Abstract
Soluble human leukocyte antigen class I (sHLA)-peptide complexes have been suggested to play a role in the modulation of immune responses and in immune evasion of cancer cells. The set of peptides eluted from sHLA molecules could serve as biomarker for the monitoring of patients with cancer or other conditions. Here, we describe an improved sHLA peptidomics methodology resulting in the identification of 1816 to 2761 unique peptide sequences from triplicate analyses of serum or plasma taken from three healthy donors. More than 90% of the identified peptides were 8-11mers and 74% of these sequences were predicted to bind to cognate HLA alleles, confirming the quality of the resulting immunopeptidomes. In comparison to the HLA peptidome of cultured cells, the plasma-derived peptides were predicted to have a higher stability in complex with the cognate HLA molecules and mainly derived from proteins of the plasma membrane or from the extracellular space. The sHLA peptidomes can efficiently be characterized using the new methodology, thus serving as potential source of biomarkers in various pathological conditions.
Keywords: sHLA, HLA peptidomics, immunopeptidomics, body fluids, antigens, peptidomics, biomarker
The human leukocyte antigen (HLA) class I alpha chain is expressed by all nucleated cells of the human body and forms a complex with beta-2-microglobulin and a peptide. The presented peptides are deriving from proteasomal degradation products of eight to twelve amino acids and are anchored in a binding cleft of the alpha chain through specific residues [1]. Interaction between immune cells and the HLA complex on the cell surface facilitates the detection of malignant transformation or of infection with intracellular pathogens. It has been observed that HLA complexes, which are naturally anchored to the cell membrane through a transmembrane domain, can be released (i.e. shed) through proteolysis. While it has been suggested that such a shedding may represent a mechanism of cancer cells to evade immune recognition [2], soluble HLA complexes (sHLA) can be found in the blood of every person and may have a more general role in immune modulation [3, 4]. Since diseased cells may shed HLA complexes to an increased extent, analysis of sHLA-eluted peptides has been suggested as an interesting tool for biomarker discovery [3, 5].
In previous work it was demonstrated that 80-179 peptides per sample could be sequenced starting with five ml of plasma from healthy donors [5], while it is possible to identify thousands of peptides from lysates of cultured cells [6, 7]. Our previous study indicated that the number of peptides identified from sHLA complexes of healthy donors is small but may increase in samples from melanoma patients [7].
In this work, we optimized an experimental procedure for the confident identification of sHLA-bound peptides resulting in 1500 to 2500 peptides per sample from three different donors as opposed to 300 to 600 peptides using the original procedure [7]. Applying a single step purification of HLA-eluted peptides using C18 resin as opposed to the protocol previously presented featuring a molecular weight filtering step [5] was responsible for this drastic increase in identifications.
