Abstract
The characterization of peptides bound to HLA (human leukocyte antigen) class I is of fundamental importance for understanding CD8+ T cell-driven immunological processes and for the development of immunomodulatory therapeutic strategies. However, until now, the mass spectrometric analysis of HLA-bound peptides has typically required billions of cells, still resulting in relatively few high-confidence peptide identifications. Capitalizing on the recent developments in mass spectrometry and bioinformatics, we have implemented a methodology for the efficient recovery of acid-eluted HLA peptides after purification with the pan-reactive antibody W6/32 and have identified a total of 27,862 unique peptides with high confidence (1% false discovery rate) from five human cancer cell lines. More than 93% of the identified peptides were 8 to 11 amino acids in length and contained signatures that were in excellent agreement with published HLA binding motifs. Furthermore, by purifying soluble HLA class I complexes from sera of melanoma patients, up to 972 high confidence peptides could be identified, including melanoma-associated antigens already described in the literature. Knowledge of the HLA class I peptidome should facilitate multiplex tetramer technology-based characterization of T cells, and allow the development of patient selection, stratification and immunomodulatory therapeutic strategies.
Keywords: HLA class I, immunopeptidomics, melanoma, sHLA tumor associated epitopes
1. Introduction
Immunotherapy is revolutionizing the treatment of cancer, particularly of metastatic melanoma, one of the malignancies with highest mutational rate [1]. Evidence has shown that melanoma cells present tumor rejection antigens on HLA class I molecules which can be recognized by cytotoxic T cells [2]. This knowledge has led to clinical investigations utilizing various immunostimulatory strategies, including cancer vaccines [3] and monoclonal antibodies directed against the immunoregulatory CTLA-4 and PD1 proteins. The latter have recently gained marketing authorization for the treatment of metastatic melanoma [4]. With the knowledge of melanoma associated peptides [5], it is possible to profile patient derived T cells for their reactivity towards melanoma lesions and gain insights into the molecular mechanism of tumor rejection [6–8]. Atlases of melanoma rejection antigens have been compiled for the last 20 years, but are still limited to the most common HLA alleles [5]. A thorough characterization of the immunopeptidome recognized by cytotoxic CD8+ T cells is urgently needed for the study of the tumor rejection process, particularly for indications other than melanoma.
Since the first report on the mass spectrometric identification of HLA-eluted peptides in 1992 [9], enormous efforts have led to the characterization of HLA-specific motifs for a variety of alleles revealing astonishing diversity in terms of peptide consensus sequences [10, 11]. Mass spectrometry was also able to identify e.g. HLA peptides recognized by melanoma-specific cytotoxic T cells [12] or peptides conferring protective immunity against vaccinia virus [13], significantly contributing to immunology [14].
Advances in mass spectrometry have made it possible to identify a substantial number of HLA-bound peptides from cultured cell lines [15–17]. Expression of recombinant soluble HLA class I molecules allowed the characterization of HLA-specific motifs and the demonstration that secreted and membrane-bound forms of HLA class I have similar peptide repertoires [18–20].
There is additional experimental evidence suggesting that the level of soluble HLA class I complexes is elevated in the blood of patients with cancer, autoimmunological disorders, allergy or viral infections and that the HLA-peptide complex may remain intact in circulation [21]. This led to the investigation of peptides eluted from soluble HLA (sHLA) complexes, recovered from the blood of patients with hematological malignancies, such as multiple myeloma, acute lymphoblastic leukemia or acute myeloid leukemia [21]. Furthermore, soluble HLA molecules complexed with disease-associated epitopes found in the blood of breast cancer patients were proposed to serve as possible biomarkers and companion diagnostics [22].
In this article, we describe the immunocapture of HLA class I complexes from pools of 100 million cells, resulting in a preparation of acid-eluted HLA class I peptides with high purity, as demonstrated by their length and consensus motifs. The methodology is based on a single purification step of the acid-eluted peptides, allowing for high peptide recovery and thereby facilitating the identification of a total of 27’862 unique peptides with high confidence from five cell lines.
By purifying sHLA class I complexes from the serum of healthy volunteers and melanoma patients, we provide evidence that melanoma-associated peptides can be recovered from patients’ sera. The analysis revealed up to 972 peptides at high confidence from 5 ml of serum. Several HLA peptides derived from tumor-associated antigens were among the identified peptides, providing evidence that HLA complexes from solid tumors reach the circulation and can be recovered with immunocapture reagents. We anticipate that the serum-based analysis of the HLA class I peptidome may facilitate the development of multiplex tetramer reagents to characterize the therapeutic activity of immunostimulatory agents or the development of personalized medicine approaches, e.g. interventional strategies based on cancer vaccines or engineered T cells [3, 23].
2. Material & Methods
2.1. Cell lines and Antibodies
RPMI8226, HL-60, THP-1 and HEK293 cells used in this study were obtained from ATCC. CA46 cells were obtained from Sigma (Sigma Aldrich). MAVER-1 cells [24] were a kind gift of Prof. Alberto Zamo. MAVER-1, THP-1 (TIB-202) and RPMI8226 cells (CCL-155) were cultivated in RPMI 1640 medium supplemented with 10% heat-inactivated FCS, whilst HL-60 cells (CCL-240) were supplemented with 20% heat-inactivated FCS, and CA46 cells were cultivated in RPMI 1640 medium supplemented with 10% heat-inactivated FCS and 2 mM glutamine. HEK293 cells (CRL-1573) were cultivated in Freestyle 293 Expression medium with 0.1% Pleuronic F-68. W6/32 hybridoma cells (HB-95) were cultivated in CD Hybridoma medium with 2 mM glutamine. All cell lines were cultivated in shaking incubators at 37°C, 5% CO2 and 120 RPM. The W6/32 antibody was purified from HB-95 hybridoma supernatant using Protein-A Sepharose and coupled to AminoLink Plus Coupling Resin (Thermo Scientific) following the manufacturer`s instructions. Cell lines were HLA typed by sequence-specific oligonucleotide (SSO) and sequence-specific primer (SSP) technologies [Supporting Information Table 1].
2.2. Cell lysis and processing of human sera
Cells were harvested, washed trice in PBS and lysed on ice at a density of 5x107 cells per ml with 0.5% IGEPAL CA-630, 0,25% sodium deoxycholate, 1 mM EDTA, 0.2 mM iodoacetamide, 1 mM PMSF, Roche Complete Protease Inhibitor Cocktail. Lysates were cleared by centrifugation at 21’000g for 30 min, 4°C.
HLA haplotypes, disease stage and tumor burden for the analyzed melanoma patients are listed in Table 1. Sera from patients and healthy volunteers were cleared by centrifugation at 3’345g for 5 min, 4°C prior to HLA purification. All patients and control subjects were from the Department of Dermatology (Tübingen, Germany) and provided written informed consent for usage of serum and clinical data for scientific purposes. This project was approved by the ethic committee of Tübingen (ethic vote 243/2015BO2).
