Abstract
We have recently described a mass spectrometry-based methodology, which enables the confident identification of hundreds of peptides bound to murine major histocompatibility complex class II3 (MHCII) molecules. Here, we describe its application to the characterization of MHCII-bound peptides isolated from lymph nodes4 (LNs) of C57BL/6 mice. More than 1000 peptides could be identified in individual analyses, allowing a direct comparison of the MHCII peptidome in different types of normal LNs, or in animals with colitis. The peptide length distribution and consensus sequences in axillary, brachial, inguinal and mesenteric LNs were virtually identical and a substantial portion of identified peptides corresponded to proteins simultaneously found in all LNs. However, skin-specific proteins Sbsn, and Dmkn, and intestine-specific proteins Dmbt1, Krt19 and Maoa, among others, were exclusively identified in skin-draining or mesenteric LNs, respectively. Differences in peptide presentation patterns were also observed when comparing healthy mice and mice with dextran sodium sulfate5 (DSS)-induced colitis. Peptides derived from a subset of proteins (including IgE, Bank1, Chpf, Cmip, and Fth1) were exclusively identified in mice with colitis, revealing changes in the peptidome associated with the inflammatory process, and activation and clonal expansion of B cells.
Keywords: Autoimmunity, Antigens/Peptides/Epitopes, Antigen Presentation/Processing, Inflammation, Spleen & Lymph Nodes
Introduction
Lymph nodes (LNs) are secondary lymphoid organs, which are important for many immunological functions, including the stimulation of immune responses against pathogens and induction of peripheral tolerance (1). Migratory dendritic cells6 (DCs) collect antigen in surrounding tissue for presentation in draining LNs. Without additional danger signals (such as single stranded DNA recognized by the DC’s toll like receptors), presentation of antigens by DCs induces tolerance (2). The importance of these events can be exemplified by the observation that mice lacking CD4+ CD25+ regulatory T cells develop a destructive autoimmune gastritis in which CD4+ T cells recognize a tissue specific ATPase presented by DCs in gastric LNs (3). Until now, knowledge of which antigens are presented in lymph nodes has been mainly derived from the study of model antigens (such as ovalbumin in transgenic mice), or antigens of pathologic relevance (i.e. tumor rejection antigens, or auto-antigens in model systems of autoimmunity). An alternative approach consists in the direct identification of peptides bound to the major histocompatibility complex class II (MHCII, the MHCII peptidome) by mass spectrometry7 (MS). MHCII-bound peptides have first been sequenced by MS 25 years ago (4). However, a comparison of the MHCII peptidomes of different cell types has so far been difficult because of the large amounts of cells (typically 1x109) required, essentially prohibiting the direct analysis of mouse material without further cell expansion steps (5, 6).
Recently, we have described an experimental procedure, based on the immuno-capture of MHCII complexes followed by mass spectrometric analysis of eluted peptides, which enabled the confident identification of thousands of MHCII-bound peptides from hundred million murine cells or spleens (7). However, the characterization of the murine MHCII peptidome from LNs, without further cultivation of isolated cells, has not yet been reported. As highlighted in a recent survey, “analysis of MHC class I and II peptide ligands from cells isolated in mouse primary tissue were reported with limited success given the high number of mice needed to perform an experiment” (8).
Here, we describe an improved experimental methodology, which routinely leads to the confident identification of more than 1000 peptides from six pooled mouse LNs, while as little as two LNs are sufficient to detect 400 peptides. A detailed comparison of LNs from healthy C57BL/6 mice revealed a high similarity in the MHCII peptidomes in axillary, brachial and inguinal LNs. By contrast, presentation of tissue-specific proteins could be demonstrated comparing skin-draining LNs and mesenteric LNs. Similarly, analysis of the MHCII peptidome in healthy mice and animals with DSS-induced colitis (9) revealed differences in the corresponding peptidomes. In particular, peptides derived from immunoglobulins, Bank1, Chpf, Cmip, and Fth1 were exclusively found in mice with colitis.
Materials and Methods
Animals
Inguinal, axial, brachial, and mesenteric LNs were taken from three healthy C57BL/6 mice. Similarly, the three sets of six mesenteric LNs were obtained from different individual healthy C57BL/6 mice. For the induction of experimental colitis, female SOPF C57BL/6 mice were purchased from Janvier Labs (Laval, France). All experiments described here were performed under the project license “Dextran Sodium Sulfate (DSS)-induced colitis mouse model for diagnostics and therapy” (Bew. Nr. 26/2013), issued to Prof. Dario Neri by the local cantonal authority (Veterinäramt des Kantons Zürich).
Induction of experimental colitis in mice
Induction, monitoring and collection of samples from the DSS-colitis mice was performed as described (9). Briefly, 8 week old mice were administered between 2.5% and 3.0% (wt/vol) DSS (40,000 g/mol, TdB Consultancy) in drinking water for 5 consecutive days ad libitum. Thereafter, mice received water plus 5% glucose and 0.25% NaHCO3 for additional 7 days. From day 9, mice received normal drinking water. At day 25, mice with colitis were sacrificed and mesenteric LNs were collected. LNs were collected from healthy animals upon sacrificing them at 12 weeks of age.
Affinity purification of the MHCII complexes and peptides
The purification of MHCII complexes from tissue lysates was essentially performed as described (7). Due to enlargement of mesenteric LNs of animals with chemically-induced colitis and variation in the weight of these LNs, 20 to 25 mg of total tissue were pooled for the analysis of inflamed LNs, corresponding to one to two LNs. In brief, LNs were disrupted with a tissue lyser (Quiagen) for 2 mins at 25 Hz in lysis buffer (0.5% deoxycholate, 1% Igepal CA-630, 50 mM Tris, 150 mM NaCl, 0.1% SDS, 0.2 mM iodoacetamide, 1 mM EDTA, 1 mM PMSF, Roche Complete Protease Inhibitor Cocktail pH 7.4). MHCII complexes were purified from tissue lysates with M5/114 antibody coupled to solid support. After a two hour incubation at 4°C, unbound protein was removed by washing with 10 ml of lysis buffer, 10 ml of buffer A (150 mM NaCl, 20 mM TrisHCl pH 7.4), 10 ml of buffer B (400 mM NaCl, 20 mM TrisHCl pH 7.4), 10 ml of buffer A, and 10 ml of buffer C (20 mM TrisHCl pH 8.00). MHCII complexes were eluted after the last wash in 1 ml of 10% acetic acid. Eluates were subjected to C18 purification using Macro SpinColumns (Harvard Apparatus). Peptides were eluted in 25% acetonitrile, 0.1% trifluoroacetic acid, and dried with a vacuum concentrator (Christ Alpha RVC). The dried peptides were stored at -20°C until MS analysis.
