Abstract
Indoor warm-water therapy pool workers in a Midwestern regional hospital were diagnosed with non-tuberculosis pulmonary hypersensitive pneumonitis and Mycobacterium avium infections. In response, we conducted a multiseason survey of microorganisms present in this therapy pool water, in biofilms associated with the pool containment walls, and in air immediately above the pool. The survey used culture, microscopy, and culture-independent molecular phylogenetic analyses. Although outfitted with a state-of-the-art UV-peroxide disinfection system, the numbers of bacteria in the therapy pool water were relatively high compared with the potable water used to fill the pool. Regardless of the source, direct microscopic counts of microbes were routinely ≈1,000 times greater than conventional plate counts. Analysis of clone libraries of small subunit rRNA genes from environmental DNA provided phylogenetic diversity estimates of the microorganisms collected in and above the pool. A survey of >1,300 rRNA genes yielded a total of 628 unique sequences, the most common of which was nearly identical to that of M. avium strains. The high proportion of clones with different Mycobacterium spp. rRNA genes suggested that such organisms comprised a significant fraction of microbes in the pool water (to >30%) and preferentially partition into aerosols (to >80%) relative to other waterborne bacteria present. The results of the study strongly validate aerosol partitioning as a mechanism for disease transfer in these environments. The results also show that culture protocols currently used by public health facilities and agencies are seriously inadequate for the detection and enumeration of potential pathogens.
Keywords: 16S rRNA genes, bioaerosols, hypersensitive pneumonitis, molecular survey, Mycobacterium
Public warm-water therapy pools and hot tubs are implicated as environments with high exposure to common waterborne and airborne pathogens, such as Legionella and Mycobacterium spp. (1-5). Monitoring and regulation of the microbiology relevant to public health in warm-water recreational pools and other environments has relied primarily on culture-based analyses that specifically target classical indicator organisms. Although culture can be successful for assessment of some microbes, a large body of gene sequence-based studies shows that standard enrichment techniques significantly underestimate the actual quantity and diversity of microorganisms in a wide variety of environments (6). Further, some waterborne pathogens (e.g., Vibrio cholera and Legionella pneumophila) are documented to remain viable for extended periods of time, but are unrecoverable by otherwise successful culture protocols (7, 8). Microbes associated with warm-water recreational and therapy pool environments have been studied only by culture-based methods and, because of the limitations of the culture approach, remain poorly characterized.
Microorganisms and other microbiological materials can become airborne and then are termed “bioaerosols.” The enrichment and partitioning of waterborne microorganisms into bioaerosols may be an important factor in disease transmission associated with warm-water pool use. Adverse health effects can be caused by inhalation of viable airborne pathogens and also by inhalation of inactive microorganisms and/or their component parts (9, 10). For example, toxic or inflammatory pneumonia, bronchitis, and asthma are of growing public health concern and do not depend on viability of microbes to produce effect. Although there is abundant evidence to suggest that airborne microorganisms or their components can cause serious respiratory illness in indoor environments (11-13), there have been few systematic investigations of bioaerosols generated by warm-water pools and their appurtenances.
During March 2000, an indoor warm-water therapy pool at a Midwestern regional hospital was closed in response to employee complaints of extended respiratory problems. In the months preceding the closure, as many as nine employees that worked in the pool area reported the following symptoms: shortness of breath, wheezing, coughing, and night sweats. At least two of the employees were diagnosed by clinical culture with pulmonary infection by Mycobacterium avium. In response to this situation, we conducted and report here the results of a multiseason survey to characterize the identity, distribution, and abundance of bacteria in different niches of the therapy pool environment, including the aerosol immediately above the pool. We analyzed the microbial components of samples with standard culture techniques and also a culture-independent method in which small subunit (SSU) rRNA genes are amplified from samples and sequenced for identification. A main attribute of the rRNA sequence approach for characterization of microbial contents is that all organisms, not only specific targets, are detected and identified, and identification does not depend on viability. Molecular techniques have been used previously for water and bioaerosol research, but only for detection of specific microorganisms [e.g., L. pneumophila and Mycobacterium tuberculosis (14, 15)]
This study of a therapeutic swimming pool is a general molecular analysis of public pool bioaerosols and of a public health application in general. We report that conventional disinfection systems may not inhibit the growth of pathogenic microorganisms at elevated operating temperatures common to therapy pools and hot tubs, and that potential pathogens can preferentially partition into aerosols. We also specifically identify in this pool a host of potential pathogens that may be widespread in such settings and should be considered of public health concern.
Materials and Methods
Hospital Therapy Pool Environment. The hospital therapy pool sampled used an independent ventilation system, separated from the ventilation system of the regional hospital located in the same building complex. The pool air relative humidity was between 25% and 40%, and the temperature was between 25°C and 30°C, whereas the pool water was held at 33°C. The volume of the pool water was ≈208 m3, and the nominal volume of air immediately above the pool was 1,100 m3. The air in the pool building was exchanged 48 times daily with outdoor air by the ventilation system (16). Pool water was cycled four times daily through six parallel, pressurized sand filters (BakerHydro Filtrations, Augusta, GA) and 12 parallel UV units with a UV dose of 40 μW·s-1·cm-2 and a water residence time of 3 s (Advanced UV Systems, Glendale, CA). Immediately after UV irradiation, hydrogen peroxide was added with a residuum of ≈100 mg·liter-1.
