Abstract
We describe the construction of a DNA-encoded chemical library comprising 148’135 members, generated through the self-assembly of two sub-libraries, containing 265 and 559 members, respectively. The library was designed to contain building blocks potentially capable of forming covalent interactions with target proteins. Selections performed against JNK1, a kinase containing a conserved cysteine residue close to the ATP binding site, revealed the preferential enrichment of a 2-phenoxynicotinic acid moiety (building block A82) and a 4-(3,4-difluorophenyl)-4-oxobut-2-enoic acid moiety (building block B272). When the two compounds were joined by a short PEG linker, the resulting bidentate binder (A82-L-B272) was able to covalently modify JNK1 in the presence of a large molar excess of glutathione (0.5 mM), used to simulate intracellular reducing conditions. By contrast, derivatives of the individual building blocks were not able to covalently modify JNK1 in the same experimental conditions. The A82-L-B272 ligand was selective over related kinases (BTK and GAK), which also contain targetable cysteine residues in the vicinity of the active site.
DNA-encoded chemical libraries are collections of compounds, individually coupled to DNA fragments, serving as amplifiable identification barcodes. [1] The technology facilitates the construction and screening of very large compound libraries, in analogy to what routinely practiced using encoded combinatorial libraries of peptides, proteins or nucleic acids (e.g., phage display libraries, yeast display libraries, mRNA display libraries ribosome display libraries, aptamer libraries). [2]
A distinction is often made between “single pharmacophore” and “dual pharmacophore” chemical libraries. In the first case, individual compounds (no matter how complex) are coupled to a DNA fragment, which serves as amplifiable identification barcode. In the second case, two building blocks are simultaneously connected to the extremities of complementary DNA strands, thus enabling the formation of combinatorial libraries by the self-assembly of oligonucleotide conjugates.[3] We have recently described a strategy for the encoding of dual-pharmacophore libraries (also called “encoded self-assembling chemical libraries”, or ESAC libraries), which was compatible with library decoding procedures, based on high-throughput DNA sequencing. [4] ESAC libraries may facilitate the identification of synergistic building blocks, which recognize adjacent pockets on target proteins of interest. These chemical moieties need to be subsequently connected through a suitable chemical linker, in order to display protein binding in the absence of DNA. [4, 5, 6]
Encoded chemical libraries have previously been used for the discovery of covalent protein binders. For example, Nicolas Winssinger and coworkers previously reported the identification of reversible and irreversible covalent binders of bromodomain, [7] kinases, [8] proteases and phosphatases [9] from both DNA and PNA encoded chemical libraries. Recently, Cuozzo et al. published the discovery of a potent covalent inhibitor of Bruton’s tyrosine kinase (BTK), with picomolar IC50 value from a single DNA encoded library. [10]
Here, we describe the construction of a DNA-encoded self-assembling chemical library, formed by combination of 265 x 559 building blocks, yielding an ESAC library with 148’135 members. The library was designed to incorporate building blocks, which were potentially capable of forming covalent interactions with certain amino acid residues. Selections performed against JNK1 (a protein kinase containing seven cysteine residues, of which one conserved Cys-116 located in close proximity to the ATP binding site) [11] led to the identification of two synergistic building blocks (A82 = 2-phenoxynicotinic acid; B272 = 4-(3,4-difluorophenyl)-4-oxobut-2-enoic acid). The conjugate A82-L-B272, obtained by joining the two carboxylic acids with a polyethyleneglycol homobifunctional cross-linker, was able to potently and selectively modify JNK1 through Michael addition with 1:1 stoichiometry, in the presence of a large molar excess of glutathione, used to simulate the thiol-rich intracellular environment.