Analysis of MHCII peptides by liquid chromatography-coupled MS
Peptides were analyzed with a Q Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo-Fisher) coupled to an EASY-nLC 1000 (Thermo-Fisher). The peptides were resolved using an Acclaim PepMap RSLC analytical column (Thermo-Fisher) with a linear gradient from 0% to 20% acetonitrile in 45 min. All buffers contained 0.1% formic acid. Spectra were collected in the Orbitrap mass analyzer using full ion scan mode over the m/z range 380–2’000, with a resolution of 70’000 with a maximum injection time of 80 ms and a MS/MS resolution of 17’500 with a maximum injection time of 240 ms. The most intense ten masses from each full mass spectrum were selected for fragmentation by higher-energy collisional dissociation in the C-trap. Spectra were processed with Proteome Discoverer (Thermo Scientific, Version 1.4.1.14) and searched with SEQUEST against a database consisting either of the murine reference proteome (52’639 entries) downloaded from the UniProt homepage on 20th January 2015, or a database consisting of the murine reference proteome (52’639 entries) concatenated with bacterial proteins of the following UniProt proteome IDs: UP000012594, UP000012542, UP000027129, UP000013570, UP000017429, UP000012445, UP000012582, and UP000012589, corresponding to the following bacterial strains: Lactobacillus sp. ASF360, Lactobacillus murinus ASF361, Lactobacillus animalis, Parabacteroides sp. ASF519, Mucispirillum schaedleri ASF457, Clostridium sp. ASF356, Clostridium sp. ASF502, and Eubacterium plexicaudatum ASF492. The total of 31’960 protein entries of the eight proteomes were downloaded from the UniProt homepage on 21th March 2016. Furthermore, following analysis settings were used for the identification of peptides and proteins: i) no-enzyme (unspecific) (ii) precursor mass tolerance 4 ppm, (iii) fragment mass tolerance 0.02 Da, (iv) one variable modification (oxidation of methionine), (v) Percolator peptide validation 1% false discovery rate. Peptide to protein annotation was performed with the DeepQuanTR software (10). All peptides were annotated to either mouse of bacterial origin. To ensure high confidence of identification, peptides were further filtered for a length of nine to 25 amino acids, typically found for MHCII peptides. Further, peptides with an XCorr value of below 2 were discarded, as these derive typically from spectra with few informative fragments. However, a manual BLAST search was performed for all bacterial high confidence identifications to rule out the probability of ambiguous annotation due to isoleucine – leucine substitutions. No homologous rodent sequences were reported for the five bacterial peptides.
For the identification of MHCII specific motifs, peptides were aligned with the GibbsCluster-1.0 server (11) using the default settings, except for the motif length, which was set to 9. A trash cluster was used to remove outliers keeping the threshold for discarding to trash at zero. MHCII specific motifs were visualized after Gibbs clustering with the Seq2Logo 2.0 server (12).
Gene expression profiling
The mouse mRNA expression dataset “GeneAtlas MOE430, gcrma” (13), accessible through BioGPS (14), was interrogated for expression data of the 20 genes identified exclusively in intestine, or skin, displayed in Table I. Data from probes demonstrating a standard deviation below five over all analyzed samples were ignored to remove transcripts with very low signal-to-noise. Remaining expression data for all probes in epidermis and intestine (including both small and large intestine) was averaged and the ratio between epidermis and intestine was calculated.
Table I.
Selection of proteins for which at least two peptides were identified in healthy axillary (aLN), brachial (bLN), inguinal (iLN) and mesenteric (mLN) lymph nodes.
| UniProt Accession Number | Protein Name | Gene Name | iLN | aLN | bLN | mLN 1 |
mLN 2 |
mLN 3 |
Peptides skin-draining LNs | Peptides mLNs |
|---|---|---|---|---|---|---|---|---|---|---|
| P04441 | H-2 class II histocompatibility antigen gamma chain | Cd74 | 16 | 28 | 21 | 50 | 15 | 18 | 32 | 52 |
| A0A075B5P6 | Ig mu chain C region | Ighm | 19 | 24 | 19 | 52 | 21 | 20 | 25 | 52 |
| P14483 | H-2 class II histocompatibility antigen, A beta chain | H2-Ab1 | 27 | 30 | 26 | 38 | 20 | 22 | 33 | 38 |
| O88307 | Sortilin-related receptor | Sorl1 | 12 | 19 | 15 | 34 | 8 | 6 | 22 | 35 |
| P13020 | Gelsolin | Gsn | 16 | 24 | 15 | 17 | 3 | 4 | 29 | 17 |
| P08226 | Apolipoprotein E | Apoe | 25 | 25 | 22 | 28 | 17 | 16 | 27 | 29 |
| P11911 | B-cell antigen receptor complex-associated protein alpha chain | Cd79a | 20 | 25 | 20 | 27 | 20 | 18 | 25 | 28 |
| P07724 | Serum albumin | Alb | 19 | 21 | 18 | 25 | 11 | 10 | 24 | 25 |
| Q62312 | TGF-beta receptor type-2 | Tgfbr2 | 7 | 9 | 5 | 19 | 6 | 11 | 13 | 26 |
| P28665 | Murinoglobulin-1 | Mug1 | 16 | 17 | 10 | 22 | 16 | 10 | 19 | 24 |
| E9QPG8 | Deleted in malignant brain tumors 1 protein | Dmbt1 | 12 | 6 | 7 | 0 | 14 | |||
| P19001 | Keratin, type I cytoskeletal 19 | Krt19 | 11 | 3 | 3 | 0 | 11 | |||
| Q64133 | Amine oxidase [flavin-containing] A | Maoa | 6 | 6 | 5 | 0 | 7 | |||
| P00329 | Alcohol dehydrogenase 1 | Adh1 | 5 | 3 | 2 | 0 | 5 | |||
| P14094 | Sodium/potassium-transporting ATPase subunit beta-1 | Atp1b1 | 5 | 1 | 1 | 0 | 5 | |||
| Q07417 | Short-chain specific acyl-CoA dehydrogenase, mitochondrial | Acads | 3 | 1 | 1 | 0 | 3 | |||
| Q9R111 | Guanine deaminase | Gda | 3 | 2 | 2 | 0 | 3 | |||
| P01634 | Ig kappa chain V-V region MOPC 21 | n/a | 1 | 2 | 1 | 0 | 2 | |||
| Q66K08 | Cartilage intermediate layer protein 1 | Cilp | 1 | 2 | 1 | 0 | 2 | |||
| P07309 | Transthyretin | Ttr | 1 | 1 | 1 | 0 | 2 | |||
| E9QPB2 | Suprabasin | Sbsn | 14 | 12 | 9 | 15 | 0 | |||
| Q8VBX4 | C-type lectin domain family 4 member K | Cd207 | 13 | 12 | 13 | 14 | 0 | |||
| E9Q2P1 | Dermokine | Dmkn | 11 | 10 | 7 | 13 | 0 | |||
| P11087 | Collagen alpha-1(I) chain | Col1a1 | 6 | 4 | 8 | 8 | 0 | |||
| P14602 | Heat shock protein beta-1 | Hspb1 | 3 | 3 | 7 | 7 | 0 | |||
| Q8BH34 | Semaphorin-3D | Sema3d | 6 | 4 | 3 | 7 | 0 | |||
| P70275 | Semaphorin-3E | Sema3e | 4 | 5 | 6 | 6 | 0 | |||
| E9PWQ3 | Protein Col6a3 | Col6a3 | 3 | 4 | 3 | 5 | 0 | |||
| O35054 | Claudin-4 | Cldn4 | 4 | 3 | 4 | 5 | 0 | |||
| Q9D708 | Protein S100a16 | S100a16 | 3 | 4 | 2 | 5 | 0 |
Matrix-based scoring of peptide sequences and peptide-MHCII binding prediction
Data from Sofron et al. (7) was the basis for the calculation of position-specific scoring matrices8 (PSSM). Peptide sequences identified either from spleens of BALB/c or C57BL/6 mice were submitted to GibbsCluster-1.1 Server (11) analysis using the standard parameters except for the motif length (set to 9 amino acids) and that the use of a trash cluster was enabled. Core sequences identified from the clustering analysis of the I-Ad, or I-Ab cluster were submitted to Seq2Logo 2.0 server (12), using the following parameters: Logo type: Kullback-Leibler, Clustering method: Hobohm1, Threshold for clustering: 0.63, Weight on prior: 200, Information content: Bits, in the advanced settings, Blossum62 matrix and respective background frequencies were enabled. From the resulting logo, the corresponding PSSM was downloaded and used for all scorings (see Supplemental Material Figure 1A-B). Scoring was performed applying a sliding window of nine amino acids to each peptide sequence, saving the highest score for each peptide, assuming that this is the most likely MHCII-binding region. For the false positive dataset, we assumed that the majority of sequences of the mouse proteome are not binding to a given MHCII allele. This assumption is supported by the fact that we only observe peptide sequences around one to ten central core sequences for any protein. Therefore, all theoretical 15mer sequences of the murine reference proteome (52’639 protein entries) downloaded from the UniProt homepage on 20th January 2015 were calculated and scored using the PSSM-based scoring function. True positive data (the scores for peptides eluted from BALB/c or C57BL/6 spleens), and the false positive data (the scores of the 15mer sequences from the murine reference proteome) were subjected to a ROC curve analysis, and the cut-off value (for which the distance to the diagonal, i.e. the random distribution, is largest) deduced from the ROC curve was defined as the minimum score to predict binding to the respective MHCII alleles.
