Abstract
The ability to generate potent immunotherapies locally and transiently for the treatment of cancers is a promising strategy to improve efficacy and decrease off-target toxicities. Here, we explored an alternative approach for the delivery of immunotherapeutic agents, in which we deliver the pDNA of an engineered PD-L1 trap and/or CXCL12 trap to the nucleus of liver hepatocytes via a lipid calcium phosphate nanoparticle. This strategy greatly increased the concentrations of immunotherapeutic agents in the local tissue, allowing the therapy to inhibit the accumulation of immune suppressive cells and liver metastasis. Furthermore, we find that the lipid calcium phosphate nanoparticles containing the pCXCL12 trap resolved the formation of immune suppressive ectopic lymphoid structures, while the pPD-L1 trap promoted T-cell survival and migration into the liver following vaccination against tumor antigens (>180% increase in survival). This approach showed superior efficacy in the treatment of the liver metastasis compared to free protein immunotherapies. This strategy should be considered as an approach to support liver metastasis therapies as well as for future research interested in manipulating the chemokine/cytokine immune factors within the liver.
INTRODUCTION
In the United States alone approximately 381,150 patients were diagnosed with colorectal cancer, resulting in over 89,640 deaths in 2015. These numbers make colorectal cancer the third most common type of cancer worldwide, and is the second most deadly.1 The cause of death is rarely due to the primary cancer burden, as local resection is quite efficient. The establishment of metastasis is the leading cause of death, in which at early stages of cancer detection, before metastases form, the five-year survival rate is approximately 90%. Once liver metastasis has been established, this rate drops drastically to less than 12%.1 In the United States and Europe, liver metastasis is much more common than primary liver cancers such as hepatocellular carcinoma (HCC). Although resection and varying chemotherapeutic treatment regimens yield improved efficacy over other traditional treatments, the off-target toxicities along with a minimal increase in median survival, 12–20 months, presents a less than desirable outcome.2 Therefore, researchers aim to improve the treatment of cancers through boosting the patient’s own immune system against cancer specific antigens. Numerous anti-cancer vaccines for colorectal liver metastasis are being investigated in clinical trials. However, no anti-cancer vaccine for colorectal liver metastasis has reached the market due to lack of efficacy or systemic toxicities. The liver and tumors immune suppressive microenvironment is one major obstacle which must be overcome before these vaccines will have a significant impact on cancer patients. Recently, a study by Finkin et. al reported that the formation of immune suppressive ectopic lymphoid structures (ELS) in patients correlated with reduced survival and increased recurrence of liver cancer.3 Therefore, targeting central factors that are critical for the formation and establishment of the immune suppressive tumor microenvironment and ELS is one such strategy to decrease tumor burden.
A crucial factor, chemokine CXCL12, has been shown to increase tumor progression through promoting the immune suppressive environment that suppresses CD8+ T-cell activation. The hepatic stellate cells are resident perisinusoidal cells that have been shown to produce high levels of endogenous CXCL12 for recruitment of immunosuppressive lymphocytes (i.e. regulatory T cells) and myeloid derived suppressor cells (MDSC) to areas of inflammation. In the presence of high levels of CXCL12, CXCR4+ cancer and immune cells accumulate to establish an immune suppressive tumor niche.4–5 Further clinical analysis on human colorectal cancer biopsies have found that increased liver metastasis correlates with increasing CXCR4 expression.6–7
Additionally, major progress has been made in recent years aimed at increasing CD8+ T-cell activation through blocking the programmed cell death receptor 1/programmed cell death ligand 1 (PD-1/PD-L1) receptor/ligand pair. It has been well documented that the immune suppressive cell populations such as T-regulatory (Treg), MDSC, and numerous other immune cell types express high levels of PD-L1, which directly deactivates PD-1+ T-cells. Therefore, this PD-L1/PD-1 has become a promising target to increase tumor specific CD8+ T-cell activation and killing. However, while PD-1/PD-L1 targeting in cancer has shown beneficial responses in some cancer types such as melanoma, it has not shown great efficacy in immune suppressive cancers such as liver metastasis or solid tumors. Furthermore, a variety of side effects termed immune-related adverse events (irAEs) have been observed following systemic delivery of PD-1/PD-L1 antibodies, which has limited their therapeutic applications in clinical trials.8
Therefore, we developed a new anti-cancer strategy centered on the liver-specific delivery of genes encoding small affinity proteins to entrap such factors as CXCL12 and/or PD-L1. We recently demonstrated this strategy by trapping the CXCL12 in the liver.9 However, through these studies we found that trapping CXCL12 can only decrease the incidence of liver metastasis for a limited time due to low CD8+ T-cell recruitment and activation in the metastatic niche. Therefore, we hypothesize that it is necessary to utilize a combination of therapies aimed at reducing the immune suppressive environment while simultaneously increasing CD8+ T-cell recruitment and activation.
In the current study, we utilize the LCP platform to deliver two engineered plasmid DNAs (pCXCL12 trap/pPD-L1 trap) encoding proteins trapping both CXCL12 and PD-L1 to treat colorectal liver metastasis. The small CXCL12 protein trap, CXCL12 trap, was designed based on an anti-CXCL12 antibody sequences, by fusing a VH and a VL domain. The PD-L1 protein trap, PD-L1 trap, was developed based on the ability to convert the extracellular PD-L1 binding domain from endogenous PD-1 into a trivalent ligand through genetic fusion with a trimerization domain. The resulting trivalent PD-L1 trap achieved approximately 219 pM binding affinity to mouse PD-L1, 82,000 times higher affinity compared to endogenous PD-1 and PD-L1 (Miao et al., publication pending). Furthermore, to increase the CD8+ T-cell activation and recruitment to the metastatic niche, we delivered a previously reported LCP vaccine formulation loaded with a RIG-1 ligand, 5’pppdsRNA, and a phosphorylated model cancer specific antigen, p-AH1-A5.
The establishment of an aggressive murine colorectal liver metastasis via hemi-splenic inoculation of CT-26FL3 (RFP/Luc) allows us to investigate our therapies ability to alter the liver microenvironment and the tumor/immune cell profile. Furthermore, this model has minimal primary tumor growth, due to surgical resection, which mirrors the clinical scenario of colorectal liver metastasis patients in which the primary tumors are resected. The use of this syngeneic model also mirrors the clinical cases of the liver metastasis immune environment in which numerous immune cell ELS aggregates are established. Therefore, investigating strategies to resolve the establishment of these aggregates as well as promoting CD8+ T-cell recruitment to the metastatic niche will be further elucidated.
RESULTS
Formulation and characterization of galactose-LCP pDNA-mc-CR8C nanoparticles
Hu et al. first reported the formulation and delivery of the galactose-LCP with pDNA-mc-CR8C cargo to the liver hepatocytes of mice.10 The formulation of the pCXCL12 trap LCP was reported by Goodwin et al. and the methods used in the cited paper were used for all pTrap LCP formulations used in this study.9 Furthermore, the 5’pppdsRNA p-AH1-A5 LCP vaccine formulation was first reported by Goodwin et al.11 which is based on the phosphorylated peptide LCP vaccine formulation first reported by Xu et al.12 The core structure can be visualized under transmission electron microscopy (TEM) (Figure 1B). The DOPA monolayer surrounding the Ca3(PO4)2 core allows for the addition of the cationic outer leaflet lipids (1,2-dioleoyl-3-trimethylammonium-propane (DOTAP), helper lipid cholesterol, and galactose or mannose conjugated to 1,2-distearoyl-sn-glycero-3-phosphatidylethanolamine-N-[succinyl(polyethylene glycol)-2000 (DSPE-PEG2000) to assist in RES evasion and dendritic cell (by mannose) or hepatocyte (by galactose) uptake. In this report, the hydrodynamic diameter and the surface charge of the LCP particles were found via dynamic light scattering zetasizer analysis to be approximately 45 nm and 10 mV, respectively (Figure 1B). The pDNA encapsulation efficiency was found to be approximately 50–60%. The p-AH1-A5 and 5’pppdsRNA encapsulation efficiency was found to be approximately 70%, which corresponds to the reported efficiency by Goodwin et al.11
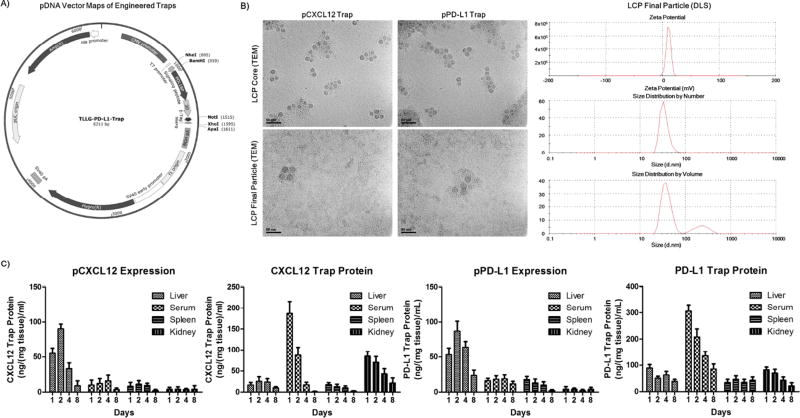
Figure 1. Formulation, characterization, and liver-specific expression profile of the engineered pPD-L1 and pCXCL12 traps.