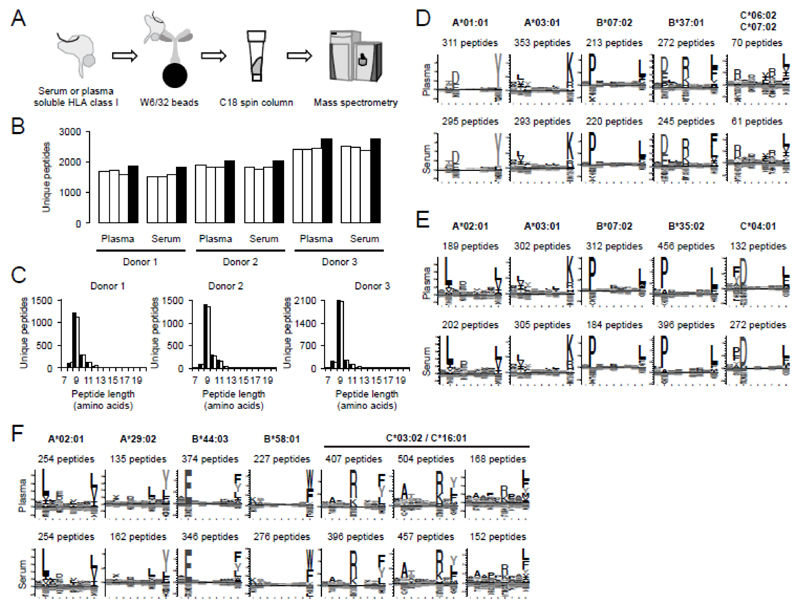
Based on this dataset, we could compare sHLA peptidomes deriving from serum or plasma, and study the differences between HLA peptidomes of cell lines and blood derived sHLA peptidomes. Plasma and serum samples were taken from three healthy donors with EDTA and serum tubes, respectively (S-Monovette K3E, plasma or S-Monovette Z, serum; both from Sarstedt). Tubes were incubated for 30 minutes at room temperature before centrifugation at 2’000 x g for 10 minutes to generate cell-free serum or plasma. All samples were snap-frozen in liquid nitrogen and stored at -80°C prior to submitting them to the purification of HLA class I complexes. The affinity purification procedure is outlined in Figure 1A and was initially described for analysis of HLA peptides recovered from cell lysates [7]. In brief, HLA complexes were purified from serum or plasma deriving from 9 ml of whole blood with the pan-HLA class I specific antibody W6/32, eluted from the affinity resin with 0.1 M acetic acid after stringent washing. HLA peptides were further purified with a single C18 step before MS analysis. Studying samples in triplicate, peptides were analyzed with a Q Exactive mass spectrometer (Thermo Fisher) coupled to an Easy-nLC 1000 equipped with a 15 cm Acclaim PepMap RSLC C18 (both Thermo Fisher). Peptides were eluted over a linear gradient from 0% to 24% ACN over 60 minutes. All buffers contained 0.1% formic acid. Peptide identification was essentially performed as previously described [7] but detailed methods can be found in the Supporting Material and Methods. In brief, peptides were searched with SEQUEST and Andromeda and filtered to 1% FDR. Peptide identifications were further annotated to MaxQuant peak lists with DeepQuanTR before retention time alignment of all replicates. Serum and plasma samples were processed individually and were not aligned to allow for a comparison of peptide identifications not influenced by bioinformatics processing. The number of peptides identified from each replicate of serum or plasma for the three healthy donors ranges between 1500 and 2500 [Figure 1B; all peptide identifications can be found in Supporting Information Table 1-6]. These numbers were significantly higher than in previous reports from our group and others [5, 7]. The peptide length distributions for both serum and plasma centered around nine amino acids [Figure 1C] and revealed that between 90.7 and 97.2% of all identified peptides were eight to eleven amino acids long [Table 1]. A Gibbs cluster analysis of the identified 9mers was performed and the resulting clusters were annotated to respective alleles according to their motifs presented in the motif viewer database [8] or in previous publications [6, 7] This resulted in clearly distinguishable motifs [Figure 1 D-F] for the three healthy donors, revealing the HLA-specific patterns expected for their respective HLA alleles [i.e. Donor 1: HLA-A*01:01:01, HLA-A*03:01:01, HLA-B*07:02:01, HLA-B*37:01:01, HLA-C*06:02:01, HLA-C*07:02:01; Donor 2: HLA-A*02:01:01, HLA-A*03:01:01, HLA-B*07:02:01, HLA-B*35:02:01, HLA-C*04:01:01, HLA-C*07:02:01; Donor 3: HLA-A*02:01:01, HLA-A*29:02:01, HLA-B*44:03:01, HLA-B*58:01:01, HLA-C*03:02:01, HLA-C*16:01:01; HLA typing was performed with next generation sequencing at the Zentrum für Humangenetik und Laboratoriumsdiagnostik (MVZ, Martinsried, Germany)]. The similarity of (i) HLA-specific binding motifs for patients with identical alleles, and (ii) motifs derived from serum or plasma samples was striking [Figure 1D-F] and a large proportion (74.2 to 89.2%) of all identified 8-11mers was predicted to bind to the respective HLA alleles according to NetMHCpan-2.8 [9] [Table 1]. However, a significantly higher number of peptides deriving from Fibrinogen alpha, Prothrombin, Zyxin and Complement C3 were identified from the serum as compared to plasma [Supporting Information Table 7]. This indicates that a defined set of contaminants originated from the proteolytic activity during blood clotting could not be removed during washing steps. We also identified many HLA-C peptides from donor 3, which are otherwise typically underrepresented in the analysis of cell lines [10].