Prediction of peptide-MHCII binding with the NetMHCIIpan 3.1 server (15) was performed keeping all standard parameters, selecting the MHCII allele I-Ad, or I-Ab and submitting peptide sequences with at least nine amino acids. The threshold for binding was defined as a rank below 10%.
Results
MHCII peptidome analysis of murine lymph nodes
We have applied a refined procedure for the analysis of MHCII-bound peptides depicted in Figure 1A. MHCII complexes were purified from tissue homogenates using the M5/114 antibody (16) coupled to a solid support. After stringent washing, peptides were eluted in acid and purified applying a single C18 purification step. This single step after elution was crucial for improved recovery of peptides, compared to ultra-filtration, still efficiently separating peptides from proteins (7). Eluted peptides were analyzed by liquid chromatography-coupled MS, applying a stringent false discovery rate (< 1%) during database search. This procedure led to an optimal balance between total number of peptide identifications and fraction of predicted MHCII binders (see Supplemental Material Figure 1C-D).
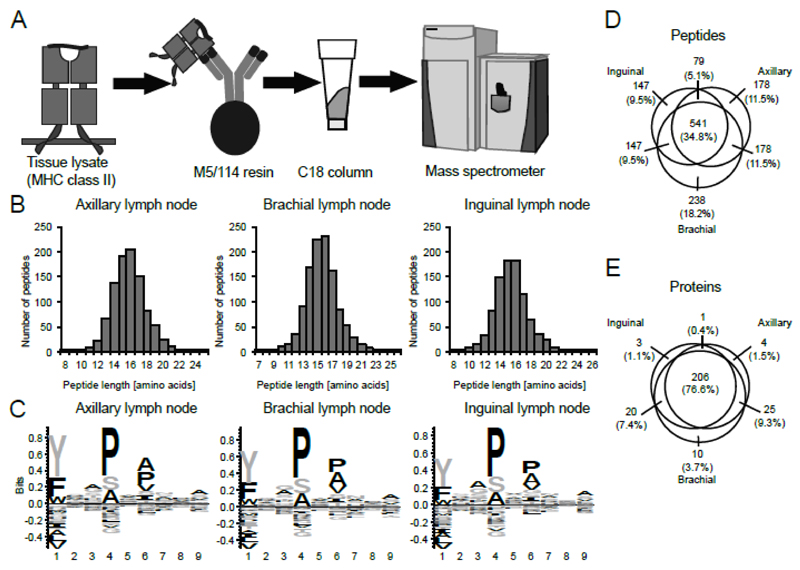
Figure 1. Characterization of the MHCII peptidome from skin-draining lymph nodes of different anatomical location.
(A) General schema for the isolation of MHCII complexes and purification of eluted peptides. MHCII complexes are purified from LN lysates with anti-murine MHCII antibody M5/114-resin. After elution of the MHCII-bound peptides with 10% acetic acid, peptides are desalted with C18 resin and analyzed by liquid chromatography-coupled MS. (B) Length distribution of peptides identified from the lysate of pools of six axillary, brachial, and inguinal LNs. (C) Consensus sequences of eluted MHCII peptides of axillary, brachial, and inguinal LNs as identified with the GibbsCluster 1.0 server (11), visualized with Seq2Logo 2.0 (12). (D-E) Peptides (D) and proteins with at least two peptides in total (E) identified from axillary, brachial, and inguinal LNs. Circles are drawn proportional to the size of the peptidomes and to the overlap between the samples, but only approximate the overlap between proteins.
Using this method, we analyzed single as well as pools of two, four and six LNs from healthy C57BL/6 animals, in an effort to determine the feasibility of the MHCII peptidome analysis of such limited amounts of sample. Peptide identifications increased from an average of 137 to 1034, when moving from one to six LNs, respectively. Throughout this paper we pooled six healthy LNs, as the quantity of identified peptides was relatively large for the limited amount of sample, and it allowed to keep the number of animals required for the analyses at a minimum.
The analysis of three pools of six LNs, derived from three C57BL/6 mice, exhibited a high level of reproducibility [Figure 1C-E, Supplemental Material Table I] with a total number of 976, 1149, and 914 peptides identified from inguinal, axillary, and brachial LNs, respectively, and an average length between 15 and 16 amino acids [Figure 1B]. The peptide consensus motifs for all samples were virtually identical [Figure 1C] and revealed the I-Ab-specific motif, in agreement with previous reports (7, 17). Between 62 and 66% of the peptides with a length of at least 15 amino acids identified in the four LN sets were predicted to bind to I-Ab by NN-align (18). While 541 peptides (34.8% of all identified peptides) were shared between axillary, brachial, and inguinal LNs, 206 source proteins with two or more peptides (76.6%) were shared among all samples, indicating that the peptidomes of these LNs are very similar [Figure 1D,E]. The observation that the peptidome of axillary, brachial and inguinal LNs are very similar is not surprising as they drain similar tissues. At this stage, it is difficult to confidently identify any significance difference between different anatomical locations. In the following section, these LNs are therefore collectively referred to as skin-draining LNs.