A) pDNA vector map of the pPD-L1 trap first reported by Miao et al. (publication pending), the pCXCL12 trap has also been reported by Goodwin et al. using the same pDNA3.1 vector backbone B) Transmission electron microscopy (TEM) and dynamic light scattering (DLS) analysis of the LCP formulation containing pTraps, TEM analysis of the morphology of the LCP core and final LCP. DLS analysis of the hydrodynamic diameter size (~45 nm) and surface charge (~10 mV) (zeta potential). C) His (6×)-tag ELISA analysis was conducted to determine the organ distribution/expression of the pCXCL12 and pPD-L1 traps in all major LCP-accumulating organs and serum as well as the biodistribution of free CXCL12 and PD-L1 protein traps following tail vein injections, 1 mg/kg QOD × 3. The liver, serum, spleen, lungs and kidneys were collected on days 1, 2, 4, and 8 following final administration of the pTrap LCP or trap protein. Data was expressed as mean ± s.d., calculated from samples ran in triplicate and as a concentration relative to the standard His (6×) tag protein calibration curve. Scale bar: 50 µm.
In vivo expression profile of pDNA (pPD-L1 and pCXCL12 trap) via LCP formulation
Expression of the pCXCL12 trap and pPD-L1 trap was assessed through ELISA analysis via the His(6×)-tag engineered at the C-terminus of the protein traps (Figure 1A). Mice were treated via IV injection of pCXCL12 or pPD-L1 trap LCPs and compared to the free CXCL12 or PD-L1 protein traps (1 mg/kg QOD × 3). Studies demonstrated the transient expression time profile following analysis of the livers, serum, spleen, and kidneys on days 1, 2, 4, and 8 following final IV administration is shown in Figure 1C. The results indicate that the pDNA LCP vectors elicits transient liver specific expression of the CXCL12 or PD-L1 trap, yielding high expression for 4 days post final injection with minimal trap expression found on day 8 (Figure 1C). These results demonstrate that the galactose-LCP vector allows for local and transient expression in the liver hepatocytes, with minimal expression in other organs or serum compared to the IV administration of free CXCL12 or PD-L1 traps (Figure 1C).
Therapeutic efficacy and increased survival via pCOMBO traps LCP treatment
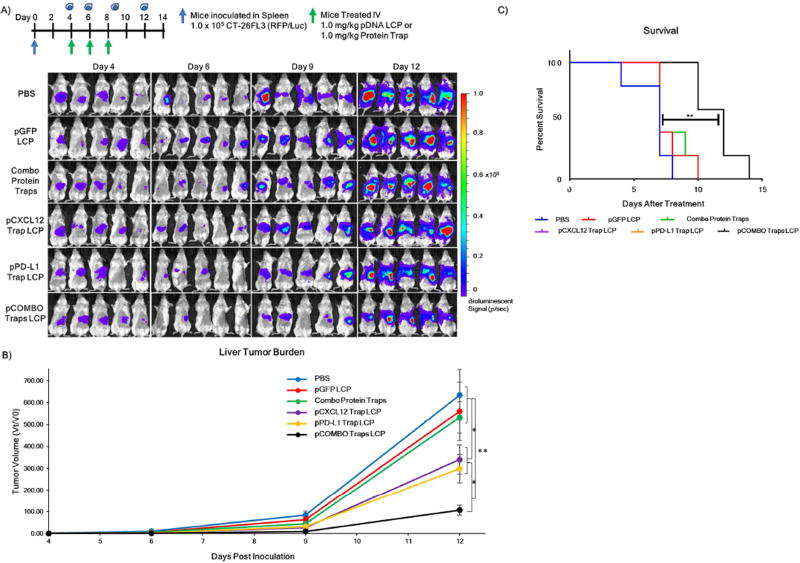
Colorectal liver metastasis was established through a hemi-spleen model to mirror clinical cases in which the primary tumor of colorectal liver metastasis patients is resected. The formation of large metastatic lesions and termination of the model due to the aggressive growth of metastasis in the liver was approximately 14 days post inoculation. Investigation of the tumor burden and survival of mice with colorectal liver metastasis was followed by IVIS bioluminescence imaging. The total tumor burden was followed through intraperitoneal (IP) administration of 100 µL of luciferin (10 mg/mL) followed by bioluminescence analysis 10 min post luciferin administration. The tumor burden was recorded on days 4, 6, 9, and 12 and quantified as tumor volume change (Figure 2A). Mice were sorted into treatment groups on day 4 when liver metastasis burden was first detected at a minimum ROI of 105 photons/sec. Mice were euthanized due to heavy liver tumor burden, in which mice became lethargic or showed signs of ascites, and survival was recorded (Figure 2B). It is clear from the bioluminescence images that mice treated with PBS, pGFP LCP, or free combo protein traps developed large metastatic tumor lesions in the liver (Figure 2A). In contrast, mice treated with pCXCL12 trap LCP, pPD-L1 trap LCP, or pCOMBO traps LCP resulted a significant (~2–6 fold) reduction in liver metastasis burden. The inability of the free combo protein traps to elicit a therapeutic response is hypothesized to be due to poor biodistribution to the liver as well as susceptibility to degradation via proteases. Interestingly, although the pCXCL12 and pPD-L1 trap LCP groups had a greater than 2-fold reduction in liver tumor burden on day 12, the median survival (7 days after final treatment) was the same as the control groups due to ascites. The pCOMBO trap LCP treatment significantly extended the median survival by 70% (12 days after treatment).
Figure 2. Decreased incidence of colorectal liver metastasis and improved survival via pCOMBO Traps LCP treatment.
A) Inoculation and treatment schedule/dose as well as bioluminescence signal and tumor burden images on day 4, 6, 9, and 12 days post inoculation. Treatment groups included PBS (n=8), pGFP LCP (n=8), combo protein traps (n=8), pCXCL12 trap LCP (n=8), pPD-L1 trap LCP (n=8), pCOMBO traps LCP (n=8). B) Tumor burden analysis and quantification (n=8 per group). C) Kaplan-Meier survival curve including all treatment groups (n=5 per group). Survival was determined by evaluating mouse weight, activity, and quality of life. Data was expressed as mean ± s.d., calculated from samples. N.S. denotes no significance. *, **, or *** denotes p < 0.05, 0.01, or 0.001, respectively, compared to corresponding PBS control and/or to nearest significant treatment. Bioluminescence signal scale minimum was set to 1×105 photons/sec to increase sensitivity on day 4. Minimum for days 6, 9, and 12 was set to 1×106 photons/sec to reduce background signal.
Decreasing the immune suppressive cell populations and metastasis in liver of colorectal cancer (CRC)
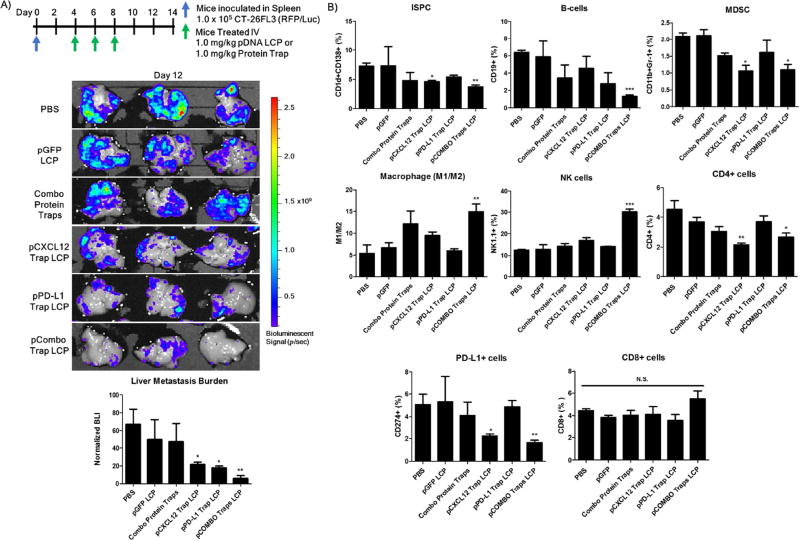
Further investigation into the changes in the immune cell populations within the liver metastasis and the therapeutic efficacy of this strategy was observed via flow cytometry and bioluminescence analysis. Mice bearing colorectal liver metastasis were treated with either single arm treatments including pCXCL12 trap LCP or pPD-L1 trap LCP, or combination treatments including the combo protein traps, or pCOMBO traps LCP. Treatment regimen and doses are shown in Figure 3A. The livers were extracted 12 days following splenic inoculation of the tumor cells and analyzed for metastatic lesions via IVIS bioluminescence imaging. The bioluminescence data clearly confirms the reduced metastatic burden following single arm treatments with pCXCL12 or pPD-L1 trap LCP and a synergistic effect following pCOMBO traps LCP treatment. Flow cytometry analysis indicates that the immune suppressive cell types, immune suppressive plasma cells (ISPC) and myeloid derived suppressor cells (MDSC), are reduced following pCOMBO traps LCP treatment (Figure 3B, Supplemental Figure 1). Interestingly, the pPD-L1 trap LCP alone shows no significant decrease in the PD-L1+ cells within the liver. However, the pCXCL12 trap and the combined pCOMBO traps therapy, which significantly reduces the recruitment and establishment of the immune suppressive cells, drastically reduces the PD-L1+ cells recruited to the liver lesions. Interestingly, the pCOMBO traps not only decreased the immune suppressive cell populations, but significantly increased the M1/M2 macrophage ratio and the natural killer (NK) cell populations found within the liver (Figure 3B, Supplemental Figure 1). The decreased immune suppressive cells along with the increases in M1/M2 and NK indicates a shift from a pro-tumor (immune suppressive) to anti-tumor environment within the liver. However, the minimal increase in the CD8+ T-cells indicates that the adaptive immune system needs a more robust signal to increase activation and invasion towards the metastatic niche.