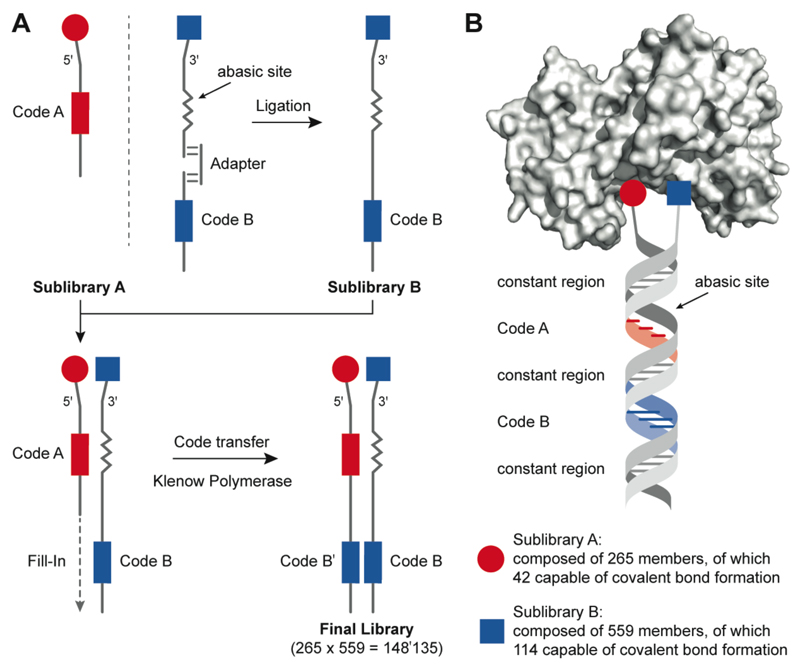
Figure 1 illustrates the procedure used for the construction and encoding of an ESAC library, featuring building blocks capable of covalent bond formation with protein targets. Two sublibraries (comprising 265 and 559 members, respectively) were synthesized by covalent modification of primary amino groups, introduced at the 5’ or 3’ extremities of partially complementary oligonucleotides. The code for oligonucleotides in Sublibrary B (carrying a chemical modification at the 3’ end) was introduced by a ligation procedure as previously described. [4] These oligonucleotides contained a short abasic site, allowing the hybridization with members of Sublibrary A. A Klenow polymerization step generated the final and fully encoded library, in which each building block pair was unambiguously identified by two oligonucleotide stretches in Sublibrary A members [Figure 1].
Figure 1. Design and encoding of the dual-display DNA-encoded chemical library.
(A) Overview of library construction. The synthesis of sub-library A (red) was performed by coupling a chemical fragment to an oligonucleotide with a distinct coding sequence that unambiguously identifies each individual library member. The conjugates were individually purified and characterized by LC-MS. The sub-library A is composed of 265 members, of which 42 are capable of covalent bond formation. The synthesis of sub-library B (blue) has been described previously (1, Wichert) and was extended with additional 359 building blocks, of which 114 are capable of covalent bond formation. With the help of an adapter, the conjugates were encoded by ligation to code B oligonucleotides. Sub-library B was hybridized with sub-library A and Klenow polymerase-assisted fill-in reaction allowed the code transfer onto the sub-library A strand to form the final library of 148’135 members. (B) Schematic representation of a member of the dual-display library binding to a target protein. The library is formed by hybridization of two individually synthesized sub-libraries A and B, resulting in a combinatorial library A × B.