Comparison of the lymph node MHCII peptidome between different anatomical locations
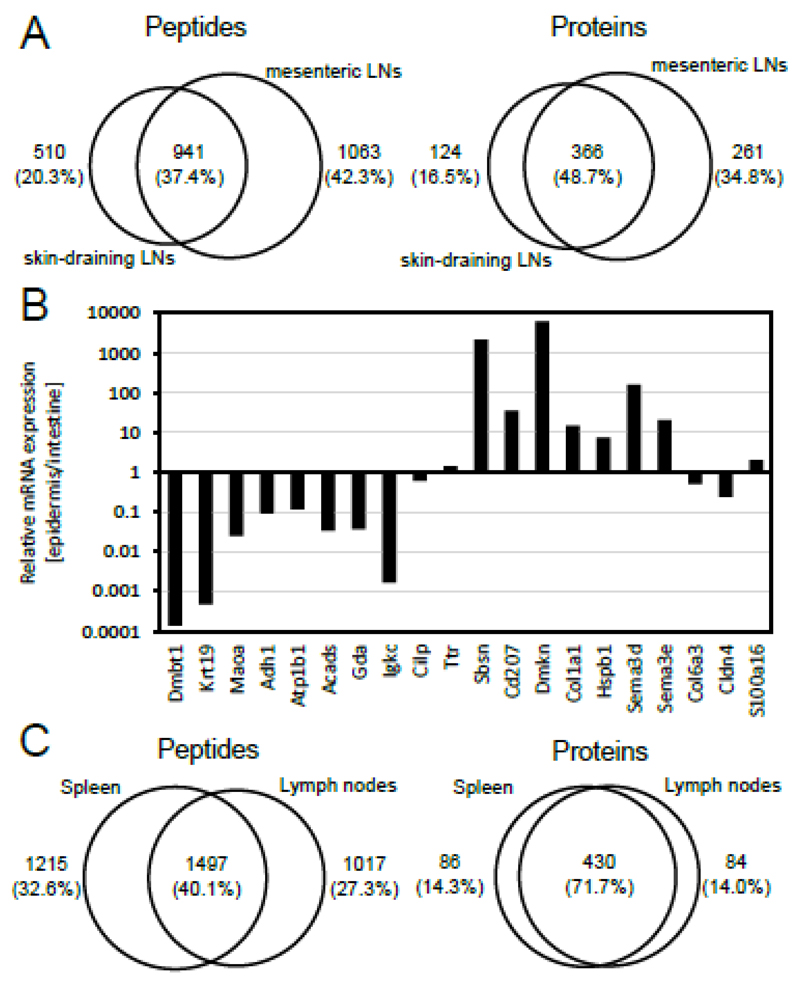
After having established the technical reproducibility of the methodology, we asked whether the MHCII peptidomes of different LNs would correspond to similar sets of peptides and proteins. We therefore collected and analyzed a triplicate set of pools of six mesenteric LNs from C57BL/6 mice. Pairwise comparisons of peptide sets identified in these LNs exhibited an overlap of 48.0 to 70.5% on the protein level. The combined mesenteric LN peptidome, resulting from the three pools, was compared to the corresponding skin-draining LN peptidome. In this case 48.7% of the source proteins were shared, while 16.5 and 34.8% were exclusively identified in skin-draining and mesenteric LNs, respectively [Figure 2A].
Figure 2. Comparative analysis of the MHCII peptidomes isolated from axillary, brachial, inguinal, mesenteric lymph nodes, and spleen.
(A) Peptides (left panel) and proteins with at least two peptides in total (right panel) identified from skin-draining, and mesenteric LNs. Circles are drawn proportional to the size of the peptidomes and to the overlap between the samples. (B) Relative mRNA expression of the proteins presented exclusively in skin-draining or mesenteric LNs (see Table I). Epidermal and intestinal mRNA expression was retrieved with BioGPS (14) averaged over all transcripts and all samples. (C) Peptides (left panel) and proteins with at least two peptides in total (right panel) identified from axillary, brachial, inguinal, and mesenteric LNs as compared with spleen. Spleen data was reported previously (7). Circles are drawn proportional to the size of the peptidomes and to the overlap between the samples.
In order to learn about differences in peptide presentation on MHCII from skin-draining and mesenteric LNs, we decided to focus on the most striking differences between the two data sets. Proteins identified with the highest number of peptides are listed in Table I, mainly corresponding to proteins of the peptide presentation machinery (MHCII alpha chain and CLIP), the B cell receptor (Cd79a) and several serum components (e.g. Apoe, Alb, and Mug1). The 38 individual peptide sequences identified from MHCII alpha chain, for example, centered around three core sequences containing signatures of the I-Ab binding motif. This observation indicates that the majority of peptides identified in this study had been eluted from MHCII and were not process-related contaminants [Supplemental Material Table I].
Cd207, a marker of epidermal Langerhans cells, was exclusively identified in axillary, brachial, and inguinal LNs. A similar observation was made for Sbsn and Dmkn, two proteins with specific expression in skin [Table I and Figure 2B]. Moreover, Col1a1, Hspb1, Sema3d, and Sema3e were found to have a higher expression in the epidermis than in the intestine [Figure 2B]. By contrast, peptide identifications in mesenteric LNs revealed Dmbt1, Krt19, Maoa, Aldh1, Atp1b1, Acads, Gda, and Igkc. The corresponding genes were preferentially expressed in the intestine [Table I and Figure 2B]. A comparison of the MHC peptidomes of LNs and spleens revealed that the tissue specific proteins, such as Dmbt1, Cd207, Sbsn, or Dmkn, were also not present in spleens. Interestingly, even though more peptides were identified from C57BL/6 spleens, a total of 1017 peptides and 84 proteins with at least two peptides were exclusively found in LN samples [Figure 2C].
Consensus motifs and scoring function
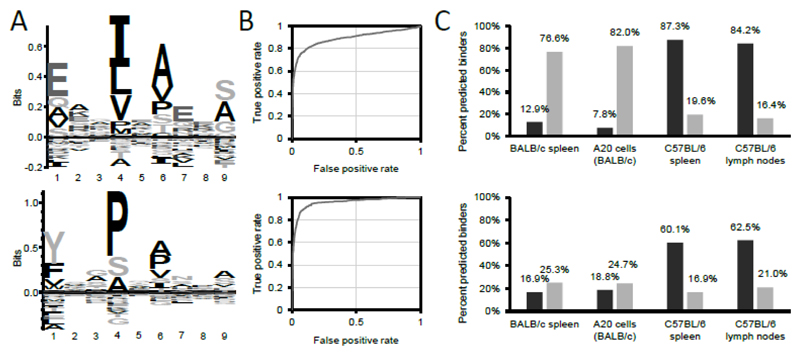
Analysis of the peptidomes derived from BALB/c and C57BL/6 spleens (the training datasets) allowed the definition of consensus motifs and of scoring functions, which could be used for the interrogation of additional biological specimens [Figure 3A].
Figure 3. Matrix-based prediction of the binding of peptide sequences to mouse MHCII alleles I-Ab and I-Ad.