Figure 1. Purification of soluble HLA class I complexes and characteristics of eluted peptides.
(A) Schematic representation of the sHLA peptidomics methodology. Serum and plasma from healthy donors (D1-D3) was subjected to affinity purification using the pan HLA class I antibody W6/32 coupled to solid support. HLA complexes were eluted and denatured with 0.1% acetic acid before isolation of eluted peptides with a single C18 purification step. HLA peptides were analyzed by LC-MS. (B) Peptide identifications for triplicate analyses of serum or plasma. Replicate runs are shown in white, total identifications in black. (C) Length distribution of peptides identified from plasma (black) or serum (white) for each of the three donors. (D-F) HLA-specific motifs deriving from 9mers identified from plasma (upper panel) and serum (lower panel) of donor 1 (D), donor 2 (E), or donor 3 (F). Motifs were created submitting all 9mers to the GibbsCluster-1.1 server [16].
Table 1. Characteristics of the sHLA peptidomes recovered from serum and plasma of healthy donors D1 to D3.
| Sample | Total number of peptides | % 8-11mers | % predicted bindersa) | Post translational modifications |
||||
|---|---|---|---|---|---|---|---|---|
| Oxidation (M) | Cysteinylation (C) | Deamidation (N) | N-terminal cyclization (Q) | Phosphorylation (S, T, Y) | ||||
| D1 plasma | 1841 | 95.4 | 76.1 | 101 | 38 | 15 | 4 | 2 |
| D1 serum | 1816 | 90.7 | 74.2 | 59 | 43 | 26 | 2 | 2 |
| D2 plasma | 2031 | 95.1 | 75.8 | 137 | 58 | 20 | 6 | 3 |
| D2 serum | 2033 | 92.1 | 75.4 | 151 | 52 | 16 | 3 | 2 |
| D3 plasma | 2741 | 97.2 | 89.1 | 285 | 106 | 16 | 2 | 6 |
| D3 serum | 2761 | 94.6 | 89.2 | 184 | 81 | 21 | 5 | 7 |
NetMHCpan-2.8 predicted affinity below 500 nM [9]
We further investigated if the sHLA peptidomes potentially contained post-translationally modified peptides by performing a separate database search with SEQUEST, taking into account cysteinylation of cysteine, deamidation of asparagine, cyclization of N-terminal glutamine and methionine oxidation [Table 1]. The levels of modification were found to be comparable to the ones previously reported for cell lines [7], with the exception of oxidation of methionine, which was found for 42.1% of all methionine-containing peptides from serum or plasma, while only for 11.6% from cells. Again, the majority of phosphorylated peptides were predicted to be modified at position P4 (62.3%), in keeping with previous reports on this position serving as an auxiliary anchor for HLA-A alleles [7, 11].