Figure 3. Improved therapeutic strategy for reducing immune suppressive cells and incidence of colorectal cancer liver metastasis via pCOMBO Traps LCP.
A) Inoculation and treatment schedule/dose in which mice were inoculated with 1×105 CT-26 FL3 cells into one-half of a separated spleen, resection of the inoculated spleen 5 min post inoculation allowed for decreased primary tumor burden. Treatment consisted of QOD, days 4–8, administered through tail vein IV for pTrap LCP. Treatment groups included PBS (n=3), pGFP (n=3), combo free protein traps (n=3), pCXCL12 trap (n=3), pPD-L1 trap LCP (n=3), pCOMBO traps (n=3). Livers were collected from mice, analyzed, and quantified for the liver metastasis burden via bioluminescence analysis on day 12 B) Liver immune cell populations (CD1d+CD138+ (ISPC), CD19+ (B-Cells), CDllb+GR1+ (MDSC), F4/80+CD206+/− (M1/M2), NK1.1+, CD4+, PD-L1+, CD8+) analysis and quantification via flow cytometry (n=3 per group). Data was expressed as mean ± s.d., calculated from samples in triplicate. N.S. denotes no significance. *, **, or *** denotes p < 0.05, 0.01, or 0.001, respectively, compared to corresponding PBS control.
Modifying the immune cell populations in the liver of colorectal liver metastasis model
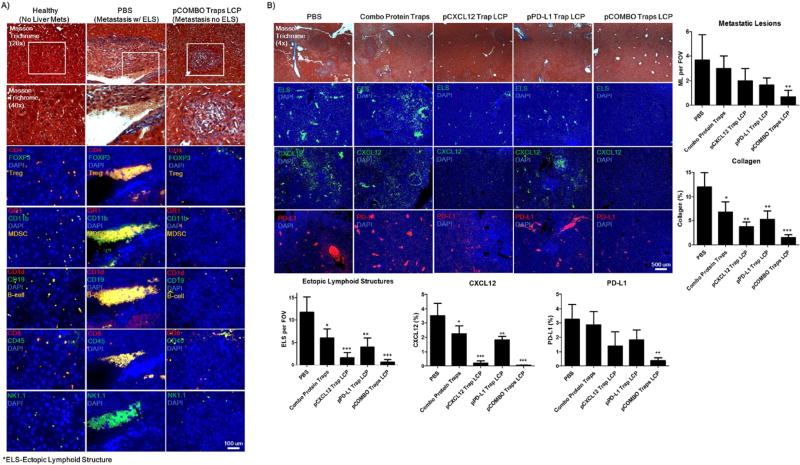
The changes in the immune cell profile within the liver metastasis was further investigated through microscopy analysis. Adjacent paraffin embedded liver sections were stained with Masson trichrome or immunofluorescent tagged antibodies (Figure 4A/B). The results clearly demonstrate that the pCOMBO traps LCP treatment significantly reduces the establishment of immune cell aggregates, ELS, in the livers of CRC mice to a level observed in a healthy liver control (Figure 4A). The expression levels of endogenous CXCL12 and PD-L1, along with the establishment of ELS in the liver of colorectal cancer bearing BALB/c mice were also investigated in all treatment groups (Figure 4B). Interestingly, similar to the flow cytometry analysis, the pPD-L1 trap shows minimal decrease in the PD-L1 expression within the liver. We hypothesize that this is due to the majority of PD-L1+ cells within the liver residing in the ELS. Therefore, the pPD-L1 trap is unable to extravagate to the center of the established ELS, in which the ELS acts as a binding site barrier. However, the pCXCL12 trap and pCOMBO traps, which significantly reduces the recruitment and establishment of the ELS, also reduces the PD-L1+ cells recruited to the liver lesions. In comparing the PBS or free combo protein traps to the pCOMBO traps LCP group, the pCOMBO traps therapy greatly resolves the establishment of ELS and metastatic lesions. The mice treated with PBS develop numerous metastatic lesions, with the majority forming near liver blood vessels and sinusoids. Interestingly, most of these metastatic lesions display a high number of large immune cell ELS aggregates proximal to the blood vessels and sinusoids (Figure 4A). The results indicate that there is a drastic decrease of all immune cells following pCOMBO traps LCP treatment. Staining of MDSC, Treg, Breg, CD8+, and NK+ cell populations show increased populations aggregated in the PBS treated mice compared to the healthy (no liver mets) and pCOMBO traps LCP groups. The pCOMBO traps LCP group showed baseline levels of immune cell aggregates similar to the healthy liver. However, a small increase was observed in the CD8+ T-cell population in the pCOMBO traps LCP group compared to the healthy group (Figure 4A). These results provide insight into the metastatic lesions’ ability to cause local inflammation, which results in increased levels of CXCL12, collagen, and subsequently an influx of PD-L1+ anti- and pro-inflammatory immune cells which aggregated to form ectopic lymphoid structures surrounding the metastatic lesions (Figure 4A/B). Although the PBS group has the highest influx of CD8+ T-cells, the high level of immunosuppressive PD-L1+, MDSC and Treg populations surrounding the NK and CD8+ T-cells, is likely to neutralize cancer specific killing.
Figure 4. Biological trapping of endogenous CXCL12 and PD-L1 reduces immune suppressive ectopic lymphoid structures consisting of numerous aggregated immune cell types.
A) Masson trichrome and fluorescently stained paraffin-sections of liver tissues from colorectal liver metastasis BALB/c mice sacrificed 12 days after inoculation of tumor cells as well as a control healthy liver with no treatment and void of colorectal cancer (No Liver Mets). Immune-fluorescent stain against CD4, FOXP3, GR-1, CDllb, CDld, CD19, CD8, CD45, NK1.1, CXCL12, and PD-L1. All sections had nuclear staining with DAPI (blue). All colors are indicated in the top-left of the section image. Three groups were studied, including healthy (No Liver Mets), PBS (Metastasis w/ ELS), and pCOMBO Traps LCP (Metastsis no ELS). B) Additional sections were stained to determine changes in ELS, CXCL12, and PD-L1 distribution throughout the liver. Immune-fluorescent stain against ELS (Immune cell aggregates, overlapping signal from Immune-fluorescent stains), CXCL12, and PD-L1. All sections had nuclear staining with DAPI (blue). All colors are indicated in the top-left of the section image. Five groups were studied, including PBS, combo protein traps, pCXCL12 trap LCP, pPD-L1 trap LCP, and pCOMBO Traps LCP. The trichrome sections are also shown to distinguish normal liver versus diseased liver metastasis morphology and collagen content. Data was expressed as mean ± s.d., calculated from samples ran in triplicate. N.S. denotes no significance, N.D. denotes under detection limit, *, **, or *** denotes p < 0.05, 0.01, or 0.001, respectively, compared to corresponding PBS control. Scale bars: A) 100 µm, B) 500 µm.