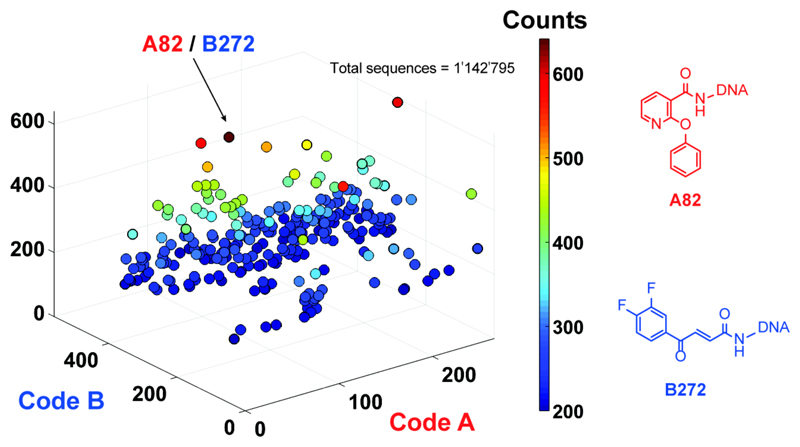
Figure 2 shows the results of a selection performed against biotinylated recombinant JNK1, immobilized on streptavidin-coated magnetic beads. [12] A plot of sequence counts for the individual library members (identified through the combination of codes A and B), revealed a preferential enrichment of the A82/B272 pair (641 counts, compared to an average count of 7.7). By contrast, the unselected library displayed an even distribution of sequence counts, while selections performed against “positive control” proteins (e.g., carbonic anhydrase IX, horseradish peroxidase) revealed a preferential enrichment of known ligands, which had been introduced as building blocks [Supplementary Figure S1]. Building block B272 (4-(3,4-difluorophenyl)-4-oxobut-2-enoic acid) is a Michael acceptor, which can potentially form a covalent bond with cysteine residues or other thiol-containing compounds. We joined the A82 and B272 moieties through a short polyethylene glycol linker, yielding the A82-L-B272 compound, whose binding properties were further characterized and compared to the ones of the individual building blocks [Figure 3].
Figure 2. Selection results from the 148’135-member dual-display library.
High-throughput DNA sequencing (HTDS) plot of selections against JNK1. The x–y plane represents library member barcodes of sub-library A and sub-library B, and the z axis shows the sequence counts (cutoff level = 200). The most enriched fragment pair corresponds to A82 (in sub-library A) and B272 (in sub-library B).
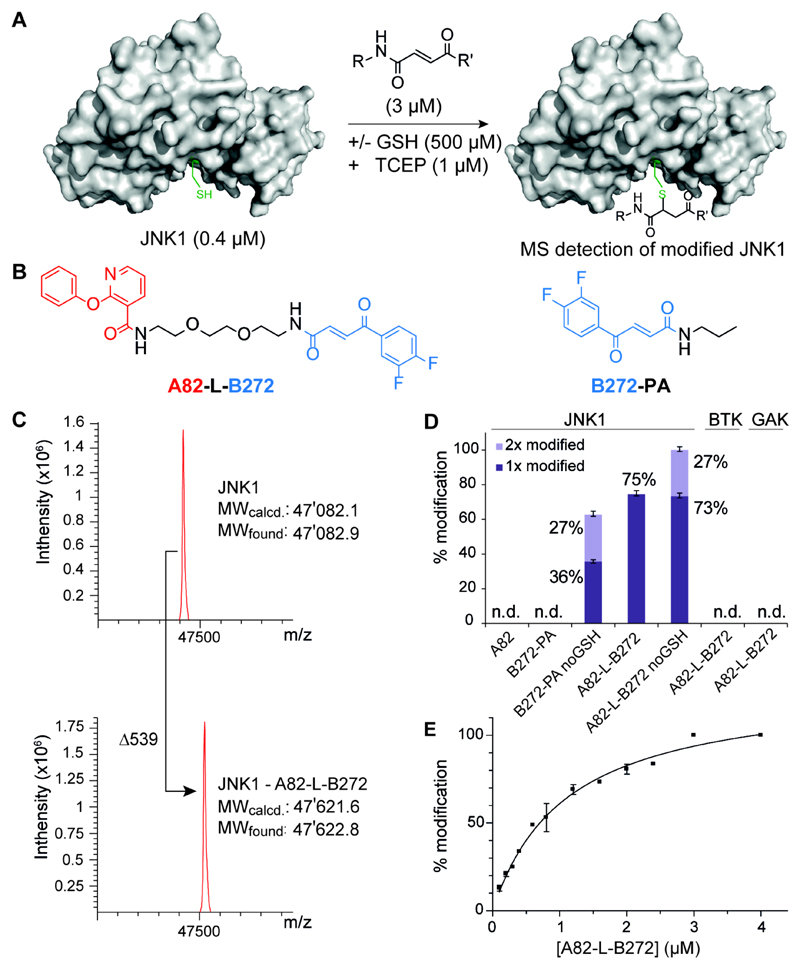
Figure 3. Mass analysis assay for covalent JNK1 binder.
