Abstract
Although all-oral direct-acting antiviral (DAA) therapy for hepatitis C virus (HCV) treatment is now a reality, today's HCV drugs are expensive, and more affordable drugs are still urgently needed. In this work, we report the identification of the 2-phenyl-4,5,6,7-Tetrahydro-1H-indole chemical scaffold that inhibits cellular replication of HCV genotype 1b and 2a subgenomic replicons. The anti-HCV genotype 1b and 2a profiling and effects on cell viability of a selected representative set of derivatives as well as their chemical synthesis are described herein. The most potent compound 39 displayed EC50 values of 7.9 and 2.6 µM in genotype 1b and 2a, respectively. Biochemical assays showed that derivative 39 had no effect on HCV NS5B polymerase, NS3 helicase, IRES mediated translation and selected host factors. Thus, future work will involve both the chemical optimization and target identification of 2-phenyl-4,5,6,7-Tetrahydro-1H-indoles as new anti-HCV agents.
Keywords: Hepatitis C virus; 4,5,6,7-Tetrahydro-1H-indole; Anti-HCV agents
1. Introduction
Hepatitis C virus (HCV) infection represents a global health problem that has an associated high risk for serious liver diseases. On the basis of annual World Health Organization (WHO) reports, more than 130–150 million people are infected and more than 350,000–500,000 individuals die from HCV-related liver pathologies each year [1]. To date, at least eleven HCV genotypes (gt) have been identified. These genotypes can be divided into multiple subtypes. The global distribution of HCV genotypes varies depending on the particular geographical area. HCV gt 1 is the most common in North and South America, Europe and Australia [2]. HCV gt 2 is widespread in America and Europe, while gt 3 is common in Central Asia and Middle East. Finally, HCV gt 4 and gt 5 are found almost exclusively in Africa, and HCV gt 6 is endemic in East and Southeast Asia [2]. Gt 1 and gt 4 are the hardest to treat and are associated with a particularly aggressive form of the disease.
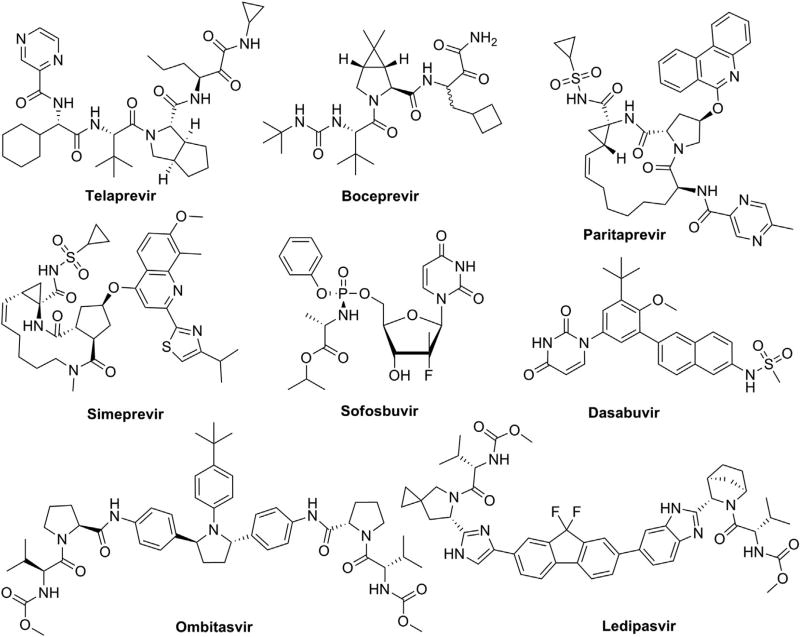
HCV was discovered in 1989, and until recently all treatments included some combination of pegylated interferon-α (pegIFN-α) and ribavirin (RBV), both of which cause debilitating side effects often worse than HCV symptoms. PEG-IFN/RBV treatment alone has been moderately successful and is genotype-dependent as only 40–50% of gt 1 and gt 4 patients have achieved a sustained virological response (SVR) indicative of a cure [3]. This treatment regimen remained the standard-of-care (SOC) until 2011 for gt 1, and until 2014 for the other genotypes. Over the past 20 years, a combination of developments of new models and tools have been able to reveal the different steps of the HCV life cycle and tremendous drug discovery efforts have allowed the development of direct-acting antivirals (DAAs) that specifically target HCV proteins. Since 2011, the new SOC for patients infected with gt 1 is based on a combination of pegIFN-α and RBV with the first-generation HCV protease inhibitors telaprevir or boceprevir (Fig. 1). Although the cure rates have improved (SVR = 60–80%), the new SOC provides only limited clinical benefit against HCV gt 2–6 and has resulted in some serious side effects in clinical trials [4,5]. Consequently, two new HCV DAAs, simeprevir and sofosbuvir (Fig. 1), have been approved in December 2013 in the United States and in the first half of 2014 in Europe [6–8]. Simeprevir is a second-generation protease inhibitor that is endowed with a broader genotypic coverage (gt 1, 2 and 4). Its combination with pegIFN-α and RBV has shown improved SVR and a better tolerance profile [6]. Sofosbuvir, the first nucleotide inhibitor of NS5B polymerase approved by FDA, has paved the way for all-oral IFN-free therapies, two of which were approved in 2014: Viekira Pak (ombitasvir, paritaprevir, ritonavir and dasabuvir), and Harvoni (ledipasvir and sofosbuvir) (Fig. 1) [9–11]. Viekira Pak and Harvoni are both approved only for adult HCV patients with gt1 infection; they have displayed >90% SVR and are also effective against other genotype in clinical trials.
Fig. 1.
DAAs – FDA approved drugs for the treatment of Hepatitis C.
There are currently many similar HCV DAAs in development, and most target the NS3 protease, NS5B polymerase and NS5A protein. They are undergoing late stages of clinical development and are close to approval. An up-to-date status of the clinical trials along with comprehensive overviews of the continued and discontinued HCV-specific DAAs have been recently described [12].
The main drawback is that the newly approved drugs and/or regimens are very expensive, thus restricting access for most HCV-infected patients to the new anti-HCV therapies. Another serious medical issue is the high mutation rate of HCV coupled with the rapid emergence of drug resistance to the DAAs [13–15]. These observations serve to encourage continuing research in the field of HCV drug discovery that will lead to the identification of new antiviral agents effective against HCV.
Thus, it is within this context that we herein report the discovery of a new chemical class of anti-HCV compounds that have a 2-phenyl-4,5,6,7-Tetrahydro-1H-indole core.