Sample Collection. Samples for the molecular surveys were collected on February 15, August 16, and December 31, 2001. The pool was occupied at the times of sampling. On Feb. 15 and Aug. 16, inside pool air was sampled with a closed-face, γ-radiated filter cassette that contained a 0.45-μm pore size GN-6 Metricel (hydrophilic mixed cellulose esters) membrane (Pall Gelman Laboratories, Ann Arbor, MI) and two swirling aerosol collectors (SACs) (Biosampler, SKC, Eighty Four, PA). At the sampling location, 20 ml of sterile 0.01 M PBS containing 0.01% Tween 80 (Sigma) was added to the SACs. The flow rate of all samplers was maintained at a constant level of 12.5 liters·min-1 for a sampling period of 1 h (February sampling event: 0.75 m3 sample volume) or 2 h 40 min (August sampling event: 2 m3 sample volume). The air above the pool was sampled by mounting the inlets of the filter and the SAC at ≈20 cm above the water surface, taking care to avoid direct splash from entering the samplers. The liquid from one SAC was filtered through an open-face, γ-radiated filter cassette, containing a 0.20-μm pore size Supor (hydrophilic polyethersulfone) membrane (Pall Gelman), which then was used for the molecular survey. The liquid from the other SAC was transferred to sterile polystyrene 50-ml vials (Corning) for culture and direct count. Approximately 100 ml of pool water sample was filtered through an open-face, γ-radiated filter cassette with a 0.20-μm pore size Supor membrane.
On August 16, in addition to pool air, the air immediately outside was sampled upwind of the pool building at a distance of 25 m, in a wooded surrounding, by using an otherwise identical sampling protocol except that air samplers used outside were covered with aluminum foil to protect them from sunlight. Biofilm associated with the side of the pool was collected by scraping with an aquarium glass scrubber (Lee's Aquarium and Pet Products, San Marcos, CA) modified to aspirate material through a closed-face, γ-radiated filter cassette with a 0.20-μm pore size Supor membrane. Approximately 250 ml of pool water was pulled through the filter while scraping the side of the pool just below the water surface.
Beginning in the spring of 2000, grab samples of pool water were collected monthly for direct microscopic counts. On Dec. 31, 2001, 1 ml of sand (and interstitial water) from the pressurized filters was collected by a random grab from a wet filter pan and immediately fixed with 4% paraformaldehyde or frozen for subsequent nucleic acid extraction.
Bacterial Colony Count and Microscopy Count. Bacterial colony counts were determined by spreading undiluted SAC liquid onto nutrient-rich tryptic soy agar (0.1 g·liter-1 cycloheximide to inhibit fungal growth), incubating at 37°C for 2-3 weeks, and counting colonies. For direct microscopic quantification, we followed the method by Hobbie et al. (17); pool or SAC liquid was filtered through a 0.45-μm pore size polycarbonate membrane filter (Osmonics, Minnetonka, MN), which was then stained with 0.1 μg/ml DAPI (Sigma), washed with sterile PBS, mounted in antifadent (CitiFluor, Leicester, England), and cells were counted with an epifluorescence microscope.
DNA Extraction and PCR Amplification. Filters for nucleic acid extraction were stored at -80°C until used. Material on filters was eluted with 2 ml 100 mM KCl, 10 mM Tris·HCl (pH 7.4), 10 mM NH4Cl, and 1% CA-630 (Igepal) (Sigma), and DNA was extracted with a bead-beating protocol (18). SSU rRNA genes were amplified by PCR from the extracted DNA samples according to Frank et al. (18). DNA extracts from the February 2001 sampling event were amplified with the universal primer pairs 515F (5′-GTGCCAGCMGCCGCGGTAA) and 907R (5′-CCGTCAATTCCTTTRAGTTT), and 515F and 1391R (5′-GACGGGCGGTGWGTRCA). DNA extracts from the August 2001 sampling were amplified with the universal primer pair 515F and 1391R, and the primer pair targeting bacterial genes 27F (5′AGAGTTTGATCCTGGCTCAG) and 1391R. DNA extracts from the December 2001 sampling were amplified with the primer pair targeting bacterial genes: 27F and 907R, and the primer pair targeting mycobacterial genes 515F and 1027R (5′-GCACACAGGCCACAAGGG), and 515F and 1037R (5′-CATGCACCACCTGCACACAG). A typical 50-μl PCR included: 17.5 μl of H2O; 5 μl of 10× PCR buffer; 5 μl of 50 mM MgCl2; 4 μl of dNTP mix (2.5 mM of each dNTP); 4 μl of 10 mg/ml BSA; 10 μl of 10 M betaine; 200 ng of forward primer; 200 ng of reverse primer; 0.5 μl of AmpliTaq Gold Polymerase (Applied Biosystems); and 2 μl of DNA sample. PCR was conducted with a Mastercycler gradient Machine (Eppendorf, Westbury, NY) by running 20 cycles with a gradient from 65°C to 45°C (92°C for 30 s; 65°C to 1°C/cycle for 30 s; 72°C for 90 s) after a hot start at 94°C for 12 min, and an additional 20 cycles at a 45°C annealing temperature (92°C for 30 s; 45°C for 90 s; 72°C for 90 s) before a final extension at 72°C for 20 min.