Table 1. Clinical characteristics and results for melanoma patients and healthy volunteers.
| Donora) | Stage | Tumor Burden | HLA-A 1 | HLA-A 2 | HLA-B 1 | HLA-B 2 | HLA-C 1 | HLA-C 2 | Identified Peptides | 8-11mers | 8-11mers (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 | stage IV | low | A*02 | A*26 | B*27 | B*44 | C*02 | C*05 | 389 | 272 | 69.9 |
| P2 | stage IV | medium | A*02 | A*03 | B*44 | B*47 | C*05 | C*06 | 633 | 502 | 79.3 |
| P3 | stage IV | high | A*02 | A*02 | B*35 | B*40 | C*03 | C*04 | 972 | 813 | 83.6 |
| P4 | stage IV | high | A*02 | A*32 | B*51 | B*57 | C*06 | C*14 | 558 | 385 | 69.0 |
| P5 | stage IV | high | A*02 | A*03 | B*15 | B*40 | C*02 | C*07 | 730 | 608 | 83.3 |
| P6 | stage III | none | A*02 | A*03 | B*51 | B*55 | C*03 | C*16 | 437 | 309 | 70.7 |
| P7 | stage III | none | A*02 | A*02 | B*52 | B*58 | C*03 | C*12 | 418 | 321 | 76.8 |
| P8 | stage IV CR | low | A*02 | A*26 | B*27 | B*44 | C*02 | C*05 | 306 | 212 | 69.3 |
| H1 | healthy | A*02 | A*03 | B*18 | B*57 | C*06 | C*07 | 568 | 457 | 80.5 | |
| H2 | healthy | A*02 | A*03 | B*39 | B*44 | C*07 | C*07 | 563 | 423 | 75.1 | |
| H3 | healthy | A*02 | A*03 | B*07 | B*15 | ntb) | nt b) | 664 | 536 | 80.7 | |
| H4 | healthy | A*02 | A*02 | B*44 | B*40 | nt b) | nt b) | 377 | 271 | 71.9 | |
P1 to P8 denote melanoma patients, H1 to H4 denote healthy volunteers
Not tested
2.3. Affinity purification of HLA class I molecules and enrichment of HLA-bound peptides
HLA class I complexes were purified from cleared lysate or human serum by incubation with W6/32 antibody-coupled resin for 2 h at 4°C. The resin was washed at 4°C once with lysis buffer, followed by buffer A (150 mM NaCl, 20 mM Tris, pH 7.4), buffer B (400 mM NaCl, 20 mM Tris, pH 7.4), a second buffer A wash and finally with buffer C (20 mM Tris, pH 8.0). Peptide-HLA complexes were eluted with 0.1 M acetic acid. The separation of peptides from extraneous protein was achieved either by centrifugation through molecular weight cutoff (MWCO) filters, or by stepwise elution from C18 resin.
For purification with MWCO filters, eluates were incubated for 10 min at 95°C, briefly cooled on ice and centrifuged for 1 min at 18’000g. The peptides were then separated from the proteins by centrifugation through 10 kDa cutoff columns (Amicon Ultra-0.5 Centrifugal Filter Unit) at 10’000g, generating an HLA fraction containing the HLA class I molecules and beta-2-microglobulin and a peptide fraction. The peptides were further concentrated and desalted on C18 resin (OMIX tips, Agilent) following the manufacturer’s instructions, dried and stored at -20°C. For purification with C18 resin, eluates were loaded on C18 Macro SpinColumns (Harvard Apparatus). Differential elution with 30 and 80% ACN generated a peptide fraction and an HLA fraction. Peptides were dried and stored at -20°C.
Purification efficiency was assessed by Western Blot analysis using the rabbit monoclonal anti-human HLA A antibody EP1395Y (ab52922, Abcam) and a goat anti-rabbit HRP-conjugated antibody (#170-6515, Bio Rad) for detection. β-Actin was probed as a loading control using the HRP-coupled mouse monoclonal anti-beta Actin antibody [AC-15] (ab49900, Abcam).
2.4. Analysis of HLA class I peptides by UHPLC MS
Peptides were analyzed by liquid chromatography–tandem mass spectrometry (LC-MS/MS) using a Q Exactive Mass Spectrometer fitted with an EASY-nLC 1000 (both Thermo Fisher). Peptides were resolved with an Acclaim PepMap RSLC C18, 50 μm x 150 mm, 2 μm analytical column (Thermo Fisher) at a flow rate of 0.3 μL/min by running a linear gradient from 0% to 24% ACN over 90 min. All buffers contained 0.1% formic acid. MS spectra were recorded in full ion scan mode from 250–2’000 m/z, with a resolution of 70’000 and a maximum injection time of 80 ms. MS/MS were recorded at a resolution of 17’500 and an injection time of 240 ms. The ten most intense masses with charges of two and three were selected for higher collisional energy dissociation fragmentation. Dynamic exclusion was set to 15 seconds.
Resulting spectra were processed and analyzed using the Proteome Discoverer software (Thermo Scientific, Version 1.4.1.14) and the MaxQuant software (Version 1.4.1.2 [25]). MS/MS data were searched against the human reference proteome databases of 89’649 human proteins downloaded from the UniProt homepage on the 20th January 2015. The following analysis settings were used with SEQUEST: i) No-Enzyme (Unspecific) (ii) precursor mass tolerance 4 ppm, (iii) fragment mass tolerance 0.02 Da, (iv) one variable modification (oxidation of methionine). False discovery rates were calculated with the Percolator plug-in. An independent SEQUEST search was performed for the assessment of potential post translationally modified peptides by allowing for additional variable modifications: (i) phosphorylation of serine, threonine and tyrosine, (ii) cysteinylation of cysteine, (iii) deamidation of asparagine, and (iv) cyclization of n-terminal glutamine. A maximum of three equal modifications was allowed for each peptide. MaxQuant parameters were set essentially as described [15]: i) digestion mode: Unspecific, (ii) first search 20 ppm; main search 4.5 ppm, (iii) fragment mass tolerance 20 ppm, (iv) one variable modification (oxidation of methionine), (v) no specific amino acids for the creation of decoy databases, (vi) peptide FDR 1%, protein FDR 100%, (vii) peptide length allowed: 7 to 20 amino acids. From the “peptide.txt” output file, reverse hits and contaminants were eliminated.
To identify melanoma associated antigens from the sHLA analysis, a list of melanoma associated antigens was retrieved from literature [5] and compared to the identified peptides at the gene level.