2. Results and discussion
2.1. Cell-based screening of EDASA compounds: hit identification
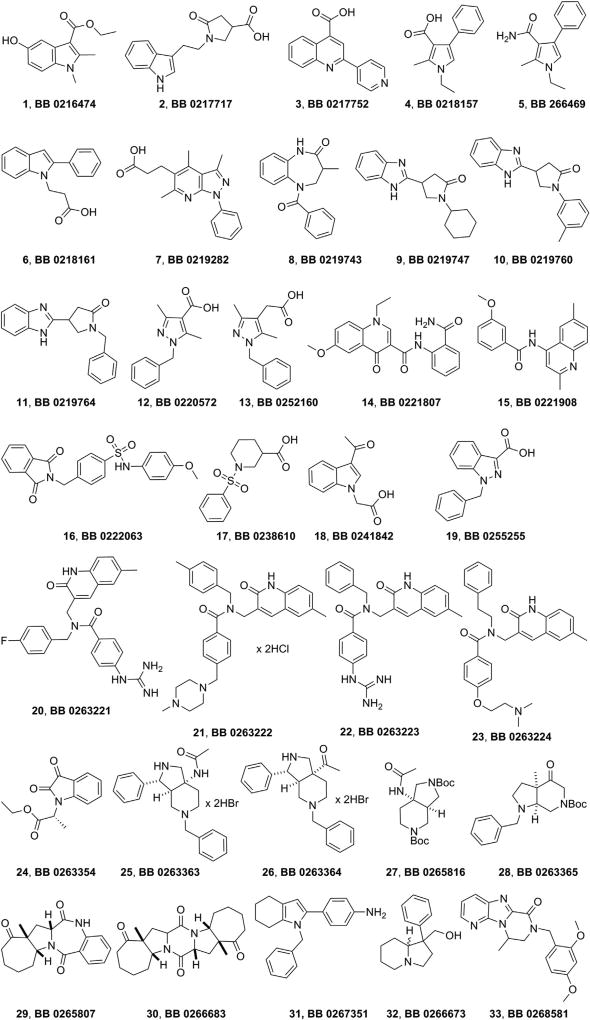
Compounds 1–33 (Fig. 2), representative chemotypes of the EDASA Scientific public compound library (http://www.edasascientific.com/page/catalogue), have been screened for their possible anti-HCV activity using HCV replicons based on the two most widely studied HCV genotypes (gt 1b and gt 2a) (Table 1). All compounds, except 24, are racemates.
Fig. 2.
Chemical structures and internal EDASA Scientific codes of the first set of compounds that underwent biological evaluation.
Table 1.
Anti-HCV activities and cytotoxicity of the first 33 EDASA Scientific compounds evaluated on gt 1b and 2a.
| Cpd | CC50a (µM) | Huh7/Rep-Feo1b
|
Huh7.5-FGR-JC1-Rluc2A
|
||||
|---|---|---|---|---|---|---|---|
| Inhibition,b % | EC50c (µM) | SId | Inhibition,b % | EC50c (µM) | SId | ||
| 1 | >200 | 19 ± 6 | ND | ND | 80 ± 5 | 11.1 ± 0.9 | >18 |
| 2 | >200 | 22 ± 9 | ND | ND | 51 ± 8 | 48.5 ± 3.9 | >4 |
| 3 | >200 | NI | ND | ND | 49 ± 6 | ND | ND |
| 4 | >200 | NI | ND | ND | 36 ± 10 | ND | ND |
| 5 | >200 | 49 ± 12 | ND | ND | 66 ± 7 | 23.6 ± 4.0 | >8 |
| 6 | ND | 26 ± 1 | ND | ND | 54 ± 4 | ND | ND |
| 7 | >200 | NI | ND | ND | 78 ± 3 | 11.7 ± 0.9 | >17 |
| 8 | >200 | 21 ± 2 | ND | ND | 77 ± 7 | 14.1 ± 1.9 | >14 |
| 9 | >200 | 19 ± 10 | ND | ND | 72 ± 3 | 15.6 ± 3.7 | >13 |
| 10 | >200 | 21 ± 2 | ND | ND | 77 ± 7 | 17.3 ± 3.2 | >12 |
| 11 | >200 | NI | ND | ND | 67 ± 5 | 21.1 ± 4.4 | >9 |
| 12 | >200 | 19 ± 8 | ND | ND | NI | ND | ND |
| 13 | >200 | NI | ND | ND | 14 ± 8 | ND | ND |
| 14 | 85.6 ± 5.9 | 17 ± 3 | ND | ND | 65 ± 5 | 20.6 ± 2.9 | 4 |
| 15 | <25 | 88 ± 2 | ND | ND | 99 ± 1 | ND | ND |
| 16 | >200 | 39 ± 4 | ND | ND | 60 ± 6 | 22.5 ± 3.8 | >9 |
| 17 | >200 | NI | ND | ND | 62 ± 1 | 28.1 ± 4.8 | >7 |
| 18 | >200 | NI | ND | ND | 55 ± 9 | ND | ND |
| 19 | >200 | NI | ND | ND | 46 ± 8 | ND | ND |
| 20 | >200 | 54 ± 8 | ND | ND | 29 ± 8 | ND | ND |
| 21 | <25 | 92 ± 1 | ND | ND | 99 ± 1 | ND | ND |
| 22 | >200 | NI | ND | ND | 44 ± 9 | ND | ND |
| 23 | >200 | 45 ± 6 | ND | ND | 61 ± 9 | 24.8 ± 4.5 | >8 |
| 24 | >200 | 19 ± 9 | ND | ND | 85 ± 9 | 7.3 ± 0.5 | >27 |
| 25 | >200 | NI | ND | ND | 73 ± 3 | 4.9 ± 0.4 | >41 |
| 26 | >200 | NI | ND | ND | 76 ± 7 | 17.4 ± 0.8 | >11 |
| 27 | >200 | NI | ND | ND | 43 ± 10 | ND | ND |
| 28 | 114.7 ± 14.6 | 73 ± 9 | 24.3 ± 1.2 | 5 | 98 ± 2 | 6.0 ± 1.0 | >19 |
| 29 | >200 | NI | ND | ND | NI | ND | ND |
| 30 | >200 | NI | ND | ND | 28 ± 4 | ND | ND |
| 31 | 109.9 ± 2.9 | 66 ± 9 | 12.4 ± 1.0 | 9 | 88 ± 8 | 8.7 ± 1.9 | 13 |
| 32 | >200 | 50 ± 4 | ND | ND | 25 ± 5 | ND | ND |
| 33 | >200 | 45 ± 2 | ND | ND | 48 ± 11 | ND | ND |
CC50 values were determined in Huh7.5 parental cells by the MTS assay. CC50 = is the concentration required to reduce the bioreduction of MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxy-phenyl)-2-(4-sulfophenyl)-2H-tetrazolium) into formazan by 50%. The reported value represents the means ± SD of data derived from three independent experiments.