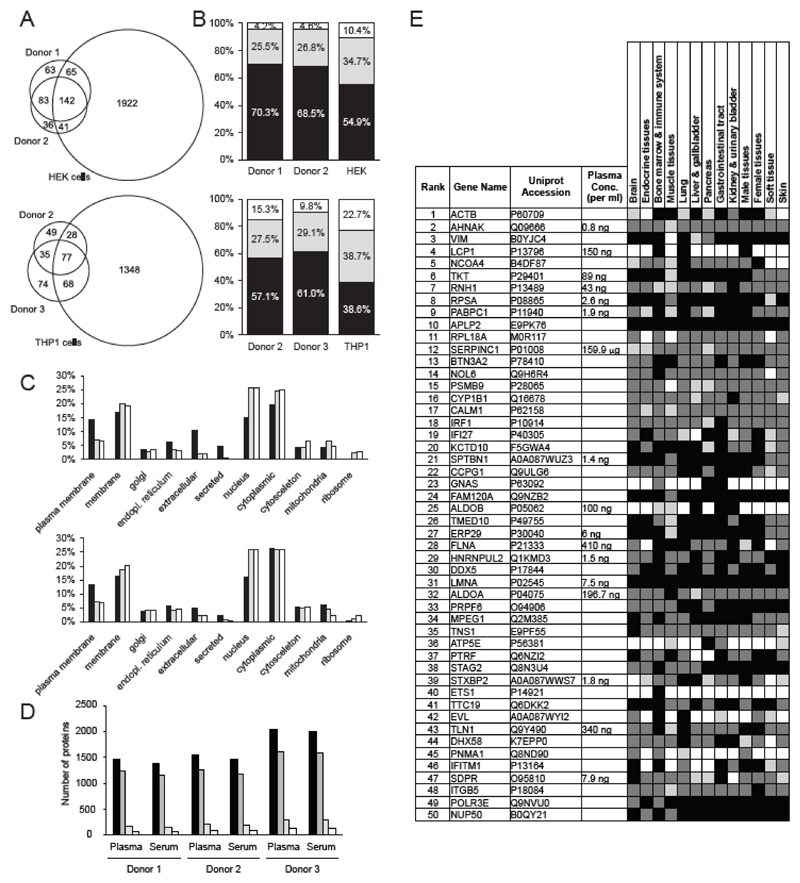
Certain peptide-sHLA complexes have been hypothesized to play a role in the modulation of a number of immune processes [2–4], urging for a better understanding of their nature and origin. Euler-Venn diagram of all 9mer sequences annotated to either HLA-A*03:01 [Figure 2A, upper panel] or HLA-A*02:01 [Figure 2A, lower panel] with the GibbsCluster-1.1 server, revealed a significant overlap between the peptidome of donors, but only a partial overlap with cell lines carrying the same alleles. A recently described neural network-based peptide-HLA complex stability prediction algorithm, NetMHCpanstab 1.0 [12], indicated a higher stability for sHLA-peptide complexes, compared to HLA-peptide complexes identified in THP1 and HEK cell lines [Figure 2B]. This finding suggests that kinetically stable complexes are more likely to survive in circulation. Furthermore, we were investigating if the HLA peptidomes were deriving from proteins of similar sub-cellular compartments. The majority of sHLA peptides originated from proteins of the plasma membrane or from extracellular proteins [Figure 2C], while cellular HLA peptidomes were more frequently derived from nuclear or ribosomal proteins. These observations indicate that sHLA peptidomes display motifs which are almost identical to the ones of the cognate HLA peptidomes derived from cultured cells. However, the peptides present in the two samples may differ in terms of stability and protein origin. These differences should be taken into consideration when investigating the role of sHLA complexes in immunological processes.
Figure 2. Source protein characterization and comparative analysis of peptides and source proteins of the sHLA peptidomes from plasma as compared to the HLA peptidomes of cultured cells.