Priming the metastatic liver microenvironment and the immune system increased survival
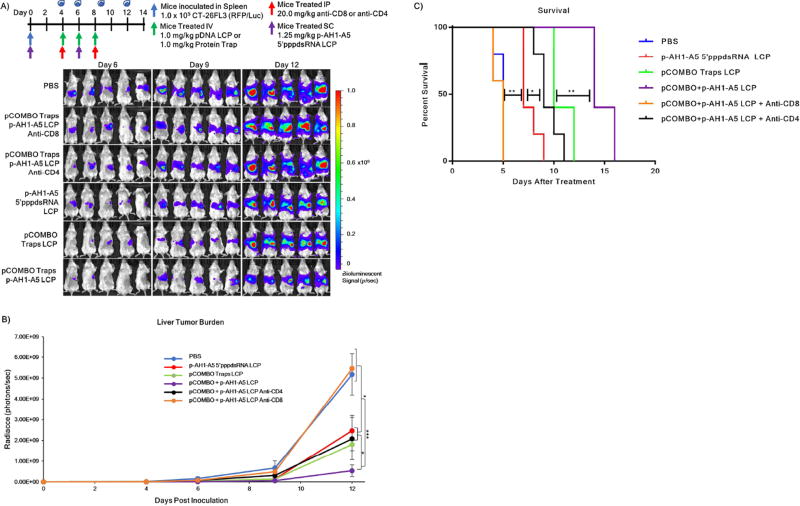
The ability to control the local immune suppressive microenvironment of metastatic lesions following pCOMBO traps LCP treatment allows for the investigation into potential synergy when combined with cancer specific vaccines. Therefore, we examined the therapeutic efficacy of a combination treatment regimen consisting of a robust cancer vaccine, p-AH1-A5+5’pppdsRNA LCP, and the pCOMBO traps LCP (pCOMBO p-AH1-A5 LCP). Furthermore, to investigate what subtype of T-cells are responsible for tumor specific killing, we studied the anti-cancer efficacy of the combination treatment in mice with depleted CD8+ or CD4+ T-cell populations. We followed a protocol reported by Harimoto et al., in which ≥95% of the CD8+ or CD4+ T-cell populations were depleted after two intraperitoneal injections of 400 µg of rat anti-CD8 or anti-CD4 (rat IgG2b).13 Mice were inoculated with CT-26FL3 cells according to the hemi-spleen model described earlier, followed by T-cell depletion on days 4 and 8. In this series of experiments, vaccine treatments were administered the same day as tumor implantation, followed by a booster vaccine on day 6. The pCOMBO traps LCP treatment began on day 4, followed by administrations on days 6 and 8. The animals were divided into six treatment groups: PBS, pCOMBO traps + p-AH1-A5 5’pppdsRNA LCP with anti-CD8, pCOMBO traps + p-AH1-A5 5’pppdsRNA LCP with anti-CD4, p-AH1-A5 5’pppdsRNA LCP, pCOMBO traps LCP, and pCOMBO traps + p-AH1-A5 5’pppdsRNA LCP. The treatment regimen, dosing, as well as the bioluminescence analysis of liver metastasis burden and quantification corresponding to each treatment on day 6, 9, and 12 are shown in Figure 5A/B. We further examined the effects of the combination therapy on the median survival. Tumor progression was followed by bioluminescence imaging (Figure 5A). Mice were euthanized when one of the follow conditions applied: drastic weight gain or loss greater than 10% within one week, or clear signs of distress were detected, such as dehydration, inactivity, lethargy. It is clear from the results that PBS mice, as well as mice depleted of CD8+ T-cells developed large metastatic tumor lesions in the liver within the 6–9 days following inoculation (Figure 5A). In contrast, mice treated with the single arm treatments, as well as the combination treatment showed a significant reduction in liver metastasis burden (3–8 fold) (Figure 5B). This reduction in liver metastasis burden significantly increased median survival in all treatment groups excluding the combination treatment with CD8+ T-cell depletion, compared to PBS (5 days after treatment). Ultimately, the combination of vaccine and pCOMBO traps therapy drastically increased survival by almost 180% in this aggressive liver metastasis model (Figure 5C).
Figure 5. Decreased incidence of colorectal liver metastasis and increased median survival via pCOMBO Trap and Vaccine LCP treatment.
A) Inoculation and treatment schedule/dose, bioluminescence signal, and tumor burden images 12 days post inoculation. Treatment groups included PBS (n=9), pCOMBO traps+p-AH1-A5 LCP+anti-CD8 (n=9), pCOMBO traps+p-AH1-A5 LCP+anti-CD4 (n=9), p-AH1-A5 5’pppdsRNA LCP (n=9), pCOMBO traps LCP (n=9), pCOMBO traps+p-AH1-A5 LCP (n=9). B) Tumor burden analysis and quantification (n=9 per group). C) Kaplan-Meier survival curve including all treatment groups (n=5 per group). Survival was determined by evaluating mouse weight, activity, and quality of life. Data was expressed as mean ± s.d., calculated from samples. N.S. denotes no significance. *, **, or *** denotes p < 0.05, 0.01, or 0.001, respectively, compared to corresponding PBS control or to nearest significant treatment.
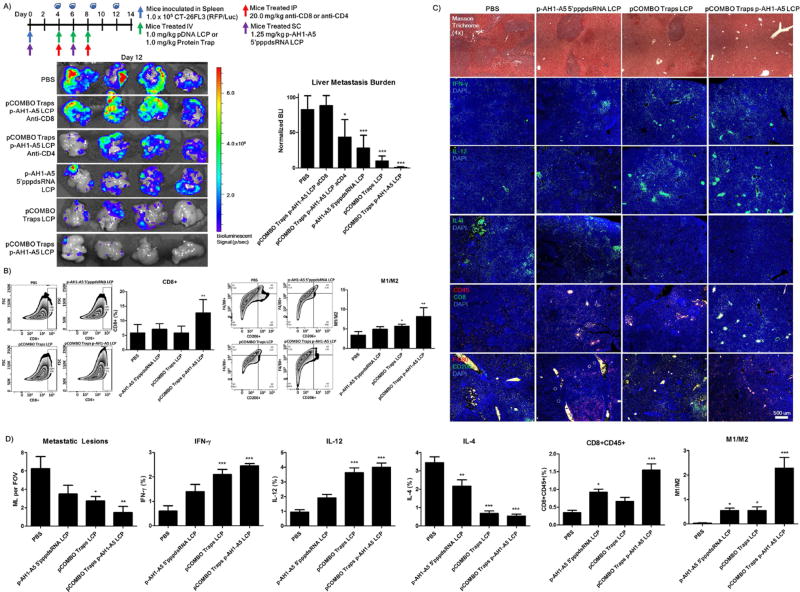
Resolving immune suppressive ELS and promoting increased CD8+ T-cell proliferation and invasion
We further examined how changes in the immune profile following the combination treatment regimen altered the survival and liver metastasis burden. The treatment regimen, dosing, as well as the bioluminescence analysis of liver metastasis burden corresponding to each treatment on day 12 is shown in Figure 6A. The results clearly show a significant reduction in both the single arm treatments of p-AH1-A5 5’pppdsRNA LCP (3 fold) and pCOMBO traps LCP (10 fold), as well as in the combination, pCOMBO p-AH1-A5 LCP with CD4+ T-cell depletion (2 fold) or most significantly without T-cell depletion (30 fold). Furthermore, the CD8+ T-cell depletion completely negates any therapeutic efficacy, resulting in comparable levels of liver metastasis found in the PBS group. The CD4+ T-cell depleted group resulted in a reduced therapeutic effect, but produced a more than 2-fold reduction in the liver metastasis burden. This may be due to CD4+ T cells critical role in the activation, expansion, and survival of primary CD8+ T cells, as well as for the establishment and maintenance of memory CD8+ T cells. Therefore, it is possible that the depletion of CD4+ T cells indirectly affects the function of CD8+ T cells resulting in reduced tumor killing effect.
Figure 6. Improved therapeutic outcome following increased M1/M2 and CD8+ cell populations and reducing immune suppressive ectopic lymphoid structure formation via combination of pCOMBO Trap+Vaccine LCP treatment.
A) Inoculation and treatment schedule/dose in which mice were inoculated with 1×105 CT-26 FL3 cells into one-half of a hemi-spleen. Treatment consisted of QOD, days 4–8, administered through tail vein IV for pTrap LCP, days 0 and 6 for SC vaccine, and days 4 and 8 for IP administration of anti-CD8 or anti-CD4. Treatment groups included PBS (n=4), pCOMBO traps+p-AH1-A5 LCP+anti-CD8 (n=4), pCOMBO traps+p-AH1-A5 LCP+anti-CD4 (n=4), p-AH1-A5 5’pppdsRNA LCP (n=4), pCOMBO traps LCP (n=4), pCOMBO traps+p-AH1-A5 LCP (n=4). Livers were collected on day 12, analyzed, and quantified for liver metastasis burden via bioluminescence analysis. B) Pro-inflammatory liver immune cell population (M1/M2 and CD8+) analysis and quantification via flow cytometry (n=4 per group). C) Masson trichrome staining for histological analysis of metastatic lesions in the liver as well as liver immune cell population (M1/M2 and CD8+CD45+) and cytokine IFN-γ, IL-12, and IL-4 analysis. D) Quantification via immunofluorescence staining (n=3). Data was expressed as mean ± s.d., calculated from samples. N.S. denotes no significance. *, **, or *** denotes p < 0.05, 0.01, or 0.001, respectively, compared to corresponding PBS control or to nearest significant treatment.