Anti-HCV activity of the compounds were carried out at 50 µM in preliminary screening.
The inhibition data from 8 to 12 quarter log dilutions were used to generate the dose response curves. EC50 = the effective concentration required to inhibit virus induced cytopathic effect by 50%. The reported values represent the means ± SD of data derived from three independent experiments.
SI: selectivity index ratio of CC50 to EC50. ND: not determined. NI: no inhibition.
Gt 1b was studied for many years because it is one of the most resistant to pegIFN-α/RBV therapy, and the gt 1b (con1) strain was used in the first subgenomic HCV replicons [16]. Gt 2a exhibits a greater sensitivity than gt 1 to pegIFN-α/RBV treatment, but it was the first to be replicated in a robust cell culture model [17]. Taking this into consideration for our discovery campaign, we decided to screen the compounds against both the HCV genotypes.
The compounds were evaluated against Huh7/Rep-Feo1b and Huh7.5-FGR-JC1-Rluc2A cells, which carry the autonomously replicating HCV RNA of gt 1b and 2a in the firefly and Renilla luciferase reporters, respectively [18]. During initial screening, the 33 EDASA Scientific compounds were assayed at 50 µM against both the HCV replicons in reporter assays. The compounds that inhibited HCV replication by > 50% in the primary assays were then further evaluated in concentration-response assays. The ability of each compound to inhibit activity in gt 1b and 2a replicons, and their effect on cell viability are shown in Table 1. The selectivity index (SI) was calculated as well to estimate the therapeutic potential of the compounds in this system. Only two compounds (28 and 31) were found to be active against gt 1b (displaying EC50 values of 24.3 and 12.4 µM, respectively), although they showed poor SI (<10). In contrast, a total of 16 compounds were active against gt 2a, with associated EC50 values in the range from 4.9 to 28.1 µM and moderate to good SI. Compound 25 was the most potent among all the tested compounds and showed EC50 value of 4.9 µM with a SI > 41. Interestingly, the only two compounds found to be active on gt 1b replicons (28 and 31) were also active against gt 2a replicons and exhibited EC50 values of 6.0 and 8.7 µM, respectively, with SI values > 10.
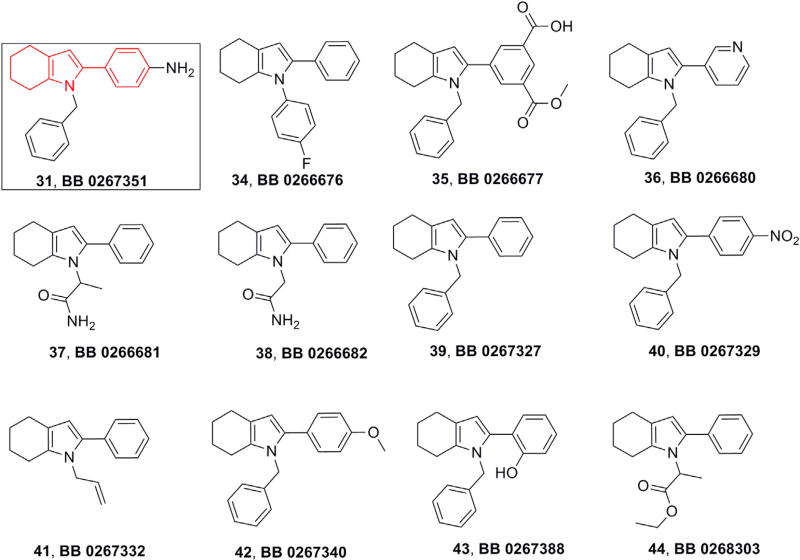
Overall, compound 31, having a 2-phenyl-4,5,6,7-Tetrahydro-1H-indole scaffold, emerged as a hit compound, displaying low cytotoxicity (CC50 = 109.9 µM) and promising anti-HCV activity in replicon reporter cells of both the genotype 1b (EC50 =12.4 µM) and 2a (EC50 = 8.7 µM). Following on from this, we had eleven more analogues of 31 available at EDASA Scientific that we decided to further evaluate for their anti-HCV activities (34–44, Fig. 3).
Fig. 3.
Structures of EDASA analogues of 2-phenyl-4,5,6,7-Tetrahydro-1H-indole 31.
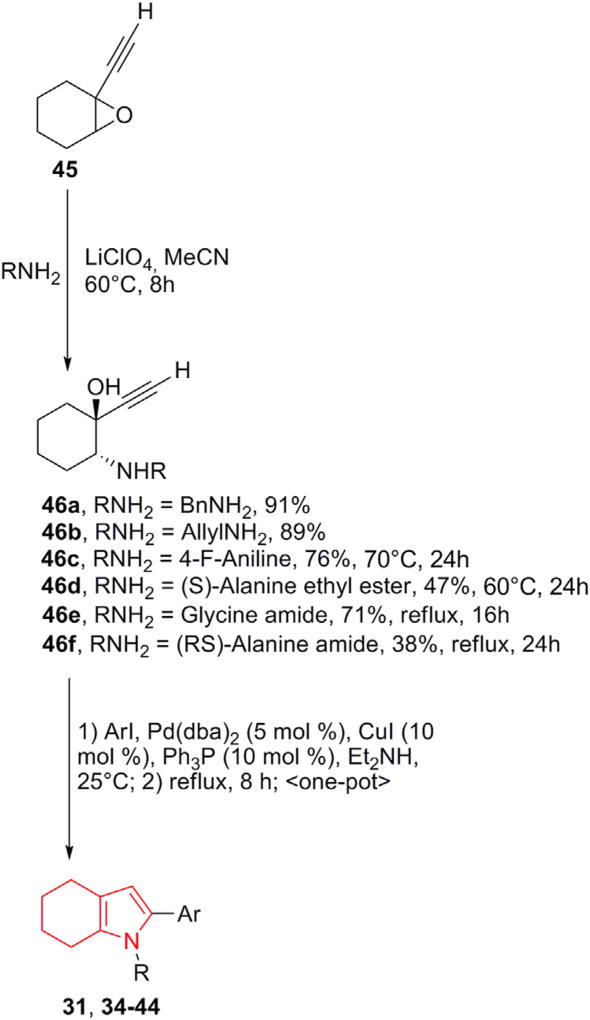
2.2. Synthesis of derivatives 31, 34–44
Recently, we have developed a two-step one-pot synthetic methodology, which leads to 4,5,6,7-Tetrahydro-1H-indoles with a wide range of substituents, including chiral moieties, both at C-2 and at the N-1 positions [19]. This synthetic sequence was successfully applied to achieve derivatives 31, 34–44 (Scheme 1).