(A) Schematic overview of the MS analysis assay for the detection of covalently modified JNK1 protein in the presence or absence of glutathion. (B) The most enriched fragment pair A82 and B272 connected with a short polyethyleneglycol-linker A82-L-B272 and the compound B-272 modified with a simple propylamine used as a monovalent control compound B272-PA. (C) A82-L-B272 treated JNK1 kinase domain. Mass spectra were obtained after deconvolution and show addition of 539 Daltons after incubation with A82-L-B282. Assay conditions: 0.4 µM JNK1 in 50 mM HEPES, 300 mM NaCl, 10 mM MgCl2, 1 µM TCEP; 0.5 mM glutathion; 3.0 µM A82-L-B282. (D) Hit validation of selected building block pair against JNK1, BTK and GAK. By MS analysis, monovalent A82 and B272-PA show no detectable (n.d.) JNK1 modification. Assay conditions: 0.4 µM JNK1/GAK in 50 mM HEPES, 300 mM NaCl, 10 mM MgCl2, 1 µM TCEP; 0.4 µM BTK in 20 mM Tris, 240 mM NaCl, 30 µM DTT; +/− glutathion (GSH, 0.5 mM); 1.6 µM binder. Error bars indicate standard deviations of three independent measurements. (E) Titration of JNK1 with increasing concentration of binder A82-L-B272. Titration conditions: 0.4 µM JNK1 in 50 mM HEPES, 300 mM NaCl, 10 mM MgCl2, 1 µM TCEP; glutathion (GSH, 0.5 mM); 0.1 - 4.0 µM binder A82-L-B272. Error bars indicate deviations of two independent measurements.
In order to simulate the thiol-rich intracellular milieu, we tested the ability of A82-L-B272 or of individual building blocks to covalently modify JNK1 (present at 400 nM concentration), in the presence of a large molar excess of glutathione (500 µM), by mass spectrometry [Figure 3A-C]. JNK1 contains seven cysteine residues, of which only one is close to the active site. At a 3 µM concentration, A82-L-B272 was able to covalently modify JNK1 with 1:1 stoichiometry and 75% yield, while no adduct was detectable for A82 or the B272-PA derivative [Figure 3D]. The formation of a 1:1 adduct between A82-L-B272 and JNK1 could be brought to completion by using a 4 µM concentration of the ligand, with an apparent dissociation constant of 1 µM [Figure 3E]. Omission of glutathione in the binding reaction led to the formation of adducts with 2:1 stoichiometry [Figure 3D]. Importantly, BTK [13] and Cyclin G-associated kinase (GAK), [14] two kinases with cysteine residues in the active site, did not exhibit any detectable covalent modification with A82-L-B272 under the same reaction conditions [Figure 3D], thus providing additional evidence for the JNK1 selectivity of that compound.