Most interestingly, following flow cytometry analysis, the combination treatment shows a clear increase in the pro-inflammatory activation, in which both the M1/M2 ratio, as well as the CD8+ T-cell population are increased (Figure 6B). These results confirm our hypothesis, in which therapeutic efficacy is dependent on both resolving the immune suppressive microenvironment as well as promoting the CD8+ T-cells via vaccination and immune checkpoint manipulation. These results clearly demonstrate a therapeutic shift to a pro-inflammatory (anti-cancer) environment. Microscopy analysis of liver sections on day 12 following inoculation further demonstrated the drastic changes in the immune populations. Immuno-fluorescence analysis of pro-inflammatory cytokines IFN-γ and IL-12, as well as immune suppressive, IL-4, were investigated (Figure 6C/D). Furthermore, M1/M2 and CD8+ T-cell populations were analyzed by immune-fluorescence microscopy (Figure 6C/D). The results clearly indicate a shift from a high IL-4 promoting TH2/M2 immune suppressive profile present in the PBS treated livers, to a high INF-γ/IL-12 TH1/M1 pro-inflammatory microenvironment following combination of the vaccine and pCOMBO traps LCP treatment. Furthermore, this shift in the cytokine profile, correlates to increased CD8+ T-cell and M1/M2 macrophage populations, yielding decreased metastatic lesions and an anti-tumor microenvironment within the liver.
Effects of pTrap LCP and free trap protein on liver, kidney, and blood (toxicity analysis)
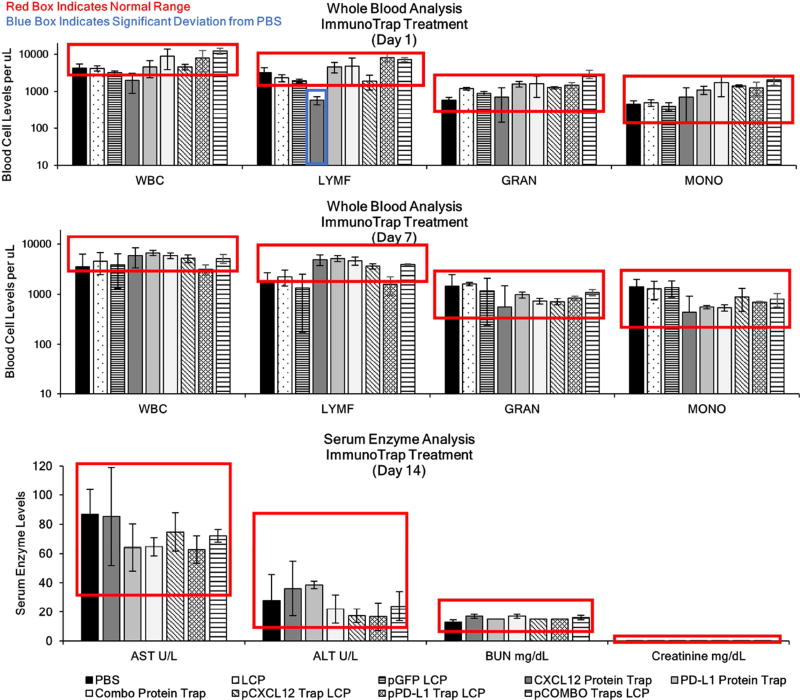
Administration of pTrap LCPs (1 mg/kg QOD × 3) showed no significant changes in the population of white blood cell levels compared to PBS mice one or seven days following final administration of the pTrap LCPs (Figure 7A). However, analysis of mouse whole blood samples one day after systemic IV injection of engineered CXCL12 trap protein (1 mg/kg QOD × 3) resulted in a transient decrease of the white blood cell and lymphocyte population. A recovery of the lymphocyte levels was observed seven days post final injection (Figure 7A). The decrease in the white blood cell populations is a clear example of the endogenous role CXCL12 plays in maintaining/recruiting the immune cells from the bone marrow to the blood and stresses the importance of avoiding toxicities through targeted and transient expression of the engineered CXCL12 trap via pDNA LCP vector. Such transient decrease of the white blood cells and lymphocytes was not observed when PD-L1 trap protein was applied.
Figure 7. Toxicological Analysis.
A) ALT, AST, creatinine, and BUN enzyme levels, blood cell levels after treatment of PBS, Blank LCP, pGFP LCP QOD, free protein traps: CXCL12 trap, PD-L1 trap or both combined, or pDNA LCP including pCXCL12, pPD-L1, or both combined pCOMBO LCP 1 mg/kg QOD × 3. Blood samples were collected via tail nick at one and seven days following final injection and analyzed for white blood cell populations. Mice were euthanized 14 days post final injection and the blood serum was collected and analyzed for liver (AST and ALT) and kidney (BUN and Creatinine) biomarkers. All LCP treatments fall within normal range. Data was expressed as mean ± s.d., calculated from samples ran in triplicate, *, **, or *** denotes p < 0.05, 0.01, or 0.001, respectively, compared to corresponding PBS control.
DISCUSSION
It is reported that the recruitment of immune cell types to establish the ELSs depends on chemoattracting chemokines CXCL12, CXCL13, CCL19, and CCL21.14 However, most of the research investigating ELS formation has investigated this mechanism in autoimmune or chronic inflammation disease. Although many cancers share several characteristics of diseases based on chronic inflammation, it is critical to investigate which factors are essential for ELS formation in liver cancers. Our ability to deliver genes expressing a trap against a specific chemokine or immune checkpoint is a promising tool to help elucidate what factors are essential for the establishment of ELSs in cancer lesions.
Here, we have provided the first evidence that an engineered pCXCL12 trap gene immunotherapy, expressed transiently and locally in the liver, can drastically resolve the establishment of immune suppressive ectopic lymphoid structures. Additional administration of an engineered immune checkpoint blockade, pPD-L1 trap, as well as a robust antigen specific vaccine, provides further evidence that increased CD8+ T-cell activation along with reduced immune suppressive tumor microenvironment elicit a promising anti-cancer response. Moreover, the ability to manipulate the liver metastasis microenvironment by transfecting hepatocytes with engineered traps, allows for future investigation into numerous other factors that may prove to be beneficial in modulating the immune response and should be considered in future applications.
Our liver metastasis cancer model displays a similar phenotype to the HCC model reported by Finkin et al.3, which revealed that liver tumorigenesis was promoted by the establishment of pro-tumor ELSs associated with chronic NF-κB activation.3 Notably, both the liver metastasis model presented in this research and Finkin et al.’s HCC model indicates that ELSs, which are commonly present in human liver cancer lesions, aid in tumor development and proliferation by promoting a protective immune suppressive microenvironment. These findings are distinctly different from the general findings in numerous other cancer types in which the establishment of ELSs within the tumor increases the pro-inflammatory and anti-tumor responses.15–19 It is clear from ours and Finkin et al’s results that the phenotype of the ELSs established near the liver lesions are vastly different compared to tumors in other organs such as the breast, lung, colon, and skin. The anti-tumorigenic effect of ELSs found in most non-liver tumor types requires a competent adaptive immune system that forms pro-inflammatory ELSs. These ELSs are rich in antibody producing B-cells and activated antigen specific CD8+ and CD4+ T-cells.20 Furthermore, these anti-tumor ELSs contain a minimal number of immune suppressive cell types such as Treg, MDSC, and Breg. However, the ELS phenotype present in the liver lesions of our liver metastasis model as well as in clinical liver cancers demonstrates a drastic contrast to the anti-tumorigenic ELSs found in most non-liver residing tumors.3 Our results clearly indicate that the ELSs formed within the liver consist of numerous immune suppressive cell types. Most of which are MDSC, Treg, and Breg. Although we do find a significant number of CD8+ T-cells and natural killer cells within the ELSs, the vast number of immune suppressive MDSC, Treg, and Breg populations drastically reduce the anti-cancer killing. This reduced killing is likely due to the high expression levels of co-inhibitory PD-L1 on the surface of these immune cells. Furthermore, most of these immune suppressive cell types have been shown to provide cytokines, as well as growth and survival factors to protect and support the invasion and proliferation of the metastatic niche.21
Encapsulation and delivery of pDNAs encoding PD-L1 and CXCL12 traps into the lipid calcium phosphate vector drives local and transient expression in the liver. Surprisingly, the expression of the CXCL12 trap reduced the formation of ectopic lymphoid structures associated with liver metastasis progression and immune evasion. Liver delivery of the lipid calcium phosphate vectors containing pPD-L1 and pCXCL12 trap achieved reduction of liver metastasis burden in murine models of colorectal liver metastasis, and prolonged survival of mice by greater than 70%. The anti-cancer immune response and survival was further boosted (>180%) through treatment of the pPD-L1/pCXCL12 traps in combination with a model cancer vaccine containing phosphorylated tumor peptide antigen and potent RIG-1 adjuvant, 5’pppdsRNA. The administration of lipid vectors capable of delivering pDNA to the liver hepatocytes of patients with liver metastasis would allow for increased local concentrations of the therapeutics, resulting in improved efficacy over traditional antibody immunotherapies. Furthermore, the transient expression profile provides clinicians the ability to closely control the expression of these therapeutic agents. This strategy will ultimately allow clinicians and researchers to manipulate the liver and immune microenvironment to resist cancer invasion, improve organ health, and prolong patient survival by using either a single or a desired combination of gene immunotherapies. Based upon these findings, we propose that the deployment of these pTrap LCPs in patients with liver cancers and established ELSs should be explored in clinical trials. Future research should combine this pTrap LCP technology which can modulate the livers immune microenvironment with other immunostimulatory agents, including immune checkpoint inhibitors or cancer vaccines. Additionally, future application of this technology as a treatment for autoimmune or chronic inflammation within the liver, such as in cases of hepatitis, may also be explored.