Cloning and Sequence Analyses. Cloning, restriction fragment length polymorphism (RFLP), and sequencing were performed according to methods described by Frank et al. (18) by using pGEM vectors (pGEM-T vector System I, Promega). pGEM vector primers equidistant from the DNA insert, pGEM forward (5′GAATACTCAAGCTATGC) and pGEM reverse (5′AGTGAATTGTAATACGACT), were used to amplify the plasmid inserts before RFLP analysis. Sequence analyses were conducted with a NEN Global IR2 DNA sequencer (Li-Cor, Lincoln, NE) and a MegaBase 1000 capillary sequencer (Amersham Pharmacia, Piscataway, NJ). In all instances, both strands were sequenced.
Phylogenetic Analyses. Approximate microbial species identifications were made by comparing sequences with those in the databases by using blastn (19). All sequences identified as mycobacterial were also aligned by using the computer application arb, which provides rRNA secondary structure information (20) and includes the Lane mask to identify conserved regions of the SSU-rRNA sequence (21). Maximum likelihood (ML), maximum parsimony (MP), and neighbor-joining (NJ) analyses were conducted by using paup* (22). Bootstrap analyses were performed by using NJ (1,000 bootstrap replicates) and MP (100 bootstrap replicates with 10 random additional heuristic searches per replicate).
Results
Quantification of Microorganisms in Outside and Inside Air. Inhalation is an obvious route for pulmonary infections and hypersensitive reactions caused by representatives of the M. avium complex. Activity and appurtenances in any pool create aerosols that entrain microorganisms in droplets, to form bioaerosols. We determined the microbial load in bioaerosols in the air enclosed above the pool and in outside air by using both standard culture-dependent colony counts and also a culture-independent method of direct microscopic counts. Results are summarized in Table 1. Regardless of the method used, inside counts were significantly higher than those immediately outside. Colony counts of all samples seriously underrepresented the actual microbial burden of the samples. Colony counts reported only 0.02-0.2% of direct microscopic counts. Thus, any realistic characterization of microbes in pool water or air requires methodologies that do not rely on conventional culture. By direct count, pool air typically contained ≈106 microbes per m3. This concentration corresponds to several hundred microbes per inhalation.
Table 1.
Heterotrophic plate counts and direct microscopic counts of bacteria in therapy pool water, air above the therapy pool, and air immediately outside therapy pool building during sampling events in 2001
| Sampling event | Environment | Sampler | Bacterial colony count, m−3 | Bacterial microscopy count, m−3 | Ratio colony and microscopy count, % |
|---|---|---|---|---|---|
| Feb. 15, 2001 | Pool air | Impinger | 1.7 × 104 | 8.1 × 106 | 0.21 |
| Pool water | Grab | 3.8 × 1011 | |||
| Aug. 16, 2001 | Pool air | Impinger | 6.1 × 102 | 7.3 × 105 | 0.08 |
| Pool water | Grab | 8.5 × 1012 | |||
| Outside air | Impinger | 9.0 × 101 | 5.9 × 105 | 0.02 |
Identification of Microorganisms in Pool Environments. To identify possible reservoirs for microbes, we analyzed DNA in samples of pool water, air above the pool, biofilm from the pool wall at the water-air interface, and filter sand through which the pool water was circulated. As detailed in Materials and Methods, DNA was purified from filtered samples, and SSU rRNA gene libraries were prepared by cloning PCR products developed by using several sets of primers with different specificities. Analysis of libraries based on universally conserved primers provided an estimate of the overall phylogenetic diversity in the samples. Bacteria-specific primers validated results obtained with universal primers; overlapping suites of sequences were expected and observed. We also used mycobacteria-specific primers to develop clone libraries with some of the samples. Overall, we screened >1,300 rRNA gene clones from 16 libraries and determined a total of 628 unique sequences.
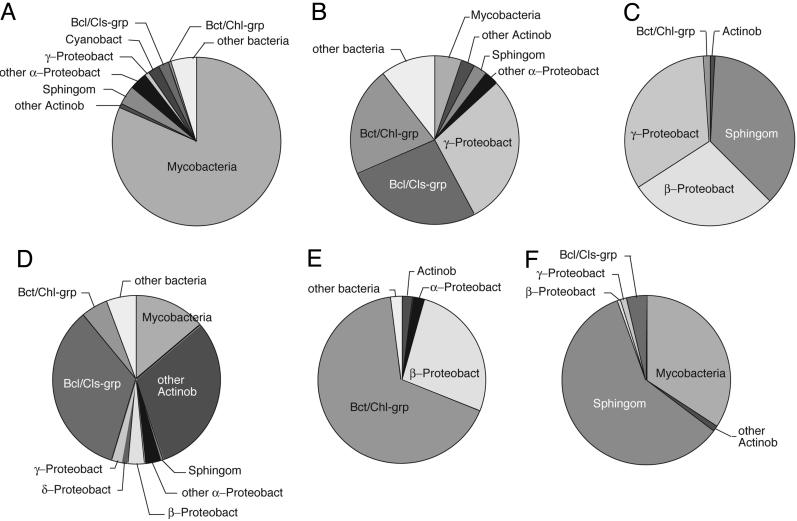
Sequences were compared with one another and sequences in the public databases, and phylogenetic assessments were conducted. Bacterial clone counts based on sequence identifications were compiled for the different PCR libraries and samples and are summarized in Fig. 1. The compositions of the different samples were highly variable. Many of the sequences in this study were similar to sequences in different samples and to database sequences, including sequences of cultivated organisms.