In conclusion, an encoded self-assembling chemical library, comprising 148’135 members, was constructed and used for the discovery of covalent ligand to JNK1, a protein of the c-Jun N-terminal protein kinase subfamily, which belongs to the mitogen-activated protein kinase (MAPK) superfamily. [15] JNKs specifically phosphorylate the transcription factor c-Jun on its N-terminal transactivation domain at two serine residues, Ser-63 and Ser-73, resulting in enhancement of c-Jun dependent transcriptional events. [16] Subsequent studies have shown that JNKs also regulate the activity of other transcription factors, including ATF2, c-Myc, JunB and p53. [17] The JNK subfamily is primarily responsive to cytokine stimulation and environmental stress ranging from osmotic shock to ionizing radiation. JNKs have shown cell type specific pro- or antiapoptotic functions, mainly depending on the duration of its activation and the activity of other signaling pathways. [18] The importance of JNKs in many pathological conditions has stimulated pharmaceutical activities, for the discovery of small molecule inhibitors. [19]
The discovery of molecules capable of selective modification of specific cysteine residues in a thiol-rich environment (such as the cell cytoplasm) represents a formidable challenge, because of the high concentration of glutathione which could potentially act as a competitor. Ibrutinib, a potent and selective covalent inhibitor of BTK, is an effective monotherapy for chronic lymphocytic leukemia (CLL), mantle cell lymphoma (MCL) and Waldenstrom’s macroglobulinemia. [20] The inhibition mechanism consists in the formation of a covalent bond between the α,β-unsaturated ketone present in the the Ibrutinib structure and Cys-481 in the active site of the target enzyme. [21]
In summary, we have shown that ESAC technology may facilitate the discovery of covalent ligands for proteins of pharmaceutical interest, in thiol-rich experimental conditions. The building block A82 facilitates the docking of the synergistic binding partner B272, thus allowing the 1:1 modification of JNK1, in the presence of a >1000-fold molar excess of glutathione. It is conceivable that additional potency gain may be obtained, replacing the flexible polyethylene glycol linker with more geometrically constrained scaffolds. Structural information could facilitate such design activities.
Supplementary Material
Experimental Section
Full experimental details are given in the Supporting Information.
Acknowledgments
Funding from ETH Zürich, the Swiss National Science Foundation (CRSII2_160699 and 310030B_163479/1), European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement No 607603), Philochem AG and is gratefully acknowledged. SK is grateful for financial support by the SGC, a registered charity (number 1097737) that receives funds from AbbVie, Bayer Pharma AG, Boehringer Ingelheim, Canada Foundation for Innovation, Eshelman Institute for Innovation, Genome Canada through Ontario Genomics Institute, Janssen, Merck & Co., Novartis Pharma AG, Ontario Ministry of Economic Development and Innovation, Pfizer, São Paulo Research Foundation-FAPESP, Takeda and the Wellcome Trust.
References