Scheme. 1.
Synthesis of 2-aryl-4,5,6,7–1H-tetrahydroindoles. The explicit structures of compounds 31 and 34–44 are reported in Fig. 3.
This one-pot Sonogashira cross-coupling/5-endo-dig cyclization procedure was used as a flexible and versatile synthetic approach. Thus, the trans-stereoselective and highly regioselective nucleophilic epoxide ring opening of 45 with different amines was followed by a subsequent one-pot Pd-catalyzed arylation/cyclization. This short sequence allowed the variation of substituents both at the nitrogen atom and at the C-2 position of the pyrrole ring, along with a judicial design and a fast preparation of the most promising tetrahydroindole derivatives. Furthermore, it utilized mild conditions and inexpensive catalysts, being highly tolerant to a range of functional groups and readily scalable to provide sufficient amounts of tetrahydroindoles on gram scales in a good to excellent yields to effectively assemble the tetrahydroindole compound array for further screening. The full report on the synthetic sequence as well as compound characterization is presented in Supporting Information.
2.3. Cell-based assays of compounds 34–44
The anti-HCV activities of the new analogues of 31 (34–44) are shown in Table 2. The cell-based assays revealed that, out of the eleven compounds tested, eight derivatives in gt 1b and ten compounds in gt 2a showed >60% inhibition during preliminary screening.
Table 2.
Anti-HCV activities and cytotoxicity of analogues of 31 evaluated on gt 1b and 2a.
| Cpd | CC50a (µM) | Huh7/Rep-Feo1b
|
Huh7.5-FGR-JC1-Rluc2A
|
||||
|---|---|---|---|---|---|---|---|
| Inhibition,b % | EC50c (µM) | SId | Inhibition,b % | EC50c (µM) | SId | ||
| 34 | >200 | 71 ± 6 | 29.2 ± 1.2 | >7 | 66 ± 10 | 12.3 ± 1.0 | >16 |
| 35 | 45.6 ± 6.1 | 81 ± 3 | 35.8 ± 3.4 | >1 | 96 ± 3 | 9.9 ± 1.6 | >5 |
| 36 | <25 | 92 ± 1 | ND | ND | 99 ± 1 | ND | ND |
| 37 | 155.6 ± 11 | 50 ± 8 | ND | ND | 83 ± 3 | 15.4 ± 3.9 | >10 |
| 38 | >200 | 37 ± 18 | ND | ND | 48 ± 11 | ND | ND |
| 39 | 80.8 ± 3.1 | 95 ± 4 | 7.9 ± 0.5 | 10 | 99 ± 1 | 2.6 ± 0.4 | 32 |
| 40 | >200 | 75 ± 7 | 15.0 ± 1.3 | 13 | 98 ± 2 | 7.3 ± 1.4 | 27 |
| 41 | 118.8 ± 2.8 | 35 ± 6 | ND | ND | 69 ± 2 | 32.1 ± 4.1 | 4 |
| 42 | 137.4 ± 1.0 | 99 ± 1 | 11.8 ± 0.6 | 12 | 96 ± 2 | 4.9 ± 0.3 | 28 |
| 43 | 48.9 ± 1.7 | 96 ± 2 | 9.2 ± 0.6 | 5 | 99 ± 1 | 6.5 ± 0.6 | 8 |
| 44 | 84.3 ± 1.3 | 74 ± 4 | 13.2 ± 1.4 | 6 | 95 ± 3 | 13.7 ± 2.1 | 6 |
CC50 values were determined in Huh7.5 parental cells by the MTS assay. CC50 = is the concentration required to reduce the bioreduction of MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxy-phenyl)-2-(4-sulfophenyl)-2H-tetrazolium) into formazan by 50%. The reported value represents the means ± SD of data derived from three independent experiments.
Anti-HCV activity of the compounds were carried out at 50 µM in preliminary screening.
The inhibition data from 8 to 12 quarter log dilutions were used to generate the dose response curves. EC50 = the effective concentration required to inhibit virus induced cytopathic effect by 50%. The reported values represent the means ± SD of data derived from three independent experiments.
SI: selectivity index ratio of CC50 to EC50. ND: not determined.
All these compounds except one were then evaluated for their EC50 and SI values; in fact, derivative 36 exerted its HCV replication inhibition at toxic concentration (CC50 < 25 µM) and thus it was not submitted to EC50 evaluation.
Taking into account the obtained biological data (Table 2), some preliminary SAR can be proposed for this new class of anti-HCV agents.
Derivatives 39, 40 and 42, all having a N-benzyl substitution at the tetrahydroindole core, showed the higher anti-HCV activities with SI values ranging from 10 to 13 for gt 1b, and from 27 to 32 for gt 2a. Among them, compound 39 was found to be the most potent in both gts displaying EC50 values of 7.9 µM (1b) and 2.6 µM (2a). Compared to 39, derivative 34, having a para-fluorophenyl group at the nitrogen atom, showed a nearly 3.7 and 4.7 fold reduction in anti-HCV activity for gt 1b and 2a replicon reporter assays, respectively. Furthermore, when the N-benzyl substituent of the tetrahydroindole nucleus was replaced with non-aromatic groups, the anti-HCV activity on gt 1b was completely lost (37, 38, and 41) or a non selective antiviral effect (i.e. low SI value) was obtained (44); the analysis on gt 2a provided similar conclusions with the exception of derivative 37 which turned out to be active.
When analyzing the biological data for the whole subset of N-benzyl derivatives (i.e. compounds 31, 35, 39, 40, 42 and 43), the key role of the aryl substituent at the C-2 position became evident. An unsubstitued phenyl (39) as well as a para-substituted phenyl (i.e. 31: NH2, 40: NO2 and 42: OCH3) were both well tolerated; conversely, the presence of either meta-disubstituents (35) or ortho-OH (43) substituent led to compounds endowed with high toxicity. Moreover, the replacement of the phenyl (39) with a 3-pyridinyl ring (36) was also responsible for the increased cytoxicity (CC50 = 80.8 µM vs CC50 < 25 µM, respectively).