(A) Euler-Venn diagram of all 9mer sequences annotated to either HLA-A*03:01 (upper panel), or HLA-A*02:01 (lower panel) with GibbsCluster-1.1 server [16]. HEK cells, donor 1 and donor 2 are positive for HLA-A*03:01, while THP1 cells, donor 2 and donor 3 are positive for HLA-A*02:01. (B) Predicted stability of either HLA-A*03:01 (upper panel), or HLA-A*02:01 (lower panel) alleles in complex with the respective 9mer sequences of healthy donors or cell lines. Stability prediction was performed with NetMHCstabpan 1.0 server [12]. Black, half-life > 6h indicating high stability; Grey, half-life < 6h and > 2h indicating medium stability; White, half-life <= 2h indicating low stability. (C) Subcellular localization of source proteins for the given sHLA peptides exclusively identified from plasma of healthy donors (black), shared between sHLA and HLA peptidomes (gray), or exclusively identified from cell lines (white). HLA-A*03:01, upper panel; HLA-A*02:01, lower panel. Subcellular localization was extracted from the UniProt entries of the source proteins, taking into account UniProt localization and GO annotations. (D) Source proteins of the peptides identified from serum and plasma. Peptide identifications were taken from Figure 1B. Total number of proteins identified in the respective triplicate analyses (black); Number of proteins identified with one (dark grey), two (light grey) or more than two peptides (white). (E) Tissue expression of the 50 proteins with the highest overall signal intensity. Expression data was extracted from the Human Protein Atlas repository and color-coded according to reported staining intensity: Protein was not detected (white); Low staining intensity (light gray); Moderate staining intensity (dark gray); High staining intensity (black). Contaminants originating from blood clotting were removed. Plasma concentration values were retrieved from the Plasma Proteome Database (http://www.plasmaproteomedatabase.org/). If several values were available for one entry, the average was taken.
In its entirety, the identified peptides [from Figure 1B] could be annotated to a total of 4128 proteins with between 1476 and 2046 proteins identified from serum or plasma of the three donors. Around 81% of the source proteins were identified with just one peptide, while an average of 5.6% of the proteins were identified with more than two peptides [Figure 2D]. Finally, we were investigating if the source proteins were deriving from the plasma proteome, or from certain tissues. For this, the tissue distribution of the 50 most intense source proteins (based on peptide intensities) was assessed with data retrieved from the Human Protein Atlas (http://www.proteinatlas.org). The majority of these proteins were found to be expressed in all tissues, while five of the 50 proteins demonstrated an expression in a maximum of four tissues [Figure 2E; see Supporting Information Table 8 for all Protein Atlas expression data]. The 50 proteins were further compared to the list of observed plasma proteins from the Plasma Protein Database (http://www.plasmaproteomedatabase.org). For a total of 17 proteins (34%), concentration values could be extracted from the database, but only SERPINC1 had a concentration above 1 μg/ml, thereby accounting for a classical plasma protein [13].
Purification of sHLA complexes from the blood of healthy donors can result in high quality peptidomes with the confident identification of thousands of HLA peptides with excellent purity (>90% of peptides being 8-11mers and most peptides being predicted to bind to cognate HLA alleles). The sHLA peptidomics methodology can be routinely applied to a large number of serum or plasma samples, thus providing high information content biomarkers. We speculate that the set of experimentally identified peptides may be used for a variety of biomedical applications, such as the monitoring of “response to therapy” in cancer patients [14] or the study of HLA class I-associated auto-antigens in ankylosing spondylitis [15].
Supplementary Material
Acknowledgements
We thank Camilla Bacci (Philogen SpA) for providing W6/32 antibody and Epameinondas Gousopoulos (ETH Zurich) for providing blood samples of the healthy donors.
Funding: This work was supported financially by ETH Zürich, the Swiss National Science Foundation, the European Union’s Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 305309 (PRIAT) and no. 305608 (EURenOmics), by the Bonveda Foundation and by the European Research Council (ERC advanced grant “ZAUBERKUGEL”).
Abbreviations
- HLA
human leukocyte antigen
- sHLA
soluble human leukocyte antigen
Footnotes
Conflicts of Interest: Dario Neri is co-founder of Philogen, shareholder and member of the board. Tim Fugmann and Danilo Ritz are employees of Philochem AG. The authors declare no additional conflict of interest.