MATERIALS AND METHODS
1,2-Distearoyl-sn-glycero-3-phosphatidylethanolamine-N-[succinyl(polyethylene glycol)-2000]-N-hydroxysuccinimide (DSPE-PEG2000–N-hydroxysuccinimide (NHS)) was purchased from NOF Corporation (Tokyo, Japan). DSPE-PEG2000-galactose and DSPE-PEG2000-mannose was synthesized through the conjugation of 10 eq. of 4-aminophenyl β-d-galactopyranoside or mannose and 1 eq. of DSPE-PEG2000-NHS in PBS buffer, followed by chloroform extraction and dialysis against water using a MWCO 1000 dialysis tube. All other lipids were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL). Peptide (mcCR8C) was purchased from Elim Biopharmaceuticals, Inc. (Hayward, CA); monocyclic abbreviated to mc. Peptides (AH1, p-AH1-A5, and β-gal) were purchased from Peptide 2.0 (Chantilly, VA). Fluorescent Cy3 cDNA labelling kit was acquired via Mirus LabelIT kit (Mirus Bio, Madison, WI). Luciferin was purchased from Promega Corporation (Madison, WI). Plasmids encoding green fluorescent protein (GFP) driven by the cytomegalovirus (CMV) promoter were custom prepared by Bayou Biolabs (Harahan, LA). IF and IHC kits as well as all antibodies were purchases through Abcam (Cambridge, MA). All other chemicals were obtained from Sigma-Aldrich (St. Louis, MO) and used without further purification. Anti-His(6x) tag ELISA, Anti-Lyt2.2 and isotype controls were purchased from BioXcell (West Lebanon, NH). Six-week-old BALB/c female mice (~18 g each) were purchased from Charles River Laboratories (Wilmington, MA). Six-week-old nude female mice (~20 g each) were purchased from the University of North Carolina (Chapel Hill, NC). All work performed on animals was in accordance with and approved by the University of North Carolina Institutional Animal Care and Use Committee.
Study design
This was a preclinical study to assess the efficacy, safety, and biological effects of two promising engineered gene immunotherapies (pCXCL12 trap and pPD-L1 trap) as well as the effect in combination with a potent vaccine (p-AH1-A5 5’pppdsRNA) delivered via a non-viral vector. In previous reported studies, we demonstrated that neutralization of the high levels of endogenous CXCL12 produced by the hepatic stellate cells in the inflamed liver decreased the migration, invasion, and proliferation of colorectal liver metastasis as well as shifted the immune environment from anti- to pro-inflammatory. However, the therapeutic efficacy was thwarted in more aggressive liver metastasis models in which increased formation of immune suppressive ectopic lymphoid structures (ELS) deactivated the tumor specific CD8+ T-cells. Therefore, our hypothesis aims to understand how to resolve the immune suppressive ELS while increasing the activation of cancer specific CD8+ T-cells to induce an improved anti-cancer response. This hypothesis was tested through an established syngeneic colorectal liver metastasis murine model in which two additional CD8+ promoting therapies (pPD-L1 Trap LCP and p-AH1-A5+5’pppdsRNA LCP) were delivered alongside the pCXCL12 Trap LCP. The number of mice used for the in vivo experiments are outlined in the figure legends. Grouping for tumor experiments was done through measuring the tumor burden via bioluminescent techniques, ranking, and distributing equally among groups or through randomized blind selection in cases where treatment was initiated on day 0. Additional study design details are also included in the statistical analysis section.
Statistical analysis
Data were expressed as the mean ± standard deviation (SD). Statistical analysis was performed via PRISM software using Students’ t-test when only two value sets were compared, and one-way analysis of variance (ANOVA) with a dunnette’s test for post hoc analysis when the data involved three or more groups. Kaplan- Meier analysis was used for determination of the survival study, in which log-rank (Mantel-Cox) test was applied to determine significance between treatment groups. *, **, ***, **** denotes p < 0.05, 0.01, 0.001, and 0.0001, respectively, and was considered significant and documented on figure or figure legend. In all statistics, the groups are compared against the PBS control if not otherwise noted.
Construction of CXCL12 and PD-L1 trap genes
The construction of pCXCL12 trap was described as we recently published9. To construct the PD-L1 trap plasmid (pPD-L1 trap), the coding sequences of the PD-1 extracellular domain (human PD-1 residues 34–150 with C93S mutation or mouse PD-1 residues 21–150) and the C-terminal trimerization domain of cartilage matrix protein (human CMP1 residues 454–496 or mouse CMP1 residues 458–500) were used for assembly of the trap gene. A flexible hinge region with optimized length was introduced between the PD-L1-binding domain and the trimerization domain. The final sequence for the monomeric PD-L1 trap codes for a secretion signaling peptide, PD-1 extracellular domain, hinge peptide, trimerization domain, E or FLAG tag, and His(6×) tag, respectively. The complete cDNA was cloned into pcDNA3.1 between NheI and Xho I sites and the accuracy was confirmed by DNA sequencing. The pPD-L1 trap map and the DNA sequence are first reported by Miao et al. (Publication Pending).
Expression and purification of recombinant trap proteins
293T cells were cultured until 70–80% confluence. To transfect the cells, 24 µg pTrap (orpcDNA3.1 negative control) and 40 µL lipofectamine were added to each 10-cm plate. The serum concentration was reduced after transfection. 293T cells were monitored each day to ascertain their survival. Ten mL supernatant was harvested after 24, 48, and 72 h, and kept at4°C for further purification. The supernatants were concentrated with 10 kDa MWCO spin filters to 200 µL and subjected to His-Mag-Ni-Sepharose beads to purify His (6×)-tagged CXCL12 or PD-L1 trap protein. The purified proteins were analyzed on 10% SDS-PAGE gel with silver stain.
Binding kinetics
Bio-layer Interferometry (BLI) analyses of the interaction between recombinant PD-L1 trap and PD-L1 or PD-L2 were performed on a fortéBIO Octet RED96 system. Assays were run at 30°C on Greiner Bio One black 96- well microplates. To measure the interaction between PD-L1 with the trimeric trap, AHC or SAX biosensors (Pall fortéBIO Corp.) were used to immobilize PD-L1 tagged with Fc or biotin. Purified trimeric PD-L1 trap was prepared in an assay buffer (1×PBS, 0.002% Tween 20, pH 7.4) and applied to a 96-well microplate in column arrangement. Various concentrations of trap (0–500 nM) were used to test the binding. To study the disruption of preformed PD-1/PD-L1 complex, PD-L1 immobilized on the SAX biosensor was first saturated with 200 nM of PD-1, followed by incubation with a mixture of 200 nM PD-1 with or without trimeric PD-L1 trap. Assays were run in triplicate and all data were acquired and analyzed with fortéBIO Data Acquisition 6.4 software. Data processing was performed by averaging the reference biosensors, applying Savitzky-Golay filtering, and fitting binding curves. All binding kinetics for the PD-L1 trap are reported by Miao et al. (publication pending). All binding kinetics for the CXCL12 trap are reported by Goodwin et al9.