2.5. DeepQuanTR analysis
For a graphical representation of the bioinformatics pipeline see Supporting Information Figure 1. The DeepQuanTR [26] software suite was modified to accept MaxQuant feature lists as input. MaxQuant uses a highly sophisticated feature detection algorithm, including the calculation of feature intensity, and is therefore ideally suited as a basis for peptide quantification. MS data for each cell line was individually processed within DeepQuanTR. Andromeda and SEQUEST identifications were annotated to features according to their respective m/z ratio and retention time. Samples were aligned and sequence identifications removed if different sequences were annotated to aligned features, improving data quality.
2.6. In Solution digestion and MS analysis of HLA fraction
The HLA fraction containing the purified proteins was subjected to tryptic digestion and LC-MS/MS analysis. From the MaxQuant software analysis, only those identified peptides which could be annotated to the respective HLA alleles were taken into account. MaxQuant intensity was used for the relative quantification of HLA alleles.
2.7. HLA binding prediction of identified peptides
Unique peptides with a length between 8 and 11 amino acids were subjected to binding prediction analysis using NetMHCpan 2.8 [27]. A predicted IC50 of less than 500 nM as determined by NetMHCpan was used to classify the peptides as putative HLA binders. The analysis was performed on a cell line by cell line basis. For the binding prediction analysis of CA46 cells, peptides with a BindLevel of SB or WB were annotated as putative HLA binders.
2.8. Gibbs clustering of HLA class I peptides
Nine-mers identified from the cell lines were subjected to GibbsCluster-1.0 Server [28] analysis using the default settings without alignment (1-9 clusters, “use trash cluster to remove outliers” enabled and Blossum90 matrix (MAVER-1, HL-60, RPMI8229 and THP-1) or Blossum30 matrix (HEK293) selected). The cluster with the highest Kullback–Leibler distance was selected for each cell line and resulting motifs were compared to previously described HLA class I motifs [29, 30].
3. Results
3.1. Isolation, identification and analysis of HLA I complexes and cognate peptidomes from cell lines
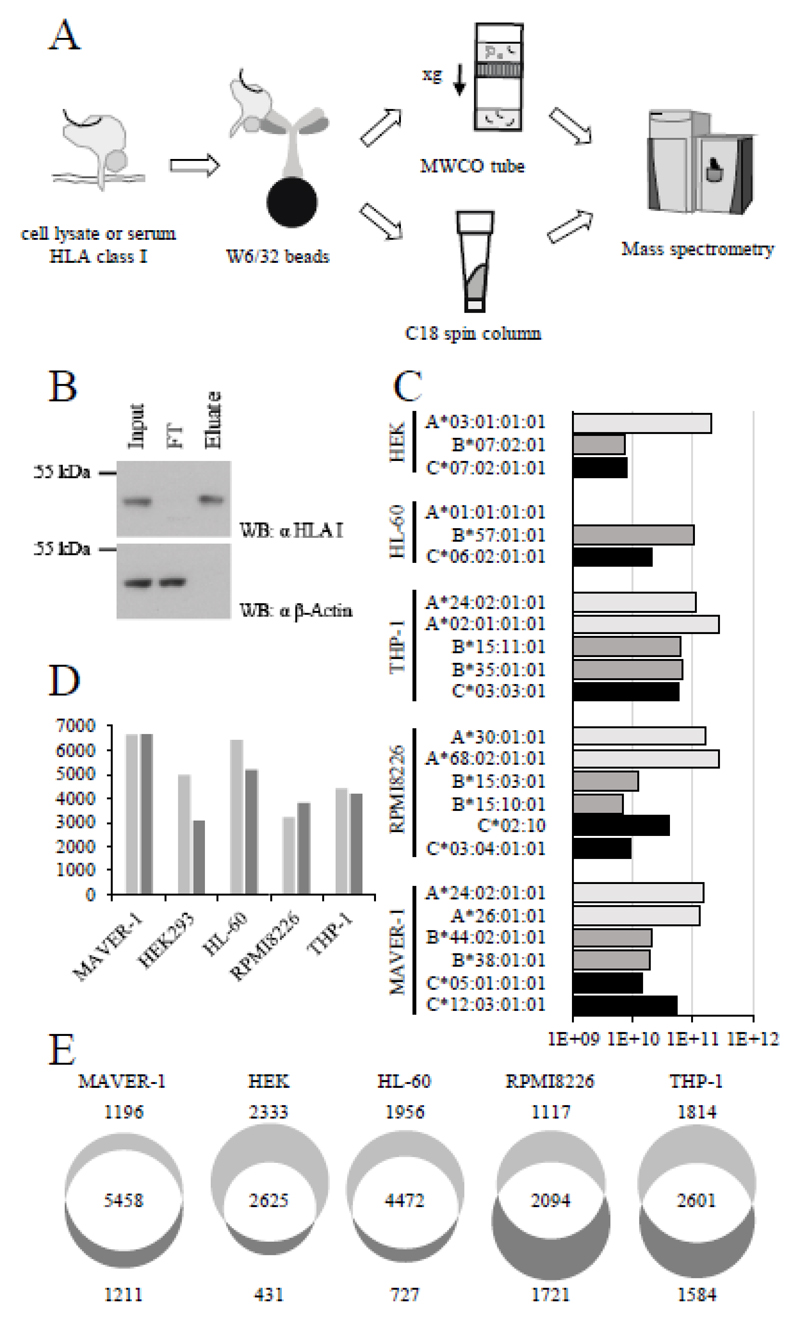
A schematic depicting the protocol used for affinity-purification of membrane-bound HLA class I complexes from cells or soluble complexes from sera is shown in Figure 1A. The procedure, which relied on the W6/32 monoclonal antibody [31], is compatible with a large variety of different HLA class I molecules [32]. Additionally, protocols which featured a selective peptide recovery were explored, either through the use of MWCO filters or a direct peptide purification on C18 spin columns followed by differential elution. The single step purification over C18 spin filters was found to yield the largest number of peptides and was therefore applied to purify HLA peptides from cell lines. HLA class I complexes and cognate peptides presented by human embryonal kidney cells (HEK293), mantle cell lymphoma cells (MAVER-1), acute monocytic leukemia (THP-1), acute myeloid leukemia cells (HL-60) and multiple myeloma cells (RPMI8226) were efficiently isolated from cell lysates of as few as 108 cells [Figure 1B]. Consecutive elutions from C18 spin columns with 30 and 80% acetonitrile yielded a peptide fraction and an HLA fraction containing HLA class I alpha chain, allowing for the probing of the HLA fraction for recovered HLA alpha chain molecules [Figure 1C and Supporting Information Table 1], while avoiding contamination of the peptide fraction with protein, which might interfere with downstream peptidome analysis.
Figure 1. Isolation of HLA class I complexes and mass spectrometric identification of HLA-eluted peptides.
