A., Bey, M., Burglin, T. R., Saedler, H. & Theissen, G. (2002) Dev. Genes Evol. 212, 452-457. [DOI] [PubMed] [Google Scholar]
- 30.Wagner, A. (2002) Mol. Biol. Evol. 19, 1760-1768. [DOI] [PubMed] [Google Scholar]
- 31.Ziegelbauer, J., Shan, B., Yager, D., Larabell, C., Hoffmann, B. & Tjian, R. (2001) Mol. Cell 8, 339-349. [DOI] [PubMed] [Google Scholar]
- 32.Lucas, W. J., Bouche-Pillon, S., Jackson, D. P., Nguyen, L., Baker, L., Ding, B. & Hake, S. (1995) Science 270, 1980-1983. [DOI] [PubMed] [Google Scholar]
- 33.Kim, J. Y., Yuan, Z. & Jackson, D. (2003) Development (Cambridge, U.K.) 130, 4351-4362. [DOI] [PubMed] [Google Scholar]
- 34.Kim, J. Y., Yuan, Z., Cilia, M., Khalfan-Jagani, Z. & Jackson, D. (2002) Proc. Natl. Acad. Sci. USA 99, 4103-4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.