- [1].a) Lerner RA, Brenner S. Angew Chem Int Ed. 2017;56:1664–1165. doi: 10.1002/anie.201612143. [DOI] [PubMed] [Google Scholar]; b) Yuen LH, Franzini RM. Chembiochem. 2016 doi: 10.1002/cbic.201600567. [DOI] [Google Scholar]; c) Zimmermann G, Neri D. Drug Discov Today. 2016;21:1828–1834. doi: 10.1016/j.drudis.2016.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Kleiner RE, Dumelin CE, Liu DR. Chem Soc Rev. 2011;40:5707–5717. doi: 10.1039/c1cs15076f. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Shi B, Zhou Y, Huang Y, Zhang J, Li X. Bioorg Med Chem Lett. 2017;27:361–369. doi: 10.1016/j.bmcl.2016.12.025. [DOI] [PubMed] [Google Scholar]; f) Zambaldo C, Barluenga S, Winssinger N. Curr Opin Chem Biol. 2015;26:8–15. doi: 10.1016/j.cbpa.2015.01.005. [DOI] [PubMed] [Google Scholar]; Blakskjaer P, Heitner T, Hansem NJV. Curr Opin Chem Biol. 2015;26:62–71. doi: 10.1016/j.cbpa.2015.02.003. [DOI] [PubMed] [Google Scholar]; h) Clark MA, Acharya RA, Arico–Muendel CC, Belyanskaya SL, Benjamin DR, Carlson NR, Centrella PA, Chiu CH, Creaser SP, Cuozzo JW, Davie CP, et al. Nat Chem Biol. 2009;5:647–654. doi: 10.1038/nchembio.211. [DOI] [PubMed] [Google Scholar]; i) Petersen LK, Blakskjær P, Chaikuad A, Christensen AB, Dietvorst J, Holmkvist J, Knapp S, Kořínek M, Pedersen AE, Röhm S, Sløk FA, et al. Med Chem Commun. 2016;7:1332–1339. [Google Scholar]
- [2].a) McCafferty J, Griffiths AD, Winter G, Chiswell DJ. Nature. 1990;348:552–554. doi: 10.1038/348552a0. [DOI] [PubMed] [Google Scholar]; b) Kang AS, Barbas CF, Janda KD, Benkovic SJ, Lerner RA. Proc Natl Acad Sci USA. 2001;98:3750–3755. [Google Scholar]; c) Boder ET, Wittrup KD. Nature Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]; d) Wilson DS, Keefe AD, Szostak JW. Proc Natl Acad Sci USA. 2001;98:3750–3755. doi: 10.1073/pnas.061028198. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Hansen J, Pluckthun A. Proc Natl Acad Sci USA. 1997;94:4937–4942. doi: 10.1073/pnas.94.10.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Ellington AD, Szostak JW. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]; g) Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- [3].a) Melkko S, Scheuermann J, Dumelin CE, Neri D. Nature Biotechnol. 2004;22:568–574. doi: 10.1038/nbt961. [DOI] [PubMed] [Google Scholar]; b) Melkko S, Dumelin CE, Scheuermann J, Neri D. Chem Biol. 2006;13:225–231. doi: 10.1016/j.chembiol.2005.12.006. [DOI] [PubMed] [Google Scholar]
- [4].Wichert M, Krall N, Decurtins W, Franzini RM, Pretto F, Schneider P, Neri D, Scheuermann J. Nat Chem. 2015;7:241–249. doi: 10.1038/nchem.2158. [DOI] [PubMed] [Google Scholar]
- [5].Melkko S, Zhang Y, Dumelin CE, Scheuermann J, Neri D. Angew Chem Int Ed. 2007;46:4671–4674. doi: 10.1002/anie.200700654. [DOI] [PubMed] [Google Scholar]
- [6].a) Daguer J-P, Zambaldo C, Ciobanu M, Morieux P, Barluenga S, Winssinger N. Chem Sci. 2015;6:739–744. doi: 10.1039/c4sc01654h. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Barluenga S, Zambaldo C, Ioannidou HA, Ciobanu M, Morieux P, Dauguer J-P, Winssinger N. Bioorg Med Chem Lett. 2016;26:1080–1085. doi: 10.1016/j.bmcl.2015.11.102. [DOI] [PubMed] [Google Scholar]
- [7].Daguer J-P, Zambaldo C, Abegg D, Barluenga S, Tallant C, Mueller S, Adibekian A, Winssinger N. Angew Chem Int Ed. 2015;54:6057–6061. doi: 10.1002/anie.201412276. [DOI] [PubMed] [Google Scholar]
- [8].Zambaldo C, Daguer J-P, Saarbach J, Barluenga S, Winssinger N. Med Chem Commun. 2016;7:1340–1351. [Google Scholar]
- [9].Dabaene F, Da Silva JA, Pianowski Z, Duran FJ, Winssinger N. Tetrahedron. 2007;63:6577–6586. [Google Scholar]