Preparation of LCP loaded with pDNA
LCP was prepared using a modified protocol 9,10. Two separate microemulsions (60 mL each) were prepared of Igepal 520 and cyclohexane (3:7 v/v) and placed under stirring. A pDNA (800 µg) solution was prepared and added to 1,800 µL of 2.5 M CaCl2. To this solution, octaarginine peptide (mc-CR8C) was added at an N:P ratio of 2:1 (~850 µg), this solution was immediately added to the microemulsion before precipitation could occur. A (NH4)2HPO4 solution (1,800 µL, 50 mM) was also prepared and added to the other microemulsion. Each microemulsion was stir for 20 min. The microemulsion containing (NH4)2HPO4 was added to the microemulsion containing the DNA/Peptide/CaCl2. This solution was stirred for 5 min before addition of 1,400 µL of 20 mM DOPA (in CHCl3). After addition of DOPA the microemulsion was stirred for an additional 30 min. An equal volume of 100% EtOH (120 ml) was added to disrupt the emulsion. The mixture was centrifuged at 10,000g for 20 min. After decanting the supernatant, the precipitate was washed, including vortex and sonication, twice thereafter with 100% EtOH to remove traces of Igepal and/or cyclohexane. The precipitate was then suspended in CHCl3. This solution was centrifuged at 14,000 rpm for 10 min for the removal of large aggregates, and the supernatant containing the LCP “cores” (DNA and peptide entrapped within a calcium phosphate nanoprecipitate, supporting and surrounded by a lipid monolayer of DOPA) was recovered. A single batch typically produces approximately 45 mg of cores, encapsulating approximately 450 µg pDNA. The ratio of cores to outer leaflet lipids for optimal final particle formulation was found to be 3.0 mg core: 225 µl DOTAP (20 mM): 225 µl Cholesterol (20 mM): 200 µl DSPE-PEG2000 (20 mM). Therein, 35 mol % DOTAP, 35 mol % cholesterol, and 30 mol % DSPE-PEG2000 (or 25 mol % DSPE-PEG and 5 mol % DSPE-PEG-Gal) were utilized as outer leaflet lipids. Addition of 0.1% (w/w) Tween 80 was added to the chloroform solution containing the cores and outer leaflet lipids. Preparation of final particles for injection followed formation of lipid film through drying off chloroform with nitrogen gas, followed by resuspension in 10% aqueous sucrose solution and sonication. Zeta potential and particle size of LCP were measured using a Malvern ZetaSizer Nano Series (Westborough, MA). TEM images of LCP were acquired using a JEOL 100CX II TEM (JEOL, Japan).
Characterization of LCP loaded with pDNA
DNA entrapment was characterized through several methods. Firstly, pDNA was labeled with Cy3 (Mirus LabelIT kit, Mirus Bio, Madison, WI) according to manufacturer instructions. Cy3-DNA was formulated into the LCP cores, after which recovery was assessed via fluorescence spectrometry. Additional encapsulation studies followed tritium labelling of the pDNA. The pDNA was dissolved in 100 uL of tritium water (5 µCi) and heated to 80 degrees Celsius for 6 h. pDNA was purified through sequential washing with cold DI water and filtration via 3000 MWCO centrifuge filter tubes. Standard curves were generated using a scintillation counter, along with known concentrations of peptide/pDNA complexes and used to calculate pDNA encapsulation efficiency (50–60%).
In vivo gene biodistribution and expression time profile
Formulation of galactose targeted LCPs containing pCXCL12 or pPD-L1 Trap DNA, which contains a His(6x)-Tag at the C-terminal end were injected (0.2 mL, balanced in osmolarity with the addition of sucrose) into 8-week-old BALB/c female mice (1 mg DNA/kg, 3 mice utilized for each group) through the tail vein. Mice were euthanized 1, 2, 4, and 8 days after administration. Liver, spleen, lungs, kidney, heart, and blood were collected and homogenized in RIPA buffer. Total protein concentration in the lysate was determined through a bicinchoninic acid protein assay kit (BCA Protein Assay Kit, Pierce, Rockford, IL). Subsequently, His-tag ELISA kit was also used in which samples were loaded for expression analysis. The kit provided standard proteins containing a His (6×)-tag to be used as a standard calibration control. Therefore, quantification of protein expression can be measured through ELISA analysis following BioXcell protocol. The CXCL12 and PD-L1 trap proteins were also injected and analyzed to compare with the pDNA LCP expression profile.
Orthotopic allografting in mice
Sub-confluent cells were harvested and washed in phosphate buffered saline (PBS) just prior to spleen implantation. Eight-week-old male BALB/c mice were anesthetized by inhalation of isoflurane (2%) via a vaporizer and placed in supine position. For splenic inoculation, an incision located below the left rib cage was made to exteriorize the spleen. The spleen was tied and cut into two parts each containing intact vascular pedicle for each half of the spleen. The distal section of the spleen was inoculated with 1×105 CT-26 (RFP/Luc) cells in 100 µL. The hemi-spleen containing inoculated cells was resected 5 mins after inoculation allowing the cancer cells to enter the portal vein. The hemi-half containing inoculated cells was resected to model primary tumor resection. The other half of the spleen was returned to the cavity and the abdominal wall and skin were closed with 4-0 polyglycolic acid sutures.
Luciferase imaging of whole animal and ex vivo tissues
Luciferase imaging of tissue and animals was performed as follows in which D-luciferin of 1.0 mg/ 10g body weight was injected intra-peritoneally into mice. Following 10 min post administration, luciferase imaging (IVIS-Kinetic) was applied on whole-mouse bodies. For ex vivo imaging, mice were dissected 10 min after luciferin injection. Livers were rinsed in PBS and placed in diluted luciferin solution (2 mg/ml) for 1 min. Organs were then placed in IVIS in which luciferase imaging was applied immediately. Imaging and quantification were performed at the UNC Biomedical Research Imaging Center (BRIC) using the IVIS kinetic optical imaging system. Quantification of bioluminescent signal was reported as either the raw average intensity, tumor volume fold change, or normalized.
Immunohistochemistry
Histology and immunohistochemistry were performed on paraffin-embedded or frozen sections from major organs and organs containing metastatic lesions. All paraffin-embedded tissue was resected, rinsed in PBS, and placed in 4% paraformaldehyde for 48 h at 4°C. Following 4% PFA, tissues were rinsed in water and placed in 70% ethanol solution until paraffin-embedded. All tissues for frozen sections were resected and rinsed in PBS, and placed in 2% paraformaldehyde overnight at 4°C. Following 2% PFA, tissues were placed in 30% sucrose/PBS solution overnight at 4°C. Tissues were snap frozen in O.C.T. (Fisher Scientific, Pittsburgh, PA). Immunofluorescence staining was processed through deparaffination, antigen retrieval, permeabilization, and blocking in 1% bovine serum albumin at room temperature for 2 hour. Primary antibodies conjugated with fluorophores (BD) were incubated overnight at 4°C and the nuclei were counterstained with 4',6-diamidino-2-phenylindole containing mounting medium (Vector Laboratories, Burlingame, CA). All antibodies were diluted after optimization usually ranging from 0.5 to 5.0 µg/mL. Images were taken using fluorescence microscopy (Nikon, Tokyo, Japan). A minimum of three randomly selected microscopic fields were quantitatively analyzed using Image J software.
Flow cytometry
Liver/Metastasis cells and splenocytes were made into single-cell suspensions. Immune lymphocytes were quantitatively analyzed by flow cytometry via immunofluorescent staining. Tissues were harvested and digested with collagenase A and DNase at 37 °C for 45 min. After red blood cell lysis via addition of ACK buffer, cells centrifuged at 2,000 g for 10 min. Following centrifugation, samples were washed and resuspended in 1 mL of phosphate-buffered saline. Cells were counted and diluted to 5×106 cells/mL. Cells were then stained by addition of the fluorescent-conjugated antibodies. For intracellular FOXP3 staining, the cells were permeated via penetration buffer (BD, Franklin Lakes, NJ) following the manufacturer’s instructions. Following staining cells were fixed and analyzed via FACS.
Supplementary Material
Flow cytometry analysis of liver metastasis immune cell populations
SIGNIFICANCE.
Our approach results in transient liver specific expression of gene immunotherapies with improved efficacy and reduced off-target toxicities over traditional systemically administered immunotherapies. This approach would allow clinicians to manipulate the liver and immune microenvironment to resist cancer invasion, improve organ health, and prolong patient survival.
Acknowledgments
We thank Dr. Maria Marjorette O. Peña for providing the highly metastatic CT-26(FL3) cell line. We thank the histology core for help with the paraffin embedded sections and trichrome stains, CHANL for allowing us to use the transmission electron microscopy, as well as the small animal imaging core at the University of North Carolina Chapel Hill for help with training on the IVIS bioluminescence imaging. Funding: This work was supported by NIH grants DK100664, CA151652, CA149387 (to L.H.) and CA157738, CA151652 (to R.L.), the Eshelman Institute of Innovation Tier III grant (to L.H. and R.L.), AFPE foundation pre-doctoral fellowship (to T.J.G.).