Fig. 1.
Phylogenetic distribution of pool environment bacterial rRNA genes. Clone sequences were grouped phylogenetically to accompany the text discussion, and percentages of sequences associated with the indicated groups are shown as pie charts. (A) Pool air, February. (B) Pool water, February. (C) Sand filter, December. (D) Pool air, August. (E) Outside air, August. (F) Pool water and side biofilm, August. Mycobacteria, Mycobacterium spp.; Actinob, Actinobacteria; other Actinob, Actinobacteria excluding mycobacteria; Sphingom, Sphingomonadaceae; α-Proteobact, α-proteobacteria; other α-Proteobact, α-proteobacteria excluding Sphingomonadaceae; β-Proteobact, β-proteobacteria; γ-Proteobact, γ-proteobacteria; δ-Proteobact, δ-proteobacteria; Cyanobact, Cyanobacteria; Bcl/Cls-grp, Bacillus/Clostridium group; Bct/Chl-grp, Bacteroidetes/Chlorobi group; other bacteria, bacteria not included in other groups.
To correlate sequence comparisons with traditional binomial nomenclatures, we take ≥97% rRNA sequence identity to correspond to species-level relationships and ≥95% sequence identity to indicate genus-level relatedness (23, 24). Among the clones sequenced, 541 (86%) showed ≥97% identity to a database sequence, indicating species-level relationship between the environmental organisms and the database reference organisms. Organisms related at the species level are expected to have common traits, so the properties of the cultured landmark organisms can be used to predict the properties of environmental organisms detected only by sequence. Eighty-six percent of the sequences corresponded to genus-level identification, and the remaining sequences were >90% identical to a database sequence, roughly the family level of identification. One particularly unusual organism was detected in this survey, a representative of Crenarchaeota, one of two archaeal kingdoms. The crenarchaeote sequence was detected only in a single sample (pool water, 4/51 clones) and was only ≈83% identical to the rRNA sequences of its closest relatives, uncultured marine Crenarchaeota. This is a very low level of sequence identity in this phylogenetic group. Thus, the sequence obtained from the therapeutic pool indicates a previously unknown and deeply divergent line of descent in the Crenarchaeota.
The distributions of organisms identified by the sequences in the PCR libraries may provide some clues to their functions in the pool microbial community. For instance, the sand filter (Fig. 1C) and pool water and side biofilm (Fig. 1F) were enriched in sequences diagnostic of Sphingomonas spp. Such organisms are often associated with water handling systems and commonly form rich biofilms. Consequently, the pool versions of these organisms likely are involved in the deposition of biofilms in the pool water system. The enrichment of mycobacterial sequences in pool water with side biofilm (Fig. 1F) may indicate that these organisms, too, preferentially associate with and perhaps deposit biofilms. We did not encounter mycobacterial sequences in the one sand filter sample analyzed with universal probes. However, we did detect mycobacterial sequences with mycobacteria-specific primers (Table 2).
Table 2. Summary of clone counts and mycobacterial sequences identified in clone libraries complied from DNA extracted from therapy pool water, air above the therapy pool, and air immediately outside therapy pool building during sampling events in 2001.
| Clones
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sampling event | Environment | Sampler | Primer pair | Mycobacterial | Bacterial | Archaeal | Fungal | Eucaryal | Total analyzed | Ratio mycobacterial and bacterial clones, % | Ratio fungal and total clones, % | Unique sequences |
| Feb. 15, 2001 | Pool air | Impinger | 515-907/1391* | 128 | 161 | 0 | 1 | 4 | 165 | 80 | 0.6 | 29 |
| Pool air | Filter | 515-907/1391* | 134 | 160 | 0 | 0 | 0 | 160 | 84 | 0 | 31 | |
| Pool water | Grab | 515-907 | 2 | 38 | 11 | 0 | 2 | 51 | 5 | 0 | 27 | |
| Aug. 16, 2001 | Pool air | Impinger | 27/515-1391† | 27 | 126 | 0 | 36 | 49 | 175 | 21 | 21 | 40 |
| Pool air | Filter | 27/515-1391† | 9 | 135 | 0 | 47 | 47 | 182 | 7 | 26 | 41 | |
| Pool sides‡ | Scrubber | 27/515-1391† | 62 | 181 | 0 | 0 | 2 | 183 | 34 | 0 | 13 | |
| Outside air | Impinger | 515-1391 | 0 | 29 | 0 | 49 | 59 | 88 | 0 | 56 | 22 | |
| Outside air | Filter | 515-1391 | 0 | 16 | 0 | 78 | 78 | 94 | 0 | 83 | 23 | |
| Dec. 31, 2002 | Sand filter | Grab | 27-907 | 0 | 88 | 0 | 0 | 0 | 88 | 0 | 0 | 22 |
| Sand filter | Grab | 515-1027/1037§ | 120 | 120 | 0 | 0 | 0 | 120 | 100 | 0 | 17 | |
| Total¶ | 482 | 1,054 | 11 | 211 | 241 | 1,306 | ||||||
Combination of data from primer pair 515-907 (universal) and 515-1391 (universal).