(A) MHC-specific motif of I-Ad (upper panel) and I-Ab (lower panel). For the creation of the motifs, I-Ad and I-Ab complexes were purified from BALB/c or C57BL/6 spleens, respectively, and eluted peptides analyzed by mass spectrometry. Peptide sequences identified with 1% FDR were clustered with GibbsCluster-1.1 server (11) and resulting I-Ad and I-Ab-specific motifs visualized with Seq2logo 2.0 (12). Seq2logo also provides positional-specific scoring matrices (PSSM, see Supplemental Material Figure 1A-B for the PSSMs) which are the basis to calculate scores. (B) ROC curves presenting the performance of the matrix-based scoring to discriminate between a true positive set (i.e. peptide sequences eluted from I-Ad (upper panel), or I-Ab (lower panel)) and a false positive set (i.e. all 15mer sequences derived from the murine reference proteome). The point at which the distance to the diagonal (i.e. the random distribution) is largest was defined as the score cut-off predicting binding to respective MHCII alleles. According to this definition, the minimum scores to predict binding were 8.0 for I-Ad, and 8.5 for I-Ab. (C) Evaluation of the matrix-based scoring (upper panel) and NetMHCIIpan 3.1 (lower panel) (15) to predict binding of peptide sequences eluted from MHCII complexes to their cognate MHCII alleles. Peptide sequences eluted from BALB/c or C57BL/6 spleens represent the training datasets used to compute the PSSM and the minimum score to predict binding. Peptide sequences from MHCII complexes of A20 cells (a B cell lymphoma cell line derived from BALB/c mice) and C57BL/6 lymph nodes represent validation data. Binding was predicted either interrogating the peptide sequences for binding to I-Ad (light gray), or to I-Ab (dark gray), with the respective PSSMs, or by selecting the respective allele on the NetMHCIIpan 3.1 server website. Peptide sequences identified from A20 cells, BALB/c and C57BL/6 spleens were taken from (7).
To define a score threshold discriminating between binders and non-binders, true positives (i.e. peptides eluted from spleen) and false positives (i.e. all possible 15mer sequences of the murine reference proteome) were subjected to a ROC curve analysis to define the cut-off point in which the distance to the diagonal (i.e. the random distribution) is largest [Figure 3B].
The scoring function and the score threshold, obtained from this training set of peptide identifications, allowed a better description of potential binders compared to NetMHCIIpan-3.1 (15) in a validation data set, based on the peptidome of A20 cells (derived from BALB/c mice) and C57BL/6 lymph nodes [Figure 3C].
Analysis of mesenteric lymph node MHCII peptidome during experimental colitis
We further applied the methodology to study MHCII peptidome changes during DSS-induced colitis, a model routinely used to chemically induce a condition, which mimics human ulcerative colitis (19). In total, two sets of normal mesenteric LNs were compared to five pools of inflamed LNs, 25 days after induction of colitis. The MHCII peptidome of animals with colitis again was reproducible, with 223 proteins (63.5%) with at least 2 peptide sequences identified in all five samples [Supplemental Material Table II].
Prominent proteins identified exclusively or with significantly more peptides from inflamed LNs include immunoglobulins, such as IgA (Igha) and IgE (Ig epsilon chain C region), Bank1, Chpf, Cmip, Fth1, Slamf6, Tlr9, Itgb7, and Notch3 [see Table II].
Table II.
Selection of proteins identified in healthy or inflamed mesenteric lymph nodes and corresponding number of peptides.
| UniProt Accession | Protein Name | Gene Name | M11 | M21 | C12 | C22 | C32 | C42 | C52 |
|---|---|---|---|---|---|---|---|---|---|
| P14483 | H-2 class II histocompatibility antigen, A beta chain | H2-Ab1 | 31 | 36 | 34 | 36 | 38 | 38 | 35 |
| A0A075B5P6 | Ig mu chain C region | Ighm | 21 | 32 | 28 | 35 | 36 | 33 | 37 |
| P08226 | Apolipoprotein E | Apoe | 24 | 23 | 23 | 29 | 30 | 25 | 26 |
| P11911 | B-cell antigen receptor complex-associated protein alpha chain | Cd79a | 19 | 25 | 24 | 26 | 25 | 27 | 23 |
| P04441 | H-2 class II histocompatibility antigen gamma chain | Cd74 | 14 | 24 | 21 | 29 | 29 | 20 | 16 |
| O88307 | Sortilin-related receptor | Sorl1 | 16 | 15 | 17 | 25 | 28 | 24 | 21 |
| P07724 | Serum albumin | Alb | 21 | 22 | 12 | 18 | 18 | 16 | 19 |
| Q62312 | TGF-beta receptor type-2 | Tgfbr2 | 13 | 18 | 13 | 17 | 20 | 20 | 17 |
| P28665 | Murinoglobulin-1 | Mug1 | 9 | 14 | 8 | 14 | 13 | 11 | 13 |
| O09126 | Semaphorin-4D | Sema4d | 10 | 13 | 11 | 12 | 12 | 12 | 12 |
|
| |||||||||
| A0A075B6A3 | Protein Igha | Igha | 4 | 7 | 8 | 11 | 10 | 5 | 5 |
| A0A075B5P4 | Ig gamma-1 chain C region secreted form | Ighg1 | 3 | 3 | 3 | 9 | 11 | 2 | 3 |
|
| |||||||||
| P06336 | Ig epsilon chain C region | n/a | 2 | 3 | 6 | 1 | |||
| Q80VH0 | B-cell scaffold protein with ankyrin repeats | Bank1 | 2 | 2 | 4 | 3 | 1 | ||
| D6RFA4 | Chondroitin sulfate synthase 2 | Chpf | 1 | 2 | 2 | 2 | 2 | ||
| A0A075B5M2 | Protein Igkv4-61 | Igkv4-61 | 2 | 2 | 1 | 2 | |||
| Q9D486 | C-Maf-inducing protein | Cmip | 2 | 3 | 1 | 1 | |||
| P09528 | Ferritin heavy chain | Fth1 | 1 | 2 | 2 | 1 | |||
| P18527 | Ig heavy chain V region 914 | n/a | 2 | 1 | 1 | 1 | 1 | ||
| P05366 | Serum amyloid A-1 protein | Saa1 | 2 | 2 | 1 | 1 | |||
| Q9ET39 | SLAM family member 6 | Slamf6 | 2 | 3 | 1 | ||||
| Q8BZQ2 | Cysteine-rich secretory protein LCCL domain-containing 2 | Crispld2 | 1 | 2 | 2 | ||||
| D3YUE2 | Procollagen C-endopeptidase enhancer 1 | Pcolce | 1 | 1 | 1 | 2 | |||
| P50446 | Keratin, type II cytoskeletal 6A | Krt6a | 2 | 2 | 1 | ||||
| P62774 | Myotrophin | Mtpn | 2 | 2 | 1 | ||||
| Q3UV17 | Keratin, type II cytoskeletal 2 oral | Krt76 | 3 | 2 | |||||
| Q9EQU3 | Toll-like receptor 9 | Tlr9 | 1 | 1 | 1 | 1 | 1 | ||
| P26011 | Integrin beta-7 | Itgb7 | 1 | 1 | 1 | 1 | 1 | ||
| Q61982 | Neurogenic locus notch homolog protein 3 | Notch3 | 1 | 1 | 1 | 1 | 1 | ||
|
| |||||||||
| E9PZW0 | Desmoplakin | Dsp | 5 | ||||||
| M9MMK0 | Semaphorin-3B | Sema3b | 2 | 2 | |||||
| P61164 | Alpha-centractin | Actr1a | 1 | 2 | |||||
| O08852 | Polycystin-1 | Pkd1 | 2 | ||||||
| Q8R016 | Bleomycin hydrolase | Blmh | 2 | ||||||
| F6YSS8 | Intelectin-1a (Fragment) | Itln1 | 1 | 1 | |||||
| Q5U462 | CUB domain-containing protein 1 | Cdcp1 | 1 | 1 | |||||
| P49138 | MAP kinase-activated protein kinase 2 | Mapkapk2 | 1 | 1 | |||||
| Q99PG0 | Arylacetamide deacetylase | Aadac | 1 | 1 | |||||
| Q80T21 | ADAMTS-like protein 4 | Adamtsl4 | 1 | 1 | |||||
M1-M2, mesenteric LNs from healthy mice.