In order to further validate the anti-HCV activity of these compounds, hit 39 was selected and tested as a representative candidate against a reporter free cell culture system. To achieve this, we treated MH-14 cells carrying stably replicating HCV sub genomic replicon gt 1b with compound 39 and the HCV RNA was quantitated using standard quantitative RT-PCR methods. Notably, 39 inhibited the HCV replication in a dose-dependent manner and exhibited EC50 value of 3.13 µM (Fig. 4), which was quite similar to the value obtained in the replicon reporter cells (i.e. EC50 = 7.9 µM).
Fig. 4.
Dose-dependent response of compound 39 assayed in MH-14 cells. **p < 0.01.
Overall, the results clearly indicated that promising anti-HCV activity coupled with no apparent cytotoxic effects were obtained when the 2-phenyl-4,5,6,7-Tetrahydro-1H-indole scaffold was properly functionalized.
2.4. Molecular target investigation
Next, we carried out target investigation for the most active tetrahydro-1H-indoles (i.e., 31, 34, 39, 40 and 42). Towards this end, we tested the compounds for their ability to inhibit the activity of two HCV viral proteins, i.e. NS5B polymerase and NS3 helicase. These two targets were chosen as first choice because indole derivatives have been reported in literature as both HCV NS5B polymerase and NS3 helicase inhibitors [20,21].
We utilized a standard primer-dependent elongation assay to test whether the compounds possessed anti-NS5B RNA-dependent RNA polymerase (RdRp) activity [22,23]. The compounds were investigated at 50 µM concentration in the preliminary assay. The results clearly revealed that none of the compounds was inhibitory to NS5B RdRp activity (data not shown), thus ruling out the possibility of possessing anti-HCV activity by targeting this protein.
The five compounds were also tested in HCV NS3 helicase assays as described previously [24]. None of the compounds inhibited the ability of the NS3 helicase to unwind a DNA substrate even at concentrations as high as 500 µM (data not shown). However, high concentrations of compound 31 inhibited the ability of NS3 helicase to cleave ATP in the presence of RNA. About 420 µM of 31 inhibited HCV helicase catalyzed ATP hydrolysis by 50% (see Fig. S1 Supporting Information).
Apart from targeting HCV proteins, small molecules known to interfere with HCV Internal Ribosome Entry Site (IRES)-mediated translation have been documented [25,26]. We therefore investigated if the observed anti-HCV activity of the 2-phenyl-tetrahydro-1H-indole scaffold could be due to the down-regulation of HCV IRES-mediated translation. Using compound 39 as representative, our results displayed that this compound had no effect on HCV IRES mediated translation (data not shown).
We also tested the possibility that compound 39 could function as a potential activator or suppressor of host-factor's that facilitate HCV replication. Towards this end, we carried-out cell based assays in which reporter plasmids of cyclooxygenase-2, heme oxygenase-1, interferon-stimulated response element or anti-oxidant response element were transfected, and the ability of derivative 39 to modulate the activation or suppression of the corresponding host-factors at three varying compound concentrations (5, 10 and 25 µM) were investigated. Our results revealed that 39 had no effect in these reporter mediated assays, thus ruling out the specified host factors as targets of the 2-phenyl-4,5,6,7-Tetrahydro-1H-indole core.
3. Conclusion
Overall, these results highlight the identification of 2-phenyl-4,5,6,7-Tetrahydro-1H-indole scaffold as a new anti-HCV chemotype.
Preliminary SAR highlighted the key role of both the substituents on the 2-phenyl ring and the N-1 benzyl moiety in modulating cytotoxicity and activity, respectively, with derivatives 39, 40 and 42 being the best hits within this first series of 2-phenyl-4,5,6,7-Tetrahydro-1H-indoles.
While the present study has revealed a novel chemotype worthy of further investigation, the exact mechanism by which these derivatives inhibit HCV replication remains to be clarified.
4. Experimental section
4.1. Cell culture
Huh7/Rep-Feo1b and Huh7.5-FGR-JC1-Rluc2A replicon reporter cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum, 5% antibiotic and 0.5 mg/mL G418. Huh 7.5 cells were grown similarly as above without G418. All cells were cultured at 37 °C in 5% humidified CO2.
4.2. NS5B RdRp assay
Recombinant HCV NS5B bearing hexa-histidine tag at N-terminal was expressed in Escherichia coli and purified as previously described [23,27]. The anti-NS5B RdRp activity of the compounds was evaluated by using a primer-dependent elongation assay as reported earlier [28]. In brief, the reaction buffer containing 20 mM Tris–HCl (pH 7.0), 100 mM Na-glutamate, 100 mM NaCl, 0.01% BSA, 0.01% Tween-20, 0.1 mM DTT, 5% glycerol, 20 U/mL of RNasin, 20 µM UTP, 1 µCi [α-32P]UTP, 0.25 µM polyrA/U12, 100 ng NS5BCΔ21 was incubated with compounds and the polymerase reaction was started by addition of 1 mM MnCl2 in a final volume of 20 µl. The reactions were incubated at 30 °C for 60 min, and then stopped by adding 5% trichloroacetic acid containing 0.5 mM sodium pyrophosphate, filtered through GF-B filters, and successively washed with water and ethanol. The amount of radiolabeled RNA was quantified using liquid scintillation counter. The activity of NS5B in the presence of an equal amount of DMSO was set at 100% and that in the presence of the compounds was determined relative to this control.
4.3. Huh7/Rep-Feo1b, Huh7.5-FGR-JC1-Rluc2A reporter system and cellular viability assay
The anti-HCV activity of compounds was measured using the Huh7/Rep-Feo1b and Huh7.5-FGR-JC1-Rluc2A replicon reporter cells as described earlier [29,30]. In short, approximately 1 × 104 cells were plated in 96 well plates and treated with compounds or DMSO for 48 h. The concentration of DMSO in cell culture was kept constant at 1.0%. The luciferase activities were measured by following the manufacturer's protocol (Promega Inc, USA). The activity of the compounds was evaluated as the comparative levels of the luciferase signals in compound-treated cells versus DMSO-treated controls. The cellular cytotoxicity assays were conducted in 96 well plate format using parental Huh7.5 cells. Briefly, cells treated at 6-8 doses of compounds for 48 h were evaluated employing the CellTiter 96® AQueous One Solution Cell Proliferation kit (Promega Inc, USA). The luciferase activities of the cells treated with an equal amount of DMSO served as control.