(A) Schematic representation of the purification of HLA class I peptides. HLA class I complexes are purified from cell lysates and sera using anti-HLA class I antibody W6/32-conjugated resin. After elution of HLA complexes from resin with 0.1 M acetic acid, peptides are separated from proteins either by filtration through 10 kDa MWCO filters or by stepwise elution from C18 resin. Peptides are further desalted over C18 resin after MWCO filtration. Desalted peptides are analyzed by LC-MS/MS. (B) Anti-HLA class I Western blot demonstrating the successful purification of HLA class I complexes. HLA class I complexes were purified from HEK293 cell lysates using anti-HLA class I antibody W6/32-conjugated resin. HLA class I complexes were released by addition of SDS loading buffer and incubation for 5 minutes at 95°C. Equal fractions of input, flowthrough (FT) and eluate were loaded. β-Actin was used as loading control. The original Western blots were cropped to the relevant mass range using the Photoshop software. (C) Proteomic analysis of the HLA fraction after elution of proteins from C18 spin columns with 80% acetonitrile and tryptic digestion. Accumulated mass spectrometric signal intensities from peptides corresponding to the different HLA class I alleles expressed by MAVER-1, HEK293, HL-60, RPMI8226 and THP-1 cells are plotted as an approximation of protein abundance. (D) Total number of unique HLA-eluted peptides identified from the MS data of the respective cell lines with Proteome Discoverer (light gray) or MaxQuant (dark gray). (E) Unique HLA-eluted peptides identified by either Proteome Discoverer (light gray) or MaxQuant (dark gray), or by both software tools (white).
The recovered peptides were analyzed by LC-MS/MS on a Q Exactive mass spectrometer and the resulting spectra were searched independently with Proteome Discoverer and MaxQuant, to evaluate the impact of the employed search engines on the number of HLA peptides identified, against the human UniProt reference proteome set applying a stringent 1% FDR [see Supporting Information Tables 3-7]. Figure 1D shows the number of unique peptide identifications for each cell line from both software tools. Between 3056 and 6669 peptides were identified using MaxQuant, while between 3211 and 6654 peptides could be identified with Proteome Discoverer. Figure 1E presents the overlap of the two peptide identification methods, demonstrating an excellent agreement for the two methodologies in general. A certain complementarity was observed for HEK293, RPMI8226 and THP-1 cells, with 43% of HEK293 peptides exclusively being identified with Proteome Discoverer.
3.2. Bioinformatics processing establishes reliable and exhaustive HLA peptidomes
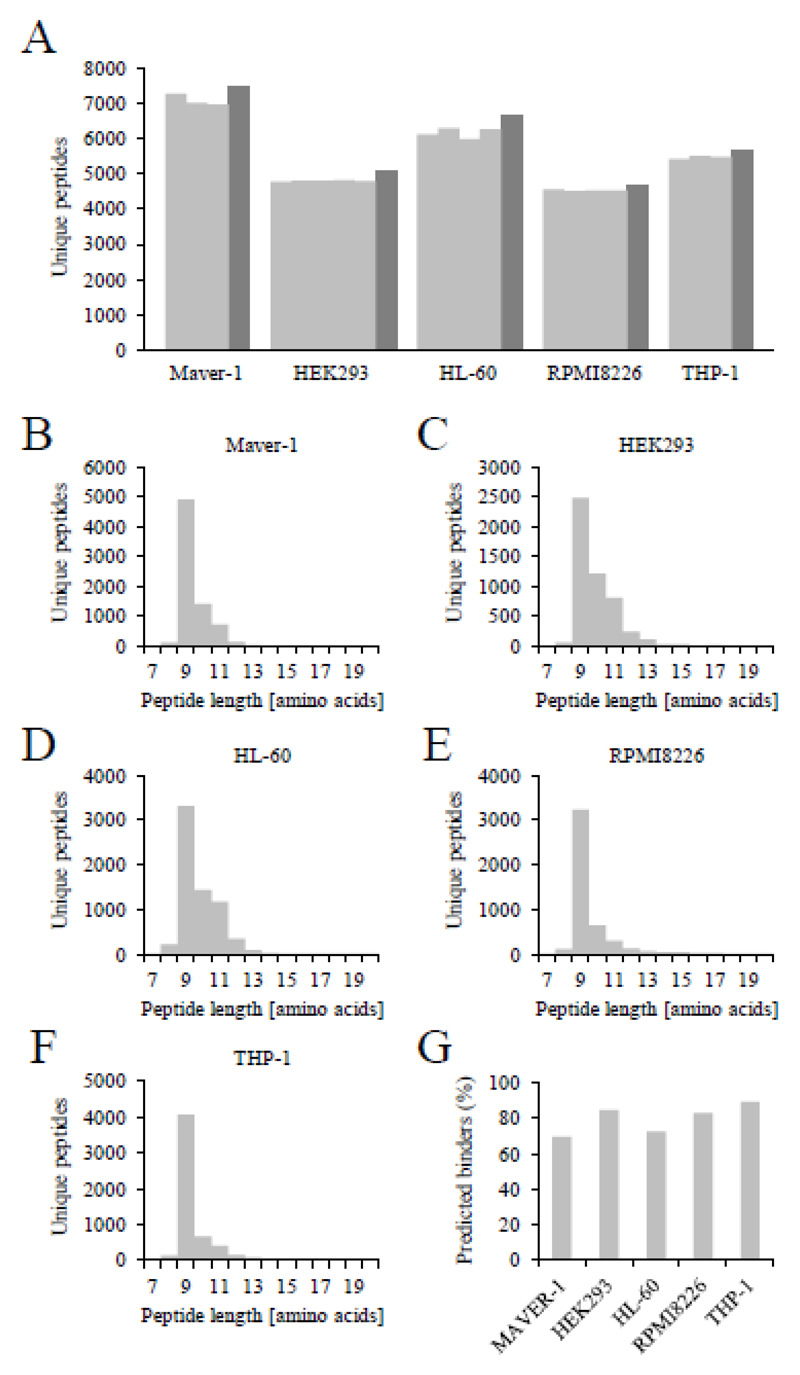
To reduce the bias of the applied search engine on peptide identifications, HLA class I peptidome identifications from Proteome Discoverer and MaxQuant were aggregated using the DeepQuanTR software suite [26] [see Supporting Information Figure 1 A-C for a graphical representation of the bioinformatics pipeline]. After alignment of the three to five replicates of a given cell line, we observed an excellent reproducibility in terms of peptide identifications [Figure 2A]. Figures 2B-F present the length distribution of identified peptides for the five cell lines. Reassuringly, peptide length peaked at nine amino acids, with the majority of peptides ranging between 8 and 11 amino acids. Futhermore, the majority of identified sequences were predicted to bind to the cognate HLA class I molecules by NetMHCpan 2.8 [27] [Figure 2G]. Consensus HLA-specific motifs [Figure 3] were produced by clustering 9-mers using the GibbsCluster-1.0 server [28], demonstrating excellent agreement with HLA binding motifs presented in the literature [29, 30].