References
- [1].Neefjes J, Jongsma ML, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nature reviews Immunology. 2011;11:823–836. doi: 10.1038/nri3084. [DOI] [PubMed] [Google Scholar]
- [2].Campoli M, Ferrone S. Tumor escape mechanisms: potential role of soluble HLA antigens and NK cells activating ligands. Tissue antigens. 2008;72:321–334. doi: 10.1111/j.1399-0039.2008.01106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Puppo F, Scudeletti M, Indiveri F, Ferrone S. Serum HLA class I antigens: markers and modulators of an immune response? Immunol Today. 1995;16:124–127. doi: 10.1016/0167-5699(95)80127-8. [DOI] [PubMed] [Google Scholar]
- [4].Tabayoyong WB, Zavazava N. Soluble HLA revisited. Leukemia research. 2007;31:121–125. doi: 10.1016/j.leukres.2006.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bassani-Sternberg M, Barnea E, Beer I, Avivi I, et al. Soluble plasma HLA peptidome as a potential source for cancer biomarkers. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:18769–18776. doi: 10.1073/pnas.1008501107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bassani-Sternberg M, Pletscher-Frankild S, Jensen LJ, Mann M. Mass spectrometry of human leukocyte antigen class I peptidomes reveals strong effects of protein abundance and turnover on antigen presentation. Molecular & cellular proteomics : MCP. 2015;14:658–673. doi: 10.1074/mcp.M114.042812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ritz D, Gloger A, Weide B, Garbe C, et al. High-sensitivity HLA class I peptidome analysis enables a precise definition of peptide motifs and the identification of peptides from cell lines and patients' sera. Proteomics. 2016;16:1570–1580. doi: 10.1002/pmic.201500445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Rapin N, Hoof I, Lund O, Nielsen M. The MHC motif viewer: a visualization tool for MHC binding motifs. Coligan John E., et al., editors. Current protocols in immunology. 2010 doi: 10.1002/0471142735.im1817s88. Chapter 18, Unit 18 17. [DOI] [PubMed] [Google Scholar]
- [9].Nielsen M, Lundegaard C, Blicher T, Lamberth K, et al. NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PloS one. 2007;2:e796. doi: 10.1371/journal.pone.0000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gloger A, Ritz D, Fugmann T, Neri D. Mass spectrometric analysis of the HLA class I peptidome of melanoma cell lines as a promising tool for the identification of putative tumor-associated HLA epitopes. Cancer Immunol Immunother. 2016 doi: 10.1007/s00262-016-1897-3. accepted manuscript. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Mohammed F, Cobbold M, Zarling AL, Salim M, et al. Phosphorylation-dependent interaction between antigenic peptides and MHC class I: a molecular basis for the presentation of transformed self. Nat Immunol. 2008;9:1236–1243. doi: 10.1038/ni.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Rasmussen M, Fenoy E, Harndahl M, Kristensen AB, et al. Pan-Specific Prediction of Peptide-MHC Class I Complex Stability, a Correlate of T Cell Immunogenicity. Journal of immunology. 2016;197:1517–1524. doi: 10.4049/jimmunol.1600582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Molecular & cellular proteomics : MCP. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- [14].Verdegaal EM, de Miranda NF, Visser M, Harryvan T, et al. Neoantigen landscape dynamics during human melanoma-T cell interactions. Nature. 2016;536:91–95. doi: 10.1038/nature18945. [DOI] [PubMed] [Google Scholar]
- [15].Bowness P. HLA-B27. Annu Rev Immunol. 2015;33:29–48. doi: 10.1146/annurev-immunol-032414-112110. [DOI] [PubMed] [Google Scholar]
- [16].Andreatta M, Lund O, Nielsen M. Simultaneous alignment and clustering of peptide data using a Gibbs sampling approach. Bioinformatics. 2013;29:8–14. doi: 10.1093/bioinformatics/bts621. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.