4.4. Target identification reporter assays
The effect of compound 39 on HCV IRES mediated translation was studied using a dual luciferase reporter construct (pClneo-Rluc-IRES-Fluc) in which Rluc was translated in a cap-dependent manner and Fluc was translated via HCV IRES-mediated initiation, as described previously [29]. Transfections were carried our using LipoD293 reagent in Huh7.5 cells. Sixteen h post-transfection, the cells were treated with compound or DMSO and Luciferase activity assay was performed using Dual-Glo Luciferase Assay Kit.
For investigation host-factors as potential targets, hepatoma cells carrying HCV subgenomic replicons (MH-14) were transfected with 300 ng of gene specific reporter plasmid pCOX-2-FLuc [31–34], pHO-1-Luc [35], pISRE-Luc [36], or p3xARE-Luc [37]. Sixteen h post-transfection, cells were treated with compound 39 or DMSO (control) for 48 h and luciferase activities were measured as described above. Transfection efficiencies were normalized by Renilla luciferase expression.
4.5. RT-PCR
Total RNA was isolated using an RNeasy mini kit (Qiagen) and quantified using NanoDrop (ND1000, NanoDrop Technologies). Approximately 500 ng of RNA was reverse transcribed using MMLV reverse transcriptase (Life Technologies) and either oligo dT18 or HCV specific primers in a final volume of 20 µl. Approximately 50 ng of synthesized cDNA's were used for PCR applications using gene specific primers and Power SYBR green PCR master mix (Applied Biosystems) in a final volume of 25 µl. The PCR was performed on Applied Biosystems 7500 Fast Dx Real-Time PCR Instrument. The forward and reverse primer sequence for β-Actin was 5′-AGCGAGCATCCCCCAAAGTT-3′ and 5′-GGGCACGAAGGCTCATCATT-3′, respectively. The HCV primer sequence was 5′-CGGGAGAGCCATAGTGG-3′ for forward and 5′-AGTACCACAAGGCCTTTCG-3′ for the reverse primer.
4.6. NS3 helicase assay
4.6.1. Chemicals and reagents
Truncated C-terminally His-tagged NS3 protein lacking the N-terminal protease (NS3h) from the con1 strain of genotype 1b [Genbank accession AB114136], was expressed and purified as previously described [38,39].
4.6.2. Molecular beacon based helicase assays
Molecular beacon-based NS3 helicase assays were performed as described by Hanson et al. [49] Reactions contained 25 mM MOPS pH 6.5, 1.25 mM MgCl2, 5% DMSO, 5 µg/ml BSA, 0.01% (v/v) Tween20, 0.05 mM DTT, 5 nM florescent DNA substrate, 12.5 nM NS3h, and 1 mM ATP.
4.6.3. ATP hydrolysis (ATPase) assays
A modified malachite green-based assay was used to measure helicase-catalyzed ATP hydrolysis (Sweeney et al., 2013). The colorimetric reagent was prepared fresh by mixing 3 volumes of 0.045% (w/v) malachite green, with 1 volume 4.2% ammonium molybdate in 4 N HCl, and 0.05 volumes of 20% Tween 20. Reactions (30 µL) were initiated by adding ATP, incubated for 15 min at 37 °C, and terminated by adding 200 µL of the malachite green reagent, followed by 30 µL of 35% sodium citrate. The color was allowed to develop for 30 min and an absorbance at 630 nm was observed.
HCV Helicase-catalyzed ATP hydrolysis in the absence of RNA was monitored in reactions containing 50 nM HCV NS3h, 25 mM MOPS pH 6.5, 1.25 mM MgCl2, 1 mM ATP, 33 µg/ml BSA, 0.07% (v/v) Tween 20, 0.3 mM DTT, and 10% v/v DMSO. Reactions in the presence of polyU RNA were performed with 4 nM HCV NS3h in the same buffer with 1 µM PolyU (Sigma, expressed and nucleotide concentration) was added to each reaction.
To determine the compound concentration, it was necessary to reduce helicase-catalyzed ATP hydrolysis by 50% (IC50). Reactions were performed in duplicate through a two-fold dilution series so that final compound concentrations ranged from 0.5 mM to 0.78 µM. Data obtained from all reactions within the linear range of the colorimetric assay as determined with a phosphate standard curve were normalized to controls lacking an inhibitor (100%) and controls lacking an enzyme (0%), and fitted to a normalized dose response equation with a variable Hill slope using GraphPad Prism (v. 6). Reactions were performed in duplicate and each titration conformed to the above concentration response equation. Average IC50 values ± standard deviations were reported. In another set of controls, 100 µM of inorganic phosphate was titrated with each compound, followed by the addition of a malachite green reagent. None of the compounds affected the absorbance of the colorimetric reaction products in these controls.
Supplementary Material
Acknowledgments
We thank Drs. Naoya Sakamoto and Hengli Tang for providing the Huh7/Rep-Feo1b and Huh7.5-FGR-JC1-Rluc2A replicon reporter cells. Plasmids pCIneo-Rluc-IRES-Fluc, pHO-1-Luc and p3xARE-Luc, were generously shared by Drs. Naoya Sakamoto, Anupam Agarwal, and Dr. Being-Sun Wung, respectively. We acknowledge and appreciate grant support from the New Jersey Health Foundation to Neerja Kaushik-Basu and the Russian Foundation for Basic Research (RFBR), Russia (Projects No. 14-03-31685, 14-03-31709, 14-03-01114).
Footnotes
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.ejmech.2015.04.022.