Figure 2. Characteristics of the HLA peptidomes from human cell lines.
(A) Unique peptide identifications per cell line after DeepQuanTR analysis. Proteome Discoverer and MaxQuant data from Figure 1D was annotated to MaxQuant feature lists in DeepQuanTR. Samples from each of the cell lines were aligned in DeepQanTR individually. Aggregation of Proteome Discoverer and MaxQuant data in DeepQuanTR removes unreliable identifications, as features annotated to different sequences after retention time alignment are discarded. It further becomes possible to use the MaxQuant intensity to quantify peptides identified with Proteome Discoverer. (B-F) Length distribution of identified peptides presented in (A). The number of identified peptides between 7 and 20 amino acids is plotted in relation to their length. Peptides were isolated from (B) MAVER-1, (C) HEK293, (D) HL-60, (E) RPMI8226 and (F) THP-1 lysates. (G) Binding prediction of identified 8 to 11-mers with netMHCpan 2.8 [27]. Peptides were annotated as being predicted to bind if the IC50 received from netMHCpan was below 500 nM for at least one of the alleles of the cell line.
Figure 3. Definition of HLA-specific motifs from the HLA peptidomes of human cell lines.
Unique peptides with a length of nine amino acids were subjected to Gibbs clustering with the GibbsCluster-1.0 server [28]. Identified motifs were annotated to HLA alleles by comparing the experimental motifs with literature data [29, 30]. HLA specific motifs are presented for (A) MAVER-1, (B) HEK293, (C) HL-60, (D) RPMI8226 and (E) THP-1.
3.3. Investigations on post translationally modified antigens presented by tumor cell lines
For some HLA alleles, it has been proposed that post-translational modifications may influence peptides binding [33–35]. To identify if this might also be the case for some of the 23 different alleles in this study, we performed a separate search with SEQUEST and allowed for (i) phosphorylation of serine, threonine and tyrosine, (ii) oxidation of methionine, (iii) cysteinylation of cysteine, (iv) deamidation of asparagine, and (v) cyclization of n-terminal glutamine [Supporting Information Table 2]. No significant bias concerning the number of identified modifications was observed for any of the alleles. However, 56.5% of all phosphorylations were found at position P4, which is in line with a recent publication indicating that a phosphate at this position can interact with arginine 65 of the HLA-A*02:01 alpha chain [34]. This is not surprising, as all of the HLA-A alleles from the investigated cell lines carry this highly conserved residue. In addition, 48 (73%) of the 65 epitopes phosphorylated at P4 carry a proline at P5. Proline at P5, however, is not a known anchor residue for any of the HLA alleles in this study, indicating this might be a direct result from the phosphorylation on the peptide sequence. Finally, 66-88% of cysteinylated residues were found at the C-terminal side of the peptides (i.e. at position P5-P8 for 9mers).
3.4. The HLA-B*27:04 peptidome and ankylosing spondylitis
Of the many diseases proposed to be associated with certain HLA variants, ankylosing spondylitis is a particular case, as 90-95% of patients present themselves with HLA-B*27 alleles [36]. Until today, it is not fully understood why alleles such as B*27:05 and B*27:04 are associated with the disease, while B*27:06 and B27:09 are weakly or not associated [37]. One theory is the presentation of specific bacterial peptides, which mimic self-peptides. Such a phenomenon could be observed for a Yersinia hsp60 peptide, which induced reactive arthritis [38].
For some B*27 alleles, specific binding motifs have been established, however, with limited numbers of peptides [37]. Only for HLA-B*27:05, a recent study identified >1200 HLA-eluted peptides with high confidence [39]. To identify if the HLA-peptidome analysis pipeline described above would also be applicable for the comparison of peptidomes from a variety of HLA-B*27 alleles, we purified HLA complexes from the CA46 cell line, which is positive for A*26:03, B*27:04 and C*12:02 [40]. After mass spectrometric characterization of eluted peptides and bioinformatics analysis, 1847 of the identified peptides were predicted to bind to B*27:04 and GibbsCluster-1.0 server analysis demonstrated excellent agreement between the experimental and literature HLA motif for B*27:04 [Supporting Information Figure 2 and Supporting Information Table 8].
3.5. Identification and analysis of the soluble HLA class I peptidome from serum of melanoma patients
Having established HLA class I peptidome analysis from cell lines, we moved our attention to study the sHLA peptidome recovered from sera of melanoma patients compared to healthy volunteers. This work was inspired by previous studies on the characterization of the sHLA peptidome of leukemia and multiple myeloma patients [21]. We purified sHLA class I complexes from 4-5 ml serum of eight melanoma patients and four healthy volunteers [see Table 1 for more information] following the procedure described in Bassani-Sternberg et al. [21]. Peptides were separated from proteins using 10 kD cutoff columns and, after acid elution and incubation for 10 min at 95°C, further purified over C18 resin before LC-MS/MS analysis. MS data was processed in analogy to the cell lines leading to the identification of between 306 peptides (patient 8) and 972 peptides (patient 3) [Table 1 and Supporting Information Table 9]. Length distribution analysis demonstrated that a majority of the peptides ranged between 8 and 11 amino acids [Table 1] indicating that a significant portion of the sequenced peptides were eluted from sHLA molecules. Interestingly, substitution of acetic acid with formic acid for peptide elution lead to the recovery of longer peptides with an increase of C-terminal aspartic acid residues under these conditions, indicating an unspecific proteolytic activity [Supporting Information Figure 3].
Analysis of identified peptides revealed the presence of three MAGE peptides, as well as a number of peptides deriving from previously reported tumor-associated antigens, such as PLIN2 and RPSA [5]. Some of these sequences are reported in Table 2 and include seven peptides, which have been patented for tumor vaccination.