Combination of data from primer pair 27-1391 (bacterial) and 515-1391 (universal).
Pool water and sides of the pool.
Combination of data from primer pair 515-1027 (mycobacterial) and 515-1037 (mycobacterial).
Summation of clone counts.
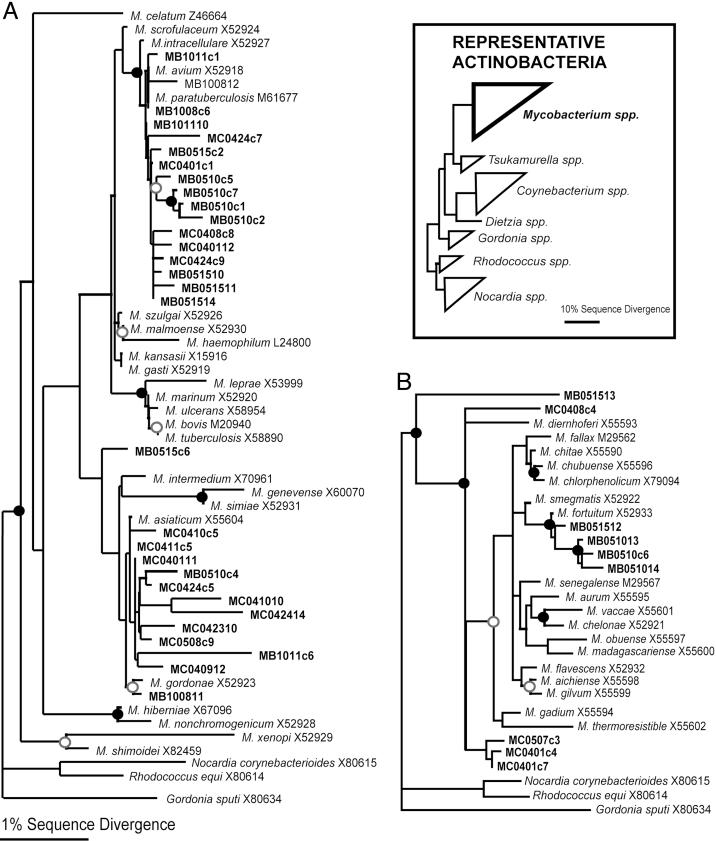
Potential Pathogens in Pool Environments. A total of 77 different mycobacterial rRNA genes, including some closely related to those of known pathogens, were detected among indoor air sequences. No mycobacteria were detected in the outside air sample. Because SSU rRNA sequences from named Mycobacterium spp. are relatively similar to one another, we conducted phylogenetic analyses to resolve the relationships between the pool mycobacteria and previously studied organisms. The alignment used for the analyses included 42 pool sequences and 42 reference sequences representing both “slow growing” and “fast growing” mycobacteria, the two large phylogenetic groups of those organisms (25, 26). One phylogenetic tree that includes the new sequences is shown in Fig. 2. Maximum likelihood, MP, and NJ analyses produced similar trees, with similar bootstrap support at resolved branches. Table 3 lists some particular pathogens to which pool sequences are closely related. The level of relatedness of the pool sequences to those of the selected pathogens, 98-99% identity, corresponds roughly to strain-level differences between the pool organisms and known pathogens, but does not necessarily indicate that the pool organisms detected by the sequences are specifically pathogenic. Nonetheless, the general distribution and relatively high abundance of mycobacterial sequences in pool water and air, particularly those associated with the M. avium complex, constitute potential health hazards.
Fig. 2.
Phylogenetic relationships of pool environment Mycobacterium spp. Shown are 16S rRNA gene sequences relative to cultured “slow-growing” (A) and “fast-growing” (B) mycobacteria. The analyses were based on an alignment of 890 nucleotide positions. Maximum likelihood, MP, and NJ algorithms resulted in identical results for the relationships supported by bootstrap analyses. NJ phylograms are shown to illustrate the phylogenetic results. Circles signify bootstrap support for nodes, with filled circles indicating MP and NJ boostrap support >70% and open circles indicating support >50%. Clones with prefixes MB and MC were obtained from the February 2001 and August 2001 sampling events, respectively. All mycobacterial clones were processed from pool air except for six clones that were processed from the pool water and sides sample (MC042310; MC0424c5; MC0424c7; MC0424c9; MC042411; and MC0508c9).