C1-C5, inflamed mesenteric LNs from mice with experimental colitis
Identification of bacterial antigens from mesenteric lymph nodes
The intestinal immune system maintains a delicate balance between defense against pathogens and tolerance to commensal bacteria (20). Essential for both processes are DC subsets, which present peptides from pathogenic and commensal bacteria on MHCII in mesenteric LNs, thereby initiating an adaptive immune response, or inducing tolerance (21). An aberrant immune response against commensal bacteria, however, may eventually lead to chronic colitis (20).
We probed the MS data for eight bacterial strains contained in the altered Schaedler flora, widely used to inoculate germfree founder animal breeding colonies (22), by performing a separate database search with SEQUEST. Bacterial peptide identifications were additionally filtered for XCorr > 2 (length of 9 to 25 amino acids) and compared to rodent proteomes using BLAST, to verify that these identifications were not false positives due to homologous murine proteins. After these stringent filtering criteria, only five peptides identified from healthy or inflamed mesenteric LNs could be unambiguously annotated to bacterial proteins, including one from Clostridium sp. ASF502, three from Eubacterium plexicaudatum ASF492, and one from Parabacteroides sp. ASF519 [Table III, Supplemental Material Table III]. Of the five peptides, the three longer ones had elements of I-Ab binding motifs (underlined in Table III). Even though none of the five peptides were predicted to bind to I-Ab by the matrix-based scoring (a score > 8.5 indicates binding to I-Ab, see Figure 3), one of the peptides had a score of 7.2 and may indeed represent a potential binder. The corresponding source protein represents an uncharacterized protein predicted to bind to DNA, as it contains an OmpR/PhoB-type DNA-binding domain. Interestingly, the proteins with accession N1ZXV2 and N2A3Y1 with unknown function were both found to contain DUF4874 and DUF4832 domains.
Table III.
Bacterial peptide sequences identified with high confidence from healthy (M1-M2) or inflamed (C1-C5) mesenteric lymph nodes.
| Peptide Sequence1 | PSMs2 | UniProt Accession | Modifications | q-Value3 | PEP4 | XCorr M15 | XCorr M25 | XCorr C15 | XCorr C25 | XCorr C35 | XCorr C45 | XCorr C55 | Matrix-based score6 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TQLLPLLPQ | 3 | N1ZXV2 | 0.002 | 0.0591 | 2.9 | -14.782 | |||||||
| ISERLDTIESI | 1 | N2A3B7 | 0.007 | 0.1623 | 2.8 | -7.491 | |||||||
| SSYANAEDMLTLTSH | 2 | N2A3Y1 | M9(Oxidation) | 0.006 | 0.1957 | 2.2 | 2.7 | 0.730 | |||||
| EDEPTIREVLKEYMT | 1 | N2A8C1 | 0.003 | 0.1105 | 2.4 | 7.198 | |||||||
| DILLMHCMTDGNWPETRKFY | 1 | UPI00029182197 | M8(Oxidation) | 0.006 | 0.0734 | 2.0 | -4.746 |
Potential I-Ab binding sites are underlined.
PSM, peptide to spectrum match
q-Value, false discovery rate calculated with percolator
PEP, posterior error probability
XCorr, SEQUEST XCorr values
Matrix-based binding prediction score. A score > 8.5 was found to predict binding to I-Ab.
UniParc accession number
Discussion
We here report the refinement and application of a high sensitive MHCII peptidome analysis (7) to the study of limited numbers of healthy and inflamed LNs. Initial observations suggested that the homogenization of six pooled healthy, skin-draining LNs pooled from the same mice with a bead mill results in reproducible MHCII peptidomes, with an acceptable number of peptide identifications (approx. 1000, Figure 1). It is possible, that other lysis methods, such as the MicroPestle-assisted pressure cycling technology applicable to the lysis of one to two mg of tissue (23) could yield acceptable numbers of peptides from single LNs. It, however, needs to be proven that the reproducibility can be improved over the bead mill and that the MHCII complex remains intact during this procedure.
The MHCII peptidome of inguinal, axillary and brachial LNs demonstrated that the peptidome of these LNs is comparable, with 67.4% of proteins with two or more peptides identified in all LN samples. Similar levels of overlap have previously been documented for technical replicates of HLA class I samples (24). Mesenteric LNs, however, revealed significant differences with exclusive presentation of the intestine-expressed genes Dmbt1, Maoa, Cps1, Fmo5, and Apoc2, and absence of the skin-expressed genes Sbsn, and Dmkn, among others. These findings, which are corroborated by mRNA expression data of murine specimens [Figure 2B] and immunohistochemical analysis in human specimens [www.proteinatlas.org], indicate that the MHCII peptidome of regional LNs reflects, at least in part, anatomical location. It has been suggested that peptides may be presented either by migratory DCs or by other antigen-presenting cells resident in LNs, which may access the peptides transported in lymph fluids (3, 25). Direct identification by MS may allow to study peptide presentation mechanisms, without limitation to model antigens (such as ovalbumin). However, differentiation between presentation of antigens by distinct cell populations will require the sorting of cells prior to peptidome analysis, which is complicated by the low number of cells that can be recovered after such procedure. The analysis of the MHC peptidome of different cell populations from in vivo derived material naturally represents the next challenge in the MHC peptidome analysis.
To further validate that the peptide sequences identified from C57BL/6 LNs could bind to the cognate MHCII allele (I-Ab), we established a motif-based peptide binding prediction algorithm [Figure 3 and Supplemental Material Figure 1A-B]. This algorithm was based on the I-Ab and I-Ad-specific motifs previously published (7), and allowed to predict binding of 76.6 to 87.3% of peptide sequences from various biological sources to the cognate alleles, while only 7.8 to 19.6% of peptide sequences were predicted to bind to the other allele. With the possibility to discriminate between I-Ab-eluted peptides and peptides derived from other sources, it was possible to prove that 1% FDR represents the filter leading to MHCII peptidomes of very high quality [Supplemental Material Figure 1C-D].