Table 2. Identified peptides from antigens previously reported to be associated with melanoma.
| Gene Name | Peptide Sequence | Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6 | Patient 7 | Patient 8 | Healthy 1 | Healthy 2 | Healthy 3 | Healthy 4 | Patenteda) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CYP1B1 | AAVGQAAHL | X | X | X | X | X | X | WO 2001035810 A2 | ||||||
| EHD2 | ALASHLIEA | X | X | X | X | X | X | X | X | X | X | X | X | CA 2736972 C |
| HMOX1 | RVIEEAKTAF | X | X | X | X | WO 2004085461 A2 | ||||||||
| KRT18 | RLESKIREH | X | X | X | X | X | X | |||||||
| MAGED1 | KEIDKEEHL | X | X | |||||||||||
| MAGED1 | RPKSAFKVQNA | X | ||||||||||||
| MAGED2 | NADPQAVTM | X | X | WO2006023598 A2 | ||||||||||
| MUC16 | STQRVTTSM | X | X | |||||||||||
| NELFA | EASRPPEEPSAP | X | X | X | X | X | X | X | X | |||||
| NPM1 | HQLSLRTV | X | ||||||||||||
| PLIN2 | AVTTTVTGAK | X | X | X | X | X | ||||||||
| PLIN2 | IARNLTQQL | X | X | X | X | X | X | X | WO 2005116051 A2 | |||||
| PLIN2 | SLLTSSKGQLQK | X | WO 2005116051 A2 | |||||||||||
| RPA1 | VTNKSQIRTW | X | X | |||||||||||
| RPSA | AEKAVTKEEF | X | X | X | X | X | ||||||||
| RPSA | NTDSPLRY | X | WO2010046469 A1 | |||||||||||
| SART1 | SMNANTITK | X | X | X | X | X | X | X | X | |||||
| SH3GLB2 | ISSTHVNHL | X | X | X | ||||||||||
| STEAP1 | KQFVWYTPPTFMIAV | X | X | |||||||||||
| TOP2A | LRNEKEQEL | X | X | X | ||||||||||
| TOP2A | SPASASRVA | X | X | X | ||||||||||
| TYMS | DAHIYLNHI | X | X | X | ||||||||||
| TOTAL | 4 | 11 | 12 | 8 | 11 | 7 | 6 | 8 | 6 | 4 | 8 | 2 |
Only one patent is displayed if more than one were filed
4. Discussion
In this study, we have shown that the immunocapture of HLA class I complexes followed by high resolution mass spectrometry analysis leads to the reproducible high-confidence identification of thousands of HLA-eluted peptides from cell lines. The methodology was also used for the experimental analysis of soluble HLA class I complexes from the serum of melanoma patients and healthy volunteers, leading to the identification of hundreds of peptides including many from previously known melanoma-associated antigens.
The identification of HLA class I-eluted peptides poses a number of additional challenges compared to shotgun proteomics experiments, including the low abundance of peptides and the absence of protease-determined N- or C-terminal specificities. Only high resolution, accurate mass and stringent FDR-based filters can ensure the quality of reported peptide identifications, particularly given the significant increase in search space. With modern mass spectrometers, it is finally possible to identify thousands of HLA-eluted peptides from 108 cells with high confidence. The high numbers of identified peptides raise questions on the complexity of the HLA peptidome presented by individual cell lines. In analogy to a recent investigation of the complexity of a tryptic digest of a cell lysate [41], we investigated the number of observed peptides eluted from MAVER-1 cells. A total of 48’064 peptidic features could be detected, indicating that the HLA peptidome of a cell is indeed extremely complex and that additional developments in the speed and sensitivity of mass spectrometers may lead to higher numbers of peptide identifications.
The physicochemical properties of HLA-bound peptides strongly depend on their cognate alleles, all of which differ significantly in the composition of their peptide binding pocket. Interestingly, we have observed that even though MaxQuant and Proteome Discoverer lead to peptidomes which are in excellent agreement with one another, on occasion one of the software tools may lead to a significantly higher number of identifications in a cell line-specific (and therefore HLA allele-specific) manner. We therefore propose combining the two approaches using software suites such as DeepQuanTR to address inherent differences in database search tools and to maximize the quality and the number of identified peptides. This is especially important in situations where a limited amount of material is available.
The HLA peptidome of the five cell lines in this study was also probed for post-translational modification of epitopes. In total, 0.5% of peptides were found to be phosphorylated, 3% were cysteinylated, 0.7% were deamidated and 0.3% carried an n-terminal pyroglutamate. This analysis is in line with prior reports on the presentation of post translationally modified peptides, which might indeed be important for T cell recognition [33–35].
The analysis of the HLA peptidome of the CA46 cell line further revealed the B*27:04-specific motif, an allele which is associated with ankylosing spondylitis [36]. It is interesting to note that the B*27:04-specific motif featured only hydrophobic residues at position P9, while the previously characterized B*27:05-specific motif demonstrates additional preference for basic amino acids at P9 [39]. This preference of basic residues has been attributed to aspartate at position 77 of the HLA alpha chain [37]. Other amino acid substitutions are unique to B*27:06 and B*27:09, which may influence the peptidomes. A more systematic characterization of the peptidomes of different B*27 alleles might proof useful in further understanding the pathological mechanisms underlying the disease.
The study of the sHLA peptidome of melanoma patients compared to healthy volunteers revealed between 306 and 972 peptides, the majority of which displayed a length compatible with HLA-bound peptides. A total of 22 peptides deriving from known tumor associated antigens were identified, 15 of them exclusively in melanoma patients’ sera. While a priori we do not know the relevance of individual HLA class I-bound peptides as tumor rejection antigens, knowledge of their sequence will allow the implementation of multiplex tetramer technology to probe for T cell reactivities [5]. Importantly, a systematic analysis of T cell specificities is now being applied to analyze response to immunostimulatory treatments [6]. The opportunity to retrieve HLA-bound peptides from serum samples dramatically expands the possibility to study patient-derived samples for peptide identification purposes. It is encouraging to see that the initial reports of the sHLA peptidome of patients with hematological malignancies could be extended to solid tumors.
In oncology, knowledge of “atlases” of HLA class I-bound peptides will not only allow the study of individual T cell specificities over the course of a disease, but might also be useful for vaccination strategies [42, 43], or for the engineering of therapeutic T cells. We and others have developed anti-cancer antibody-cytokine fusion proteins (immunocytokines), which display a potent therapeutic activity in mouse models of cancer and encouraging results in patients [44]. A detailed molecular knowledge of putative tumor rejection antigens may represent an invaluable set of biomarkers for the prediction and monitoring of response to therapy [6, 45]. Recent evidence suggests that mutated HLA peptides are important determinants of response to immunotherapy [7, 8, 46]. In a recent study, Kalaora et al., combined exome sequencing analysis of healthy and diseased tissue of a melanoma patient with HLA peptidome analysis, allowing the experimental identification of two mutated peptides that were presented on HLA class I, demonstrating that the identification of neo-epitopes by MS is possible, but is a rare event [47]. Considering the observed depth of our peptidome analysis, a combination with high throughput sequencing data of individual patients could enable the identification of mutated peptides from patient serum in the future, thus generating patient- and tumor-specific HLA peptidomes.
Supplementary Material
Significance.
Immunotherapy is increasingly being used in clinical practice with the recent approval of checkpoint inhibitors such as Ipilimumab and Pembrolizumab. The aim of such therapy is to stimulate the host’s immune system to fight cancer cells. An important mediator of this therapeutic effect is the recognition of tumor associated antigens presented on tumor cells by CD8+ cytotoxic T lymphocytes. Our molecular understanding of the recognition process is reliant upon a detailed knowledge of which peptides are presented by tumor cells, and of atlases of peptides presented in health and disease. The methodologies described in this publication allow for a rapid and detailed characterization of the HLA peptidome which will then facilitate the study of the tumor rejection process both in vitro and in vivo, e.g. by interrogation of T cell specificities with HLA tetramer reagents.