Table 3. Summary and comparison of rRNA sequence identities associated with potentially pathogenic microbes from public databases and the relative abundance of selected sequences to all sequences compiled in clone libraries.
| Nearest relative for potential pathogens* | % identity† | Clones‡ | Clone | Sample | Sampling date |
|---|---|---|---|---|---|
| M. avium complex | 99 | 108/325 | MB1008c6 | Pool air | Feb. 2001 |
| M. avium complex | 99 | 5/357 | MC0401c1 | Pool air | Aug. 2001 |
| M. avium complex | 99 | 7/183 | MC0424c9 | Pool sides§ | Aug. 2001 |
| M. asiaticum | 98 | 2/325 | MB0510c4 | Pool air | Feb. 2001 |
| M. asiaticum | 99 | 10/357 | MC0411c5 | Pool air | Aug. 2001 |
| M. asiaticum | 99 | 55/183 | MC0508c9 | Pool sides§ | Aug. 2001 |
| M. fortuitum | 99 | 34/325 | MB051512 | Pool air | Feb. 2001 |
| M. gordonae | 99 | 1/325 | MB100811 | Pool air | Feb. 2001 |
| M. diemhoferi | 99 | 7/357 | MC0610c3 | Pool air | Aug. 2001 |
| Staphylococcus hominis | 99 | 2/325 | MB050710 | Pool air | Feb. 2001 |
| Staphylococcus hominis | 99 | 8/357 | MC061013 | Pool air | Aug. 2001 |
| Streptococcus pneumoniae | 99 | 6/357 | MC0604c7 | Pool air | Aug. 2001 |
| P. melaninogenica | 99 | 1/325 | MB050715 | Pool air | Feb. 2001 |
| P. melaninogenica | 98 | 1/51 | MB1001c1 | Pool water | Feb. 2001 |
| A. otitis | 98 | 2/357 | MC040916 | Pool air | Aug. 2001 |
| Gemella morbillorum | 99 | 4/357 | MC061116 | Pool air | Aug. 2001 |
| Veillonella atypica | 99 | 2/51 | MB1003c3 | Pool water | Feb. 2001 |
| Coriobacterium glomerans | 99 | 2/51 | MB100111 | Pool water | Feb. 2001 |
Based on arb alignment and identification.
Based on blastn identification.
Number of clones per total number of clones in sample.
Pool water and sides of the pool.
Several potentially pathogenic bacteria besides mycobacteria were detected in the inside pool air and pool water (Table 3). Prevotella melaninogenica, for instance, is implicated in a number of afflictions such as vaginosis, periodontal disease, and sinusitis; Staphylococcus spp. and Streptococcus spp. detected in the pool air and water are associated with diverse human infections. Association of environmental sequences with named pathogens can be misleading as to health risk in some cases, however. Alloiococcus otitis, for instance, detected in one of the samples analyzed, is associated clinically with ear infections but also is implicated as a commensal inhabitant of the outer ear (18). We did not detect Legionella spp., which is often cited as common waterborne potential pathogens, in any of the pool samples.
Fungal rRNA sequences, possibly from spores, were abundant in August in both outdoor and indoor aerosol samples, although not in February. Fungal sources evidently were distributed inside the pool by means of the facility's ventilation system, because no fungal spores were detected in water or biofilm samples (Table 2). No known pathogenic fungi were detected. Predominant fungal rRNA gene sequences outside and inside were from the phyla Basidiomycota and Ascomycota. The most prevalent fungal sequences in the pool air sample corresponded to those of Ustilago hordei (a barleysmut fungus) and Raciborskiomyces longisetosum (phylum Ascomycota). In addition to eucaryal DNA from fungi, rRNA genes from a nematode (99% identity to the rRNA genes of Diplolaimelloides meyli) and a plant (99% identity to Atropa belladonna and Pisum sativum) were found in the pool air.
Discussion
In our daily lives we are surrounded by a complex microbiota that is mainly of unknown character. Indeed, before the advent of molecular methods for identification of organisms without culture, it was not possible to know the nature of those unknown microbes, because most evade standard culture identification. The comparison (Table 1) of colony counts with direct microscopic counts of microbes in pool water and air shows clearly the inadequacy of culture for detection of unknown microbes in the pool setting. Identification of environmental organisms by gene sequences does not require culture, and the results are unambiguous in the context of the gene sequences used in the phylogenetic analysis. The rRNA genes are particularly useful for casting a wide phylogenetic net because all organisms contain rRNA genes. Although the rRNA gene is only a rough indicator of phenotype, the rRNA sequences reveal the basic nature of the environmental organisms relative to known organisms. The census of rRNA sequences also is an assessment of the relative abundances of the different organisms represented by the sequences.