A comparative analysis of the MHCII peptidome of healthy mice and animals with colitis revealed changes during inflammation. Prominent proteins identified exclusively or with significantly more peptides from inflamed LNs include immunoglobulins, such as IgA (Igha) and IgE (Ig epsilon chain C region), Bank1, Chpf, Cmip, Fth1, Slamf6, Tlr9, Itgb7, and Notch3 [see Table II]. B cells control intestinal microbiota through secretion of IgA, identified with higher number of peptides during colitis. Moreover, some specific IgG sequences (e.g. Protein Igkv4-61 and Ig heavy chain V region 914) were identified exclusively during colitis indicating clonal expansion of certain antigen specific cells. It is possible that some of the IgG sequences may be derived from the capture antibody, a rat IgG2b of unknown sequence. Further, IgE-derived peptides were exclusively identified during disease, which is in line with reports demonstrating that in the murine T cell transfer model, B cells producing IgE were found to mediate oxazolone-induced colitis (26), and that an anti-IgE antibody ameliorated symptoms of DSS-induced colitis (27).
Other proteins exclusively presented during colitis include Notch3 (28), Slamf6 (29) and Bank1, which is involved in B cell receptor signaling but was recently also found to control CpG-induced interleukin 6 secretion, in which Tlr9 is also involved (30). The identification of the Chondroitin sulfate synthase 2 (Chpf) in LNs from mice with colitis was of interest, as the protein had been reported to ameliorate symptoms of DSS-induced colitis in rats, if administered orally (31). Together, these data suggest that many of the proteins, whose peptides are exclusively presented in inflamed mesenteric LNs, are involved in the immune response against pathogens or in the pathogenesis of colitis.
The intestinal immune system maintains a delicate balance between defense against pathogens and tolerance of commensal bacteria, while an aberrant immune response against the latter may eventually lead to chronic colitis. We therefore searched the MS data against a database with the proteomes of eight bacterial strains contained in the altered Schaedler flora, widely used to inoculate germfree founder animal breeding colonies (22). Of the 2188 peptides identified from healthy or inflamed mesenteric LNs, however, only five peptides were annotated unambiguously to bacterial proteins [Table III]. Similar results were obtained with a database including all bacterial proteomes from UniProt (data not shown). The performance of very large databases in MHC peptidomics experiments, however, is suboptimal due to the increased search space and can result in a high false negative rate. So far, our results indicate that the level of presentation of bacterial peptides in mesenteric LNs may be low. The main limitation for a proteomic investigation of peptides of bacterial origin relates to the heterogeneous nature of commensals and to the incomplete knowledge of their genomes. In most cases, 16S sequencing is performed to characterize intestinal bacteria, but this information is not sufficient for proteomic investigations (32). In the future, proteomic studies of germ-free animals inoculated with one or more sequenced bacterial strains would be desirable, in order to better characterize whether bacterial peptides are indeed presented in mesenteric lymph nodes.
In summary, we have described the purification of MHCII molecules from pools of six LNs leading to the high confidence identification of more than 1000 peptides on average. We have applied this methodology to the study of the MHCII peptidome of axillary, brachial, inguinal, and mesenteric LNs demonstrating the presentation of skin and intestine-specific proteins, respectively. We have also investigated on the MHCII peptidome of mesenteric LNs during DSS-induced colitis, revealing changes of the MHCII peptidome associated with the inflammatory process. It has previously been reported that the abundance of an auto-antigen is significantly increased in LNs of mice with destructive gastritis (3). Thus, the MS-based detection of specific peptides in regional lymph nodes, combined with knowledge of protein function and tissue distribution, should facilitate the discovery of auto-antigens in chronic inflammatory and autoimmune conditions.
Supplementary Material
Acknowledgements
We would like to thank Prof. C. Halin, Dr. Erica Russo, and Martina Vranova, ETH Zurich for scientific advice and technical support.
1 Grant support
D. Neri was supported by the Swiss Federal Institute of Technology Zurich (ETHZ) and by the European Research Council (ERC advanced grant “ZAUBERKUGEL”). F. Bootz was supported by the Swiss National Fund and KTI MedTech. A. Sofron, D. Ritz, T. Fugmann and D. Neri were supported by the European Community's Seventh Framework Program (FP7/2007-2013) under the grant agreement numbers 305309 (PRIAT). D. Ritz and T. Fugmann were supported by the European Union’s Seventh Framework Program (FP7/2007-2013) under grant agreement no. 305608 (EURenOmics).
Footnotes
MHCII, major histocompatibility complex class II
LN, lymph node
DSS, dextran sodium sulfate
DC, dendritic cell
MS, mass spectrometry
PSSM, position-specific scoring matrix
The online version of this article contains supplemental material.
Conflict of Interest Disclosure
Dario Neri is co-founder of Philogen, shareholder and member of the board. T. Fugmann, and D. Ritz are Philochem AG employees. The authors declare no additional competing financial interest.
References
- 1.Buettner M, Bode U. Lymph node dissection--understanding the immunological function of lymph nodes. Clin Exp Immunol. 2012;169:205–212. doi: 10.1111/j.1365-2249.2012.04602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turley SJ, Fletcher AL, Elpek KG. The stromal and haematopoietic antigen-presenting cells that reside in secondary lymphoid organs. Nature reviews Immunology. 2010;10:813–825. doi: 10.1038/nri2886. [DOI] [PubMed] [Google Scholar]
- 3.Scheinecker C, McHugh R, Shevach EM, Germain RN. Constitutive presentation of a natural tissue autoantigen exclusively by dendritic cells in the draining lymph node. The Journal of experimental medicine. 2002;196:1079–1090. doi: 10.1084/jem.20020991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hunt DF, Michel H, Dickinson TA, Shabanowitz J, Cox AL, Sakaguchi K, Appella E, Grey HM, Sette A. Peptides presented to the immune system by the murine class II major histocompatibility complex molecule I-Ad. Science. 1992;256:1817–1820. doi: 10.1126/science.1319610. [DOI] [PubMed] [Google Scholar]
- 5.Bozzacco L, Yu H, Zebroski HA, Dengjel J, Deng H, Mojsov S, Steinman RM. Mass spectrometry analysis and quantitation of peptides presented on the MHC II molecules of mouse spleen dendritic cells. Journal of proteome research. 2011;10:5016–5030. doi: 10.1021/pr200503g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suri A, Walters JJ, Kanagawa O, Gross ML, Unanue ER. Specificity of peptide selection by antigen-presenting cells homozygous or heterozygous for expression of class II MHC molecules: The lack of competition. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:5330–5335. doi: 10.1073/pnas.0330859100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sofron A, Ritz D, Neri D, Fugmann T. High-resolution analysis of the murine MHC class II immunopeptidome. European journal of immunology. 2016;46:319–328. doi: 10.1002/eji.201545930. [DOI] [PubMed] [Google Scholar]