Acknowledgements
We thank Camilla Bacci (Philogen SpA) for providing W6/32 antibody and Jean Villard for providing the HLA typing results for the human cell lines, Giuliano Elia for reading the manuscript and Alberto Zamo for providing an aliquot of the MAVER-1 cell line. We acknowledge funding by the European Union’s Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 305309 (PRIAT) and 305608 (EURenOmics). Additional funding was received from the ETH Zurich, the Swiss National Science Foundation (SNF), the Bovena and Majores Foundations and the European Research Council (ERC Advanced Grant “Zauberkugel”).
Grant Support
D. Neri was supported by the Swiss Federal Institute of Technology Zurich (ETHZ). D. Ritz, A. Gloger, B. Weide, C. Garbe, T. Fugmann and D. Neri were supported by the European Union’s Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 305309 (PRIAT). D. Ritz and T. Fugmann were supported by the European Union’s Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 305608 (EURenOmics).
Abbreviations
- HLA
human leukocyte antigen
- LC-MS/MS
liquid chromatography–tandem mass spectrometry
- sHLA
soluble human leukocyte antigen
- MWCO
molecular weight cutoff
Footnotes
Conflict of Interest Disclosure
Dario Neri is co-founder, shareholder and member of the board of Philogen.
Authors’ Contributions
Conception and design: D. Neri, C. Garbe
Development of methodology: D. Ritz, T. Fugmann
Acquisition of data: D. Ritz, A. Gloger, B. Weide, T. Fugmann
Analysis and interpretation of data: D. Ritz, T. Fugmann, D. Neri
Writing, review and/or revision of the manuscript: D. Ritz, B. Weide, T. Fugmann, D. Neri
Study supervision: D. Neri, C. Garbe
References
- [1].Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Heemskerk B, Kvistborg P, Schumacher TN. The cancer antigenome. The EMBO journal. 2013;32:194–203. doi: 10.1038/emboj.2012.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Haen SP, Rammensee HG. The repertoire of human tumor-associated epitopes--identification and selection of antigens and their application in clinical trials. Current opinion in immunology. 2013;25:277–283. doi: 10.1016/j.coi.2013.03.007. [DOI] [PubMed] [Google Scholar]
- [4].Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- [5].Andersen RS, Thrue CA, Junker N, Lyngaa R, et al. Dissection of T-cell antigen specificity in human melanoma. Cancer research. 2012;72:1642–1650. doi: 10.1158/0008-5472.CAN-11-2614. [DOI] [PubMed] [Google Scholar]
- [6].Kvistborg P, Philips D, Kelderman S, Hageman L, et al. Anti-CTLA-4 therapy broadens the melanoma-reactive CD8+ T cell response. Science translational medicine. 2014;6:254ra128. doi: 10.1126/scitranslmed.3008918. [DOI] [PubMed] [Google Scholar]
- [7].Lu YC, Yao X, Crystal JS, Li YF, et al. Efficient identification of mutated cancer antigens recognized by T cells associated with durable tumor regressions. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:3401–3410. doi: 10.1158/1078-0432.CCR-14-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].van Rooij N, van Buuren MM, Philips D, Velds A, et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31:e439–442. doi: 10.1200/JCO.2012.47.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hunt DF, Michel H, Dickinson TA, Shabanowitz J, et al. Peptides presented to the immune system by the murine class II major histocompatibility complex molecule I-Ad. Science. 1992;256:1817–1820. doi: 10.1126/science.1319610. [DOI] [PubMed] [Google Scholar]
- [10].Rammensee HG, Friede T, Stevanoviic S. MHC ligands and peptide motifs: first listing. Immunogenetics. 1995;41:178–228. doi: 10.1007/BF00172063. [DOI] [PubMed] [Google Scholar]
- [11].Hickman HD, Luis AD, Buchli R, Few SR, et al. Toward a definition of self: proteomic evaluation of the class I peptide repertoire. Journal of immunology. 2004;172:2944–2952. doi: 10.4049/jimmunol.172.5.2944. [DOI] [PubMed] [Google Scholar]
- [12].Cox AL, Skipper J, Chen Y, Henderson RA, et al. Identification of a peptide recognized by five melanoma-specific human cytotoxic T cell lines. Science. 1994;264:716–719. doi: 10.1126/science.7513441. [DOI] [PubMed] [Google Scholar]
- [13].Gilchuk P, Spencer CT, Conant SB, Hill T, et al. Discovering naturally processed antigenic determinants that confer protective T cell immunity. J Clin Invest. 2013;123:1976–1987. doi: 10.1172/JCI67388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hillen N, Stevanovic S. Contribution of mass spectrometry-based proteomics to immunology. Expert Rev Proteomics. 2006;3:653–664. doi: 10.1586/14789450.3.6.653. [DOI] [PubMed] [Google Scholar]
- [15].Bassani-Sternberg M, Pletscher-Frankild S, Jensen LJ, Mann M. Mass spectrometry of human leukocyte antigen class I peptidomes reveals strong effects of protein abundance and turnover on antigen presentation. Molecular & cellular proteomics : MCP. 2015;14:658–673. doi: 10.1074/mcp.M114.042812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hassan C, Kester MG, de Ru AH, Hombrink P, et al. The human leukocyte antigen-presented ligandome of B lymphocytes. Molecular & cellular proteomics : MCP. 2013;12:1829–1843. doi: 10.1074/mcp.M112.024810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sofron A, Ritz D, Neri D, Fugmann T. High-resolution analysis of the murine MHC class II immunopeptidome. European journal of immunology. 2015 doi: 10.1002/eji.201545930. [DOI] [PubMed] [Google Scholar]
- [18].Prilliman K, Lindsey M, Zuo Y, Jackson KW, et al. Large-scale production of class I bound peptides: assigning a signature to HLA-B*1501. Immunogenetics. 1997;45:379–385. doi: 10.1007/s002510050219. [DOI] [PubMed] [Google Scholar]
- [19].Scull KE, Dudek NL, Corbett AJ, Ramarathinam SH, et al. Secreted HLA recapitulates the immunopeptidome and allows in-depth coverage of HLA A*02:01 ligands. Molecular immunology. 2012;51:136–142. doi: 10.1016/j.molimm.2012.02.117. [DOI] [PubMed] [Google Scholar]
- [20].Lazarus D, Weinstein-Marom H, Fishman S, Yossef R, et al. Efficient peptide recovery from secreted recombinant MHC-I molecules expressed via mRNA transfection. Immunol Lett. 2015;165:32–38. doi: 10.1016/j.imlet.2015.03.008. [DOI] [PubMed] [Google Scholar]