References
- 1.Seeff LB. Natural history of chronic hepatitis C. Hepatology. 2002;36:S35–S46. doi: 10.1053/jhep.2002.36806. [DOI] [PubMed] [Google Scholar]
- 2.Hnatyszyn HJ. Chronic hepatitis C and genotyping: the clinical significance of determining HCV genotypes. Antivir. Ther. 2005;10:1–11. [PubMed] [Google Scholar]
- 3.Palumbo E. Pegylated interferon and ribavirin treatment for hepatitis C virus infection. Ther. Adv. Chronic Dis. 2011;2:39–45. doi: 10.1177/2040622310384308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poordad F, McCone J, Jr, Bacon BR, Bruno S, Manns MP, Sulkowski MS, Jacobson IM, Reddy KR, Goodman ZD, Boparai N, DiNubile MJ, Sniukiene V, Brass CA, Albrecht JK, Bronowicki JP. Boceprevir for untreated chronic HCV genotype 1 infection. Engl. J. Med. N. 2011;364:1195–1206. doi: 10.1056/NEJMoa1010494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeuzem S, Andreone P, Pol S, Lawitz E, Diago M, Roberts S, Focaccia R, Younossi Z, Foster GR, Horban A, Ferenci P, Nevens F, Mullhaupt B, Pockros P, Terg R, Shouval D, van Hoek B, Weiland O, Van Heeswijk R, De Meyer S, Luo D, Boogaerts G, Polo R, Picchio G, Beumont M. Telaprevir for retreatment of HCV infection. Engl. J. Med. N. 2011;364:2417–2428. doi: 10.1056/NEJMoa1013086. [DOI] [PubMed] [Google Scholar]
- 6.Lawitz E, Sulkowski MS, Ghalib R, Rodriguez-Torres M, Younossi ZM, Corregidor A, DeJesus E, Pearlman B, Rabinovitz M, Gitlin N, Lim JK, Pockros PJ, Scott JD, Fevery B, Lambrecht T, Ouwerkerk-Mahadevan S, Callewaert K, Symonds WT, Picchio G, Lindsay KL, Beumont M, Jacobson IM. Simeprevir plus sofosbuvir, with or without ribavirin, to treat chronic infection with hepatitis C virus genotype 1 in non-responders to pegylated interferon and ribavirin and treatment-naive patients: the COSMOS randomised study. Lancet. 2014;384:1756–1765. doi: 10.1016/S0140-6736(14)61036-9. [DOI] [PubMed] [Google Scholar]
- 7.Kowdley KV, Lawitz E, Crespo I, Hassanein T, Davis MN, DeMicco M, Bernstein DE, Afdhal N, Vierling JM, Gordon SC, Anderson JK, Hyland RH, Dvory-Sobol H, An D, Hindes RG, Albanis E, Symonds WT, Berrey MM, Nelson DR, Jacobson IM. Sofosbuvir with pegylated interferon alfa-2a and ribavirin for treatment-naive patients with hepatitis C genotype-1 infection (ATOMIC): an open-label, randomised, multicentre phase 2 trial. Lancet. 2013;381:2100–2107. doi: 10.1016/S0140-6736(13)60247-0. [DOI] [PubMed] [Google Scholar]
- 8.Lawitz E, Poordad FF, Pang PS, Hyland RH, Ding X, Mo H, Symonds WT, McHutchison JG, Membreno FE. Sofosbuvir and ledipasvir fixed-dose combination with and without ribavirin in treatment-naive and previously treated patients with genotype 1 hepatitis C virus infection (LONESTAR): an open-label, randomised, phase 2 trial. Lancet. 2014;383:515–523. doi: 10.1016/S0140-6736(13)62121-2. [DOI] [PubMed] [Google Scholar]
- 9.Feeney ER, Chung RT. Antiviral treatment of hepatitis C. BMJ. 2014;348:g3308. doi: 10.1136/bmj.g3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.A combination of ledipasvir and sofosbuvir (Harvoni) for hepatitis C. The Medical letter on drugs and therapeutics. 2014;56:111–112. [PubMed] [Google Scholar]
- 11.A 4-drug combination (Viekira Pak) for hepatitis C. The Medical letter on drugs and therapeutics. 2015;57:15–17. [PubMed] [Google Scholar]
- 12.De Clercq E. Current race in the development of DAAs (direct-acting antivirals) against HCV. Biochem. Pharmacol. 2014;89:441–452. doi: 10.1016/j.bcp.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 13.Wyles DL. Antiviral resistance and the future landscape of hepatitis C virus infection therapy. J. Infect. Dis. 2013;207(Suppl. 1):S33–S39. doi: 10.1093/infdis/jis761. [DOI] [PubMed] [Google Scholar]
- 14.Paolucci S, Fiorina L, Piralla A, Gulminetti R, Novati S, Barbarini G, Sacchi P, Gatti M, Dossena L, Baldanti F. Naturally occurring mutations to HCV protease inhibitors in treatment-naive patients. Virol. J. 2012;9:245. doi: 10.1186/1743-422X-9-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Svarovskaia ES, Martin R, McHutchison JG, Miller MD, Mo H. Abundant drug-resistant NS3 mutants detected by deep sequencing in hepatitis C virus-infected patients undergoing NS3 protease inhibitor monotherapy. J. Clin. Microbiol. 2012;50:3267–3274. doi: 10.1128/JCM.00838-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lohmann V, Körner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999 Jul 2;285(5424):110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 17.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. U. S. A. 2005 Jun 28;102(26):9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Itsui Y, Sakamoto N, Kakinuma S, Nakagawa M, Sekine-Osajima Y, Tasaka-Fujita M, Nishimura-Sakurai Y, Suda G, Karakama Y, Mishima K, Yamamoto M, Watanabe T, Ueyama M, Funaoka Y, Azuma S, Watanabe M. Antiviral effects of the interferon-induced protein guanylate binding protein 1 and its interaction with the hepatitis C virus NS5B protein. Hepatology. 2009;50:1727–1737. doi: 10.1002/hep.23195. [DOI] [PubMed] [Google Scholar]
- 19.Andreev IA, Belov DS, Kurkin AV, Yurovskaya MA. Synthesis of 4,5,6,7-Tetrahydro-1H-indole derivatives through successive sonogashira coupling/Pd-mediated 5-endo-dig cyclization. Eur. J. Org. Chem. 2013;4:649–652. [Google Scholar]
- 20.Sofia MJ, Chang W, Furman PA, Mosley RT, Ross BS. Nucleoside, nucleotide, and non-nucleoside inhibitors of hepatitis C virus NS5B RNA-dependent RNA-polymerase. Med. Chem. J. 2012;55:2481–2531. doi: 10.1021/jm201384j. [DOI] [PubMed] [Google Scholar]
- 21.LaPlante SR, Padyana AK, Abeywardane A, Bonneau P, Cartier M, Coulombe R, Jakalian A, Wildeson-Jones J, Li X, Liang S, McKercher G, White P, Zhang Q, Taylor SJ. Integrated strategies for identifying leads that target the NS3 helicase of the hepatitis C virus. J. Med. Chem. 2014 Mar 13;57(5):2074–2090. doi: 10.1021/jm401432c. [DOI] [PubMed] [Google Scholar]