- 8.Caron E, Kowalewski DJ, Chiek Koh C, Sturm T, Schuster H, Aebersold R. Analysis of Major Histocompatibility Complex (MHC) Immunopeptidomes Using Mass Spectrometry. Molecular & cellular proteomics : MCP. 2015;14:3105–3117. doi: 10.1074/mcp.O115.052431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bootz F, Ziffels B, Neri D. Antibody-Based Targeted Delivery of Interleukin-22 Promotes Rapid Clinical Recovery in Mice With DSS-Induced Colitis. Inflamm Bowel Dis. 2016;22:2098–2105. doi: 10.1097/MIB.0000000000000851. [DOI] [PubMed] [Google Scholar]
- 10.Fugmann T, Neri D, Roesli C. DeepQuanTR: MALDI-MS-based label-free quantification of proteins in complex biological samples. Proteomics. 2010;10:2631–2643. doi: 10.1002/pmic.200900634. [DOI] [PubMed] [Google Scholar]
- 11.Andreatta M, Lund O, Nielsen M. Simultaneous alignment and clustering of peptide data using a Gibbs sampling approach. Bioinformatics. 2013;29:8–14. doi: 10.1093/bioinformatics/bts621. [DOI] [PubMed] [Google Scholar]
- 12.Thomsen MC, Nielsen M. Seq2Logo: a method for construction and visualization of amino acid binding motifs and sequence profiles including sequence weighting, pseudo counts and two-sided representation of amino acid enrichment and depletion. Nucleic acids research. 2012;40:W281–287. doi: 10.1093/nar/gks469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lattin JE, Schroder K, Su AI, Walker JR, Zhang J, Wiltshire T, Saijo K, Glass CK, Hume DA, Kellie S, Sweet MJ. Expression analysis of G Protein-Coupled Receptors in mouse macrophages. Immunome Res. 2008;4:5. doi: 10.1186/1745-7580-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu C, Macleod I, Su AI. BioGPS and MyGene.info: organizing online, gene-centric information. Nucleic acids research. 2013;41:D561–565. doi: 10.1093/nar/gks1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andreatta M, Karosiene E, Rasmussen M, Stryhn A, Buus S, Nielsen M. Accurate pan-specific prediction of peptide-MHC class II binding affinity with improved binding core identification. Immunogenetics. 2015;67:641–650. doi: 10.1007/s00251-015-0873-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Germain RN, Bhattacharya A, Dorf ME, Springer TA. A single monoclonal anti-Ia antibody inhibits antigen-specific T cell proliferation controlled by distinct Ir genes mapping in different H-2 I subregions. Journal of immunology. 1982;128:1409–1413. [PubMed] [Google Scholar]
- 17.Zhu Y, Rudensky AY, Corper AL, Teyton L, Wilson IA. Crystal structure of MHC class II I-Ab in complex with a human CLIP peptide: prediction of an I-Ab peptide-binding motif. Journal of molecular biology. 2003;326:1157–1174. doi: 10.1016/s0022-2836(02)01437-7. [DOI] [PubMed] [Google Scholar]
- 18.Nielsen M, Lund O. NN-align. An artificial neural network-based alignment algorithm for MHC class II peptide binding prediction. BMC bioinformatics. 2009;10:296. doi: 10.1186/1471-2105-10-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wirtz S, Neurath MF. Mouse models of inflammatory bowel disease. Adv Drug Deliv Rev. 2007;59:1073–1083. doi: 10.1016/j.addr.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 20.de Souza HS, Fiocchi C. Immunopathogenesis of IBD: current state of the art. Nat Rev Gastroenterol Hepatol. 2016;13:13–27. doi: 10.1038/nrgastro.2015.186. [DOI] [PubMed] [Google Scholar]
- 21.Coombes JL, Powrie F. Dendritic cells in intestinal immune regulation. Nature reviews Immunology. 2008;8:435–446. doi: 10.1038/nri2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dewhirst FE, Chien CC, Paster BJ, Ericson RL, Orcutt RP, Schauer DB, Fox JG. Phylogeny of the defined murine microbiota: altered Schaedler flora. Appl Environ Microbiol. 1999;65:3287–3292. doi: 10.1128/aem.65.8.3287-3292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shao S, Guo T, Gross V, Lazarev A, Koh CC, Gillessen S, Joerger M, Jochum W, Aebersold R. Reproducible Tissue Homogenization and Protein Extraction for Quantitative Proteomics Using MicroPestle-Assisted Pressure-Cycling Technology. Journal of proteome research. 2016;15:1821–1829. doi: 10.1021/acs.jproteome.5b01136. [DOI] [PubMed] [Google Scholar]
- 24.Caron E, Espona L, Kowalewski DJ, Schuster H, Ternette N, Alpizar A, Schittenhelm RB, Ramarathinam SH, Lindestam Arlehamn CS, Chiek Koh C, Gillet LC, et al. An open-source computational and data resource to analyze digital maps of immunopeptidomes. Elife. 2015;4:e07661. doi: 10.7554/eLife.07661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stern LJ, Santambrogio L. The melting pot of the MHC II peptidome. Current opinion in immunology. 2016;40:70–77. doi: 10.1016/j.coi.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoving JC, Kirstein F, Nieuwenhuizen NE, Fick LC, Hobeika E, Reth M, Brombacher F. B cells that produce immunoglobulin E mediate colitis in BALB/c mice. Gastroenterology. 2012;142:96–108. doi: 10.1053/j.gastro.2011.09.044. [DOI] [PubMed] [Google Scholar]
- 27.Kang OH, Kim DK, Choi YA, Park HJ, Tae J, Kang CS, Choi SC, Nah YH, Lee HK, Lee YM. Suppressive effect of non-anaphylactogenic anti-IgE antibody on the development of dextran sulfate sodium-induced colitis. Int J Mol Med. 2006;18:893–899. [PubMed] [Google Scholar]
- 28.Sansone P, Storci G, Tavolari S, Guarnieri T, Giovannini C, Taffurelli M, Ceccarelli C, Santini D, Paterini P, Marcu KB, Chieco P, Bonafe M. IL-6 triggers malignant features in mammospheres from human ductal breast carcinoma and normal mammary gland. J Clin Invest. 2007;117:3988–4002. doi: 10.1172/JCI32533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Driel B, Wang G, Liao G, Halibozek PJ, Keszei M, O'Keeffe MS, Bhan AK, Wang N, Terhorst C. The cell surface receptor Slamf6 modulates innate immune responses during Citrobacter rodentium-induced colitis. International immunology. 2015;27:447–457. doi: 10.1093/intimm/dxv029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu YY, Kumar R, Haque MS, Castillejo-Lopez C, Alarcon-Riquelme ME. BANK1 controls CpG-induced IL-6 secretion via a p38 and MNK1/2/eIF4E translation initiation pathway. Journal of immunology. 2013;191:6110–6116. doi: 10.4049/jimmunol.1301203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hori Y, Hoshino J, Yamazaki C, Sekiguchi T, Miyauchi S, Horie K. Effects of chondroitin sulfate on colitis induced by dextran sulfate sodium in rats. Jpn J Pharmacol. 2001;85:155–160. doi: 10.1254/jjp.85.155. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen TL, Vieira-Silva S, Liston A, Raes J. How informative is the mouse for human gut microbiota research? Dis Model Mech. 2015;8:1–16. doi: 10.1242/dmm.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.