Our analyses detected and identified a broad diversity of microbes, >600 strains and species. Most of the organisms detected by the sequences probably are innocuous, but we also encountered a surprising abundance of mycobacteria closely related to known pathogens. Over several years of routine water monitoring at this site, rarely were any mycobacteria recovered by conventional culture analysis, even though the accredited laboratory used media conditions that are specific for mycobacteria. In contrast, using the molecular methods, we detected abundant mycobacteria in all pool samples. The rRNA sequences indicated substantial diversity among these mycobacteria, and a significant number of sequences had 98-99% strain-level identity to species documented to cause human disease, including M. avium sp. avium, Mycobacterium asiaticum, Mycobacterium interjectum, and M. avium sp. paratuberculosis (Fig. 2). This level of rRNA sequence variation is an indicator of potentially pathogenic character of the organisms. Particularly noteworthy is the abundant occurrence of M. avium sequences in air samples. M. avium-related organisms are adventitious pathogens and are known to elicit severe pulmonary responses. Based on clinical cultures, such organisms were implicated in some of the lifeguard syndromes that sparked this study. Cultures obtained from afflicted lifeguards had not been retained, so we were not able to compare directly sequences of the clinical isolates with the pool sequences. The specific enrichment of mycobacteria in the pool air suggested that increased air circulation could reduce the risk of exposure due to inhalation, and, since this investigation, the rate of air exchange in the pool area has been increased (16).
The abundance of mycobacterial rRNA genes relative to those of other bacteria provides strong evidence of their sustained enrichment in the pool water and associated biofilm(s). We anticipated that mycobacteria might occur in biofilms growing on containment surfaces because of their documented occurrence in chlorinated water pipes and other drinking water appurtenances (27, 28). Indeed, we observed enrichment of mycobacteria in biofilms compared with pool water (Fig. 1 B and F). The highest proportions of Mycobacterium spp., however, were observed in the pool air samples: ≈20% of bacterial clones from the summer air samples and 80% of those from the winter air samples.
Because Mycobacterium spp. were not detected in any of the outdoor air samples, the pool environment is implicated as the source of these microbes in the indoor air. A combination of several physical factors probably contributed to the partitioning of mycobacteria into an airborne state. The aerosol enrichment of Mycobacterium spp. relative to their pool water source likely was facilitated by the hydrophobic character of typical mycobacterial cell walls and their documented ability to concentrate on gas bubble surfaces (29). Several studies have suggested that air bubbles entrained in recreational aquatic facilities generate bioaerosols by a “bubble-burst, jet-drop mechanism” (30) through which bacteria that partition onto bubble surfaces can be liberated with/as droplet nuclei when they emerge from liquid surfaces (31, 32). This mechanism has been observed to aerosolize Mycobacterium spp. cells preferentially in controlled laboratory studies (33), but has not been documented in pool waters. Elevated temperatures and the presence of detergents, commonly added to therapy pools and hot tubs, are expected to enhance the aerosol partitioning potential of microbes with hydrophobic cell walls, because even small temperature increases and low surfactant concentrations markedly decrease liquid-air interface surface tension (34). Whereas several studies have implicated hot tub use in hypersensitivity pneumonitis, granulomatous lung diseases, and non-tuberculosis pulmonary infections (2, 4, 35), these results present in situ evidence for aqueous/biofilm enrichment, and the subsequent aerosol partitioning of potentially pathogenic agents.
The high abundance of Mycobacterium spp. rRNA gene sequences in the clone libraries from air is strong evidence to validate aerosol partitioning as a mechanism for acquisition of pulmonary disease in this environment. The fact that so many of the cloned sequences obtained from air samples were members of the M. avium complex indicates that an aerosol may have facilitated the hypersensitivity responses and M. avium pulmonary infections diagnosed in pool employees. The cloned sequences obtained from the pool samples also expose inadequacies in disinfection systems as well as in culture-based approaches to assess the efficiency of disinfection systems. Although many different water quality conditions may exist in public pools, inadequate disinfection may be more widespread than is indicated by conventional culture assays used for regulatory criteria. This study shows that the efficacy of disinfection methods used in these environments should be reassessed with modern molecular methods rather than with conventional culturing protocols. In general, assessment of public health in environmental settings must be done with culture-independent molecular assays that are capable of detecting a broad diversity of organisms. Culture protocols used for such assessments considerably underestimate potential pathogens.
Author contributions: L.T.A., N.R.P., and M.T.H. designed research; L.T.A., S.T.K., and A.S.A. performed research; L.T.A., S.T.K., A.S.A., and N.R.P. analyzed data; and L.T.A., S.T.K., N.R.P., and M.T.H. wrote the paper.
Abbreviations: SSU, small subunit; SAC, swirling aerosol collector; MP, maximum parsimony; NJ, neighbor-joining.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY897621-AY898248).