- [10].Cuozzo JW, Centrella PA, Gikunju D, Habeshian S, Hupp CD, Keefe AD, Sigel EA, Soutter HH, Thomson HA, Zhang Y, Clark MA. ChemBioChem. 2017 doi: 10.1002/cbic.201600573. [DOI] [PubMed] [Google Scholar]
- [11].Nakaniwa T, Fukada H, Inoue T, Gouda M, Nakai R, Kirii Y, Adachi M, Tamada T, Segawa S, Kuroki R, Tada T, et al. Biochemistry. 2012;51:8410–8421. doi: 10.1021/bi300918w. [DOI] [PubMed] [Google Scholar]
- [12].Decurtins W, Wichert M, Franzini RM, Buller F, Stravs MA, Zhang Y, Neri D, Scheuermann J. Nat Protoc. 2016;11:764–780. doi: 10.1038/nprot.2016.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Marcotte DJ, Liu Y–T, Arduini RM, Hession CA, Miatkowski K, Wildes CP, Cullen PF, Hong V, Hopkins RT, Mertsching E, Jenkins TJ, et al. Protein Sci. 2010;19:429–439. doi: 10.1002/pro.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kimura SH, Tsuruga H, Yabuta N, Endo Y, Nojima H. Genomics. 1997;44:179–187. doi: 10.1006/geno.1997.4873. [DOI] [PubMed] [Google Scholar]
- [15].Pearson G, Robinson F, Beers Gibson T, Xu BE, Karandikar M, Berman K, Cobb MH. Endocr Rev. 2001;22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- [16].a) Minden A, Lin A, Smeal T, Dérijard B, Cobb M, Davis R, Karin M. Mol Cell Biol. 1994;14:6683–6688. doi: 10.1128/mcb.14.10.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Bogoyevitch MA, Kobe B. Microbiol Mol Biol Rev. 2006;70:1061–1095. doi: 10.1128/MMBR.00025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].a) Adamson AL, Darr D, Holley-Guthrie E, Johnson RA, Mauser A, Swenson J, Kenney S. J Virol. 2000;74:1224–1233. doi: 10.1128/jvi.74.3.1224-1233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]; Davis RJ. Cell. 2000;103:239–252. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]; b) Chang L, Karin M. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]; c) Karin M. J Biol Chem. 1995;270:16483–16486. doi: 10.1074/jbc.270.28.16483. [DOI] [PubMed] [Google Scholar]; d) Liu J, Lin A. Cell Res. 2005;15:36–42. doi: 10.1038/sj.cr.7290262. [DOI] [PubMed] [Google Scholar]
- [18].a) Liu ZG, Hsu H, Goeddel DV, Karin M. Cell. 1996;87:565–576. doi: 10.1016/s0092-8674(00)81375-6. [DOI] [PubMed] [Google Scholar]; b) Verheij M, Bose R, Lin XH, Yao B, Jarvis WD, Grant S, Birrer MJ, Szabo E, Zon LI, Kyriakis JM, Haimovitz-Friedman A, et al. Nature. 1996;380:75–9. doi: 10.1038/380075a0. [DOI] [PubMed] [Google Scholar]
- [19].a) Cicenas J. MAP Kinases. 2015;4:32–35. [Google Scholar]; b) Yarza R, Vela S, Solas M, Ramirez MJ. Front Pharmacol. 2016;6:321–332. doi: 10.3389/fphar.2015.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Solinas G, Becattini B. Mol Metab. 2017;6:174–184. doi: 10.1016/j.molmet.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Roskoski R., Jr Pharmacol Res. 2016;111:784–803. doi: 10.1016/j.phrs.2016.07.038. [DOI] [PubMed] [Google Scholar]
- [20].a) Burger JA, Tedeschi A, Barr PM, Robak T, Owen C, Ghia P, Bairey O, Hillmen P, Bartlett NL, Li J, Simpson D, et al. N Engl J Med. 2015;373:2425–2437. doi: 10.1056/NEJMoa1509388. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Cinar M, Hamedani F, Mo Z, Cinar B, Amin H, Alkan S. Leuk Res. 2013;37:1271–1277. doi: 10.1016/j.leukres.2013.07.028. [DOI] [PubMed] [Google Scholar]; c) Castillo JJ, Palomba ML, Advani R, Treon SP. Ther Adv Hematol. 2016;7:179–186. doi: 10.1177/2040620716654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kim ES, Dhillon S. Drugs. 2015;75:769–776. doi: 10.1007/s40265-015-0380-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.