- 22.Golub AG, Gurukumar KR, Basu A, Bdzhola VG, Bilokin Y, Yarmoluk SM, Lee JC, Talele TT, Nichols DB, Kaushik-Basu N. Discovery of new scaffolds for rational design of HCV NS5B polymerase inhibitors. Eur. J. Med. Chem. 2012;58:258–264. doi: 10.1016/j.ejmech.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaushik-Basu N, Bopda-Waffo A, Talele TT, Basu A, Costa PR, da Silva AJ, Sarafianos SG, Noel F. Identification and characterization of coumestans as novel HCV NS5B polymerase inhibitors. Nucleic Acids Res. 2008;36:1482–1496. doi: 10.1093/nar/gkm1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li K, Frankowski KJ, Belon CA, Neuenswander B, Ndjomou J, Hanson AM, Shanahan MA, Schoenen FJ, Blagg BSJ, Aubé J, Frick DN. Optimization of potent hepatitis C virus NS3 helicase inhibitors isolated from the yellow dyes thioflavine S and primuline. J. Med. Chem. 2012;55:3319–3330. doi: 10.1021/jm300021v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paulsen RB, Seth PP, Swayze EE, Griffey RH, Skalicky JJ, Cheatham TE, 3rd, Davis DR. Inhibitor-induced structural change in the HCV IRES domain IIa RNA. Proc. Natl. Acad. Sci. U. S. A. 2010;107:7263–7268. doi: 10.1073/pnas.0911896107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soler M, McHutchison JG, Kwoh TJ, Dorr FA, Pawlotsky JM. Virological effects of ISIS 14803, an antisense oligonucleotide inhibitor of hepatitis C virus (HCV) internal ribosome entry site (IRES), on HCV IRES in chronic hepatitis C patients and examination of the potential role of primary and secondary HCV resistance in the outcome of treatment. Antivir. Ther. 2004;9:953–968. [PubMed] [Google Scholar]
- 27.Nichols DB, Leao RA, Basu A, Chudayeu M, de Moraes Pde F, Talele TT, Costa PR, Kaushik-Basu N. Evaluation of coumarin and neoflavone derivatives as HCV NS5B polymerase inhibitors. Chem. Biol. Drug Des. 2013;81:607–614. doi: 10.1111/cbdd.12105. [DOI] [PubMed] [Google Scholar]
- 28.Manfroni G, Manvar D, Barreca ML, Kaushik-Basu N, Leyssen P, Paeshuyse J, Cannalire R, Iraci N, Basu A, Chudaev M, Zamperini C, Dreassi E, Sabatini S, Tabarrini O, Neyts J, Cecchetti V. New pyrazolobenzothiazine derivatives as hepatitis C virus NS5B polymerase palm site I inhibitors. J. Med. Chem. 2014;57:3247–3262. doi: 10.1021/jm401688h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim K, Kim KH, Kim HY, Cho HK, Sakamoto N, Cheong J. Curcumin inhibits hepatitis C virus replication via suppressing the Akt-SREBP-1 pathway. FEBS Lett. 2010;584:707–712. doi: 10.1016/j.febslet.2009.12.019. [DOI] [PubMed] [Google Scholar]
- 30.Kucukguzel I, Satilmis G, Gurukumar KR, Basu A, Tatar E, Nichols DB, Talele TT, Kaushik-Basu N. 2-Heteroarylimino-5-aryliden-4-thiazolidinones as a new class of non-nucleoside inhibitors of HCV NS5B polymerase. Eur. J. Med. Chem. 2013;69:931–941. doi: 10.1016/j.ejmech.2013.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manvar D, Pelliccia S, La Regina G, Famiglini V, Coluccia A, Ruggieri A, Anticoli S, Lee J, Basu A, Cevik O, Nencioni L, Palamara AT, Zamperini C, Botta M, Neyts J, Leyssen P, Kaushik-Basu N, Silvestri R. New 1-phenyl-5-(1H-pyrrol-1-yl)-1H-pyrazol-3-carboxamides inhibit hepatitis C virus replication via suppression of cyclooxygenase-2. Eur. J. Med. Chem. 2014;90:497–506. doi: 10.1016/j.ejmech.2014.11.042. [DOI] [PubMed] [Google Scholar]
- 32.Lee JC, Chen WC, Wu SF, Tseng CK, Chiou CY, Chang FR, Hsu SH, Wu YC. Anti-hepatitis C virus activity of Acacia confusa extract via suppressing cyclooxygenase-2. Antivir. Res. 2011;89:35–42. doi: 10.1016/j.antiviral.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 33.Chen KJ, Tseng CK, Chang FR, Yang JI, Yeh CC, Chen WC, Wu SF, Chang HW, Lee JC. Aqueous extract of the edible Gracilaria tenuistipitata inhibits hepatitis C viral replication via cyclooxygenase-2 suppression and reduces virus-induced inflammation. PLoS One. 2013;8:e57704. doi: 10.1371/journal.pone.0057704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin YT, Wu YH, Tseng CK, Lin CK, Chen WC, Hsu YC, Lee JC. Green tea phenolic epicatechins inhibit hepatitis C virus replication via cycloxygenas-2 and attenuate virus-induced inflammation. PLoS One. 2013;8:e54466. doi: 10.1371/journal.pone.0054466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin D, Rojo AI, Salinas M, Diaz R, Gallardo G, Alam J, De Galarreta CM, Cuadrado A. Regulation of heme oxygenas-1 expression through the phosphatidylinositol 3-kinase/Akt pathway and the Nrf2 transcription factor in response to the antioxidant phytochemical carnosol. J. Biol. Chem. 2004;279:8919–8929. doi: 10.1074/jbc.M309660200. [DOI] [PubMed] [Google Scholar]
- 36.Peng HK, Chen WC, Lee JC, Yang SY, Tzeng CC, Lin YT, Yang SC. Novel anilinocoumarin derivatives as agents against hepatitis C virus by the induction of IFN-mediated antiviral responses. Org. Biomol. Chem. 2013;11:1858–1866. doi: 10.1039/c2ob26860d. [DOI] [PubMed] [Google Scholar]
- 37.Chen WC, Wang SY, Chiu CC, Tseng CK, Lin CK, Wang HC, Lee JC. Lucidone suppresses hepatitis C virus replication by Nrf2-mediated heme oxygenas-1 induction. Antimicrob. Agents Chemother. 2013;57:1180–1191. doi: 10.1128/AAC.02053-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lam AM, Keeney D, Eckert PQ, Frick DN. Hepatitis C virus NS3 ATPases/helicases from different genotypes exhibit variations in enzymatic properties. J. Virol. 2003;77:3950–3961. doi: 10.1128/JVI.77.7.3950-3961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frick DN, Ginzburg O, Lam AM. A method to simultaneously monitor hepatitis C virus NS3 helicase and protease activities. Methods Mol. Biol. 2010;587:223–233. doi: 10.1007/978-1-60327-355-8_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.