References
- 1.Havelaar, A. H., Berwald, L. G., Groothuis, D. G. & Baas, J. G. (1985) Zentralbl. Bakteriol. Mikrobiol. Hyg. Abt. 1 Orig. B 180, 505-514. [PubMed] [Google Scholar]
- 2.Embil, J., Warren, P., Yakrus, M., Stark, R., Corne, S., Forrest, D. & Hershfield, E. (1997) Chest 111, 813-816. [DOI] [PubMed] [Google Scholar]
- 3.Kahana, L. M., Kay, M., Yakrus, M. A. & Waserman, S. (1997) Chest 111, 242-245. [DOI] [PubMed] [Google Scholar]
- 4.Rose, C. S., Martyny, J. W., Newman, L. S., Milton, D. K., King, T. E., Jr., Beebe, J. L., McCammon, J. B., Hoffman, R. E. & Kreiss, K. (1998) Am. J. Public Health 88, 1795-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leoni, E., Legnani, P. P., Bucci Sabattini, M. A. & Righi, F. (2001) Water Res. 35, 3749-3753. [DOI] [PubMed] [Google Scholar]
- 6.Pace, N. R. (1997) Science 276, 734-740. [DOI] [PubMed] [Google Scholar]
- 7.Colwell, R. R., Brayton, P. B. & Al., E. (1985) BioTechnology 3, 817-820. [Google Scholar]
- 8.Hussong, D., Colwell, R. R., O'Brien, M., Weiss, E. & Pearson, A. D. (1987) BioTechnology 5, 947-950. [Google Scholar]
- 9.Burge, H. (1990) J. Allergy Clin. Immunol. 86, 687-701. [DOI] [PubMed] [Google Scholar]
- 10.Flannigan, B., McCabe, E. M. & McGarry, F. (1991) J. Appl. Bacteriol. Symp. Suppl. 70, 61S-73S. [PubMed] [Google Scholar]
- 11.Milton, D. K. (1999) in Bioaerosols: Assessment and Control, eds. Macher, J., Ammann, H. A., Burge, H. A., Milton, D. K. & Morey, P. R. (American Conference of Governmental Industrial Hygienists (ACGIH), Cincinnati, OH).
- 12.Sherwood, R. L. (2000) in Pulmonary Immunotoxicology, eds. Cohen, M. D., Zelikoff, J. T. & Schlesinger, R. B. (Kluwer, Boston), pp. 181-197.
- 13.Falkinham, J. O., III. (2003) Emerg. Infect. Dis. 9, 763-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mukoda, T. J., Todd, L. A. & Sobsey, M. D. (1994) J. Aerosol Res. 25, 1523-1532. [Google Scholar]
- 15.Shafer, M. P., Fernback, J. E. & Jensen, P. A. (1998) AIHA J. 59, 540-546. [DOI] [PubMed] [Google Scholar]
- 16.Kujundzic, E., Angenent, L. T., Zander, D. A., Henderson, D. E., Miller, S. L. & Hernandez, M. T. (2005) Air Waste 55, 210-218. [DOI] [PubMed] [Google Scholar]
- 17.Hobbie, J. E., Daley, R. J. & Jasper, S. (1977) Appl. Environ. Microbiol 33, 1225-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frank, D. N., Spiegelman, G. B., Davis, W., Wagner, E., Lyons, E. & Pace, N. R. (2003) J. Clin. Microbiol. 41, 295-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ludwig, W., Strunk, O., Westram, R., Richter, L., Meier, H., Yadhukumar, Buchner, A., Lai, T., Steppi, S., Jobb, G., et al. (2004) Nucleic Acids Res. 32, 1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lane, D. J., Pace, B., Olsen, G. J., Stahl, D. A., Sogin, M. L. & Pace, N. R. (1985) Proc. Natl. Acad. Sci. USA 82, 6955-6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swofford, D. (1998) paup*: Phylogenetic Analysis Using Parsimony (and Other Methods) (Sinauer, Sunderland, MA).
- 23.Stackebrandt, E. & Goebel, B. M. (1994) Int. J. Syst. Bacteriol. 44, 846-849. [Google Scholar]
- 24.McCaig, A. E., Glover, L. A. & Prosser, J. I. (1999) Appl. Environ. Microbiol. 65, 1721-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stahl, D. A. & Urbance, J. W. (1990) J. Bacteriol. 172, 116-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rogall, T., Wolters, J., Flohr, T. & Bottger, E. C. (1990) Int. J. Syst. Bacteriol. 40, 323-330. [DOI] [PubMed] [Google Scholar]
- 27.Hall-Stoodley, L. & Lappin-Scott, H. (1998) FEMS Microbiol. Lett. 168, 77-84. [DOI] [PubMed] [Google Scholar]
- 28.Falkinham, J. O., 3rd, Norton, C. D. & LeChevallier, M. W. (2001) Appl. Environ. Microbiol. 67, 1225-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de los Reyes, F. L. & Raskin, L. (2002) Water Res. 36, 445-459. [DOI] [PubMed] [Google Scholar]
- 30.Blanchard, D. C. & Syzdek. (1970) Science 170, 626-628. [DOI] [PubMed] [Google Scholar]
- 31.Woodcock, A. H. (1955) Sewage Ind. Waste 27, 1189-1192. [Google Scholar]
- 32.Ulevicius, V., Willeke, K., Grinshpun, S. A., Donnelly, J., Lin, X. & Mainelis, G. (1997) Aerosol Sci. Technol. 26, 175-190. [Google Scholar]
- 33.Parker, B. C., Ford, M. A., Gruft, H. & Falkinham, J. O., 3rd. (1983) Am. Rev. Respir. Dis. 128, 652-656. [DOI] [PubMed] [Google Scholar]
- 34.Ho, C.-F. & Jenkins, D. (1991) Water Sci. Technol. 23, 879-887. [Google Scholar]
- 35.Khoor, A., Leslie, K. O., Tazelaar, H. D., Helmers, R. A. & Colby, T. V. (2001) Am. J. Clin. Pathol. 115, 755-762. [DOI] [PubMed] [Google Scholar]