- [21].Bassani-Sternberg M, Barnea E, Beer I, Avivi I, et al. Soluble plasma HLA peptidome as a potential source for cancer biomarkers. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:18769–18776. doi: 10.1073/pnas.1008501107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Weidanz JA, Doll KL, Mohana-Sundaram S, Wichner T, et al. Detection of human leukocyte antigen biomarkers in breast cancer utilizing label-free biosensor technology. J Vis Exp. 2015 doi: 10.3791/52159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348:69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- [24].Zamo A, Ott G, Katzenberger T, Adam P, et al. Establishment of the MAVER-1 cell line, a model for leukemic and aggressive mantle cell lymphoma. Haematologica. 2006;91:40–47. [PubMed] [Google Scholar]
- [25].Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- [26].Fugmann T, Neri D, Roesli C. DeepQuanTR: MALDI-MS-based label-free quantification of proteins in complex biological samples. Proteomics. 2010;10:2631–2643. doi: 10.1002/pmic.200900634. [DOI] [PubMed] [Google Scholar]
- [27].Nielsen M, Lundegaard C, Blicher T, Lamberth K, et al. NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PloS one. 2007;2:e796. doi: 10.1371/journal.pone.0000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Andreatta M, Lund O, Nielsen M. Simultaneous alignment and clustering of peptide data using a Gibbs sampling approach. Bioinformatics. 2013;29:8–14. doi: 10.1093/bioinformatics/bts621. [DOI] [PubMed] [Google Scholar]
- [29].Rapin N, Hoof I, Lund O, Nielsen M. The MHC motif viewer: a visualization tool for MHC binding motifs. Current protocols in immunology / edited by John E. Coligan … [et al.] 2010 doi: 10.1002/0471142735.im1817s88. Chapter 18, Unit 18 17. [DOI] [PubMed] [Google Scholar]
- [30].Rasmussen M, Harndahl M, Stryhn A, Boucherma R, et al. Uncovering the peptide-binding specificities of HLA-C: a general strategy to determine the specificity of any MHC class I molecule. Journal of immunology. 2014;193:4790–4802. doi: 10.4049/jimmunol.1401689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Barnstable CJ, Bodmer WF, Brown G, Galfre G, et al. Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens-new tools for genetic analysis. Cell. 1978;14:9–20. doi: 10.1016/0092-8674(78)90296-9. [DOI] [PubMed] [Google Scholar]
- [32].Hilton HG, Parham P. Direct binding to antigen-coated beads refines the specificity and cross-reactivity of four monoclonal antibodies that recognize polymorphic epitopes of HLA class I molecules. Tissue antigens. 2013;81:212–220. doi: 10.1111/tan.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Marcilla M, Alpizar A, Lombardia M, Ramos-Fernandez A, et al. Increased diversity of the HLA-B40 ligandome by the presentation of peptides phosphorylated at their main anchor residue. Molecular & cellular proteomics : MCP. 2014;13:462–474. doi: 10.1074/mcp.M113.034314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Mohammed F, Cobbold M, Zarling AL, Salim M, et al. Phosphorylation-dependent interaction between antigenic peptides and MHC class I: a molecular basis for the presentation of transformed self. Nat Immunol. 2008;9:1236–1243. doi: 10.1038/ni.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Mommen GP, Frese CK, Meiring HD, van Gaans-van den Brink J, et al. Expanding the detectable HLA peptide repertoire using electron-transfer/higher-energy collision dissociation (EThcD) Proceedings of the National Academy of Sciences of the United States of America. 2014;111:4507–4512. doi: 10.1073/pnas.1321458111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Brown MA, Pile KD, Kennedy LG, Calin A, et al. HLA class I associations of ankylosing spondylitis in the white population in the United Kingdom. Ann Rheum Dis. 1996;55:268–270. doi: 10.1136/ard.55.4.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lopez de Castro JA, Alvarez I, Marcilla M, Paradela A, et al. HLA-B27: a registry of constitutive peptide ligands. Tissue antigens. 2004;63:424–445. doi: 10.1111/j.0001-2815.2004.00220.x. [DOI] [PubMed] [Google Scholar]
- [38].Ugrinovic S, Mertz A, Wu P, Braun J, Sieper J. A single nonamer from the Yersinia 60-kDa heat shock protein is the target of HLA-B27-restricted CTL response in Yersinia-induced reactive arthritis. Journal of immunology. 1997;159:5715–5723. [PubMed] [Google Scholar]
- [39].Ben Dror L, Barnea E, Beer I, Mann M, Admon A. The HLA-B*2705 peptidome. Arthritis Rheum. 2010;62:420–429. doi: 10.1002/art.27257. [DOI] [PubMed] [Google Scholar]
- [40].Boegel S, Lower M, Bukur T, Sahin U, Castle JC. A catalog of HLA type, HLA expression, and neo-epitope candidates in human cancer cell lines. Oncoimmunology. 2014;3:e954893. doi: 10.4161/21624011.2014.954893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Michalski A, Cox J, Mann M. More than 100,000 detectable peptide species elute in single shotgun proteomics runs but the majority is inaccessible to data-dependent LC-MS/MS. Journal of proteome research. 2011;10:1785–1793. doi: 10.1021/pr101060v. [DOI] [PubMed] [Google Scholar]
- [42].Walter S, Weinschenk T, Stenzl A, Zdrojowy R, et al. Multipeptide immune response to cancer vaccine IMA901 after single-dose cyclophosphamide associates with longer patient survival. Nature medicine. 2012;18:1254–1261. doi: 10.1038/nm.2883. [DOI] [PubMed] [Google Scholar]
- [43].Galassie AC, Link AJ. Proteomic contributions to our understanding of vaccine and immune responses. Proteomics Clin Appl. 2015;9:972–989. doi: 10.1002/prca.201500054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Pasche N, Neri D. Immunocytokines: a novel class of potent armed antibodies. Drug discovery today. 2012;17:583–590. doi: 10.1016/j.drudis.2012.01.007. [DOI] [PubMed] [Google Scholar]
- [45].Schumacher TN, Kesmir C, van Buuren MM. Biomarkers in cancer immunotherapy. Cancer cell. 2015;27:12–14. doi: 10.1016/j.ccell.2014.12.004. [DOI] [PubMed] [Google Scholar]
- [46].Gubin MM, Zhang X, Schuster H, Caron E, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515:577–581. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kalaora S, Barnea E, Merhavi-Shoham E, Qutob N, et al. Use of HLA peptidomics and whole exome sequencing to identify human immunogenic neo-antigens. Oncotarget. 2016 doi: 10.18632/oncotarget.6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.