Abbreviations
- ALT
Alanine Aminotransferase
- AST
Aspartate Aminotransferase
- BLI
Bio-Layer Interferometry
- BUN
Blood Urea Nitrogen
- CMV
Cytomegalovirus
- CRC
Colorectal Cancer
- DLS
Dynamic Light Scattering
- DAPI
4',6-diamidino-2-phenylindole
- DOPA
1,2-dioleoyl-sn-glycero-3-phosphate
- DOTAP
1,2-dioleoyl-3-trimethylammonium-propane
- DSPE
1,2-distearoyl-sn-glycero-3-phosphatidylethanolamine
- ELISA
Enzyme Linked Immunosorbent Assay
- GFP
Green Fluorescent Protein
- HCC
Hepatocellular Carcinoma
- IFN-γ
Interferon gamma
- IL-4
Interleukin 4
- IL-12
Interleukin 12
- ISPC
Immune Suppressive Plasma Cells
- LCP
Lipid Calcium Phosphate
- mc
Mono-cyclic
- MDSC
Myeloid Derived Suppressor Cells
- NHS
N-Hydroxysuccinimide
- PBS
Phosphate Buffered Saline
- PD-1
Programmed Death Receptor 1
- PD-L1
Programmed Death Ligand 1
- PEG
Polyethylene Glycol
- PK
Pharmacokinetics
- pTrap
Galactose-PEG-LCP with pDNA trap-mc-CR8C
- SDF-1
Stromal Derived Factor-1
- RFP
Red Fluorescent Protein
- TEM
Transmission Electron Microscopy
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions: T.J.G., R.L., and L.H. conceived and designed the research. J.L., R.F., and O.D. generated and characterized the PD-L1 and CXCL12 traps. T.J.G. and M.H. performed the protein expression and LCP characterization experiments. T.J.G. and M.H. performed the in vivo mouse experiments. T.J.G. and L.S. prepared, analyzed and quantified the histology and flow cytometry. T.J.G. and L.S. performed the statistical analysis. T.J.G., R.L., and L.H. analyzed the data and wrote the manuscript.
Competing interests: The authors declare U.S. Provisional Patent application no.62/232,169 filed on 24 September 2015 covering the Trap technologies described.
Data and materials availability: All data for this study are presented here, and all materials are available for noncommercial researchers via a material transfer agreement.
References
- 1.Howlader NNA, Krapcho M, Miller D, Bishop K, Altekruse SF, Kosary CL, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA. SEER Cancer Statistics Review, 1975–2013. National Cancer Institute. 2016 http://seer.cancer.gov/csr/1975_2013/
- 2.Correale P, Botta C, Ciliberto D, Pastina P, Ingargiola R, Zappavigna S, Tassone P, Pirtoli L, Caraglia M, Tagliaferri P. Immunotherapy of colorectal cancer: new perspectives after a long path. Immunotherapy. 2016;8(11):1281–1292. doi: 10.2217/imt-2016-0089. [DOI] [PubMed] [Google Scholar]
- 3.Finkin S, Yuan D, Stein I, Taniguchi K, Weber A, Unger K, Browning JL, Goossens N, Nakagawa S, Gunasekaran G, Schwartz ME, Kobayashi M, Kumada H, Berger M, Pappo O, Rajewsky K, Hoshida Y, Karin M, Heikenwalder M, Ben-Neriah Y, Pikarsky E. Ectopic lymphoid structures function as microniches for tumor progenitor cells in hepatocellular carcinoma. Nat Immunol. 2015;16(12):1235–44. doi: 10.1038/ni.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kucia M, Reca R, Miekus K, Wanzeck J, Wojakowski W, Janowska-Wieczorek A, Ratajczak J, Ratajczak MZ. Trafficking of normal stem cells and metastasis of cancer stem cells involve similar mechanisms: pivotal role of the SDF-1-CXCR4 axis. Stem Cells. 2005;23(7):879–94. doi: 10.1634/stemcells.2004-0342. [DOI] [PubMed] [Google Scholar]
- 5.Lombardi L, Tavano F, Morelli F, Latiano TP, Di Sebastiano P, Maiello E. Chemokine receptor CXCR4: role in gastrointestinal cancer. Crit Rev Oncol Hematol. 2013;88(3):696–705. doi: 10.1016/j.critrevonc.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 6.Zeelenberg IS, Ruuls-Van Stalle L, Roos E. The chemokine receptor CXCR4 is required for outgrowth of colon carcinoma micrometastases. Cancer Res. 2003;63(13):3833–9. [PubMed] [Google Scholar]
- 7.Zhang SS, Han ZP, Jing YY, Tao SF, Li TJ, Wang H, Wang Y, Li R, Yang Y, Zhao X, Xu XD, Yu ED, Rui YC, Liu HJ, Zhang L, Wei LX. CD133(+)CXCR4(+) colon cancer cells exhibit metastatic potential and predict poor prognosis of patients. BMC Med. 2012;10:85. doi: 10.1186/1741-7015-10-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byun DJ, Wolchok JD, Rosenberg LM, Girotra M. Cancer immunotherapy - immune checkpoint blockade and associated endocrinopathies. Nat Rev Endocrinol. 2017 doi: 10.1038/nrendo.2016.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodwin TJ, Zhou Y, Musetti SN, Liu R, Huang L. Local and transient gene expression primes the liver to resist cancer metastasis. Sci Transl Med. 2016;8(364) doi: 10.1126/scitranslmed.aag2306. 364ra153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu Y, Haynes MT, Wang Y, Liu F, Huang L. A highly efficient synthetic vector: nonhydrodynamic delivery of DNA to hepatocyte nuclei in vivo. ACS Nano. 2013;7(6):5376–84. doi: 10.1021/nn4012384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodwin TJ, Huang L. Investigation of phosphorylated adjuvants co-encapsulated with a model cancer peptide antigen for the treatment of colorectal cancer and liver metastasis. Vaccine. 2017;35(19):2550–2557. doi: 10.1016/j.vaccine.2017.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Z, Ramishetti S, Tseng YC, Guo S, Wang Y, Huang L. Multifunctional nanoparticles co-delivering Trp2 peptide and CpG adjuvant induce potent cytotoxic T-lymphocyte response against melanoma and its lung metastasis. J Control Release. 2013;172(1):259–65. doi: 10.1016/j.jconrel.2013.08.021. [DOI] [PubMed] [Google Scholar]
- 13.Harimoto H, Shimizu M, Nakagawa Y, Nakatsuka K, Wakabayashi A, Sakamoto C, Takahashi H. Inactivation of tumor-specific CD8(+) CTLs by tumor-infiltrating tolerogenic dendritic cells. Immunol Cell Biol. 2013;91(9):545–55. doi: 10.1038/icb.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corsiero E, Bombardieri M, Manzo A, Bugatti S, Uguccioni M, Pitzalis C. Role of lymphoid chemokines in the development of functional ectopic lymphoid structures in rheumatic autoimmune diseases. Immunol Lett. 2012;145(1–2):62–7. doi: 10.1016/j.imlet.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 15.Pitzalis C, Jones GW, Bombardieri M, Jones SA. Ectopic lymphoid-like structures in infection, cancer and autoimmunity. Nat Rev Immunol. 2014;14(7):447–62. doi: 10.1038/nri3700. [DOI] [PubMed] [Google Scholar]
- 16.Di Caro G, Bergomas F, Grizzi F, Doni A, Bianchi P, Malesci A, Laghi L, Allavena P, Mantovani A, Marchesi F. Occurrence of tertiary lymphoid tissue is associated with T-cell infiltration and predicts better prognosis in early-stage colorectal cancers. Clin Cancer Res. 2014;20(8):2147–58. doi: 10.1158/1078-0432.CCR-13-2590. [DOI] [PubMed] [Google Scholar]
- 17.Gu-Trantien C, Loi S, Garaud S, Equeter C, Libin M, de Wind A, Ravoet M, Le Buanec H, Sibille C, Manfouo-Foutsop G, Veys I, Haibe-Kains B, Singhal SK, Michiels S, Rothe F, Salgado R, Duvillier H, Ignatiadis M, Desmedt C, Bron D, Larsimont D, Piccart M, Sotiriou C, Willard-Gallo K. CD4(+) follicular helper T cell infiltration predicts breast cancer survival. J Clin Invest. 2013;123(7):2873–92. doi: 10.1172/JCI67428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Messina JL, Fenstermacher DA, Eschrich S, Qu X, Berglund AE, Lloyd MC, Schell MJ, Sondak VK, Weber JS, Mule JJ. 12-Chemokine gene signature identifies lymph node-like structures in melanoma: potential for patient selection for immunotherapy? Sci Rep. 2012;2:765. doi: 10.1038/srep00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Caro G, Marchesi F. Tertiary lymphoid tissue: A gateway for T cells in the tumor microenvironment. Oncoimmunology. 2014;3:e28850. doi: 10.4161/onci.28850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes CE, Benson RA, Bedaj M, Maffia P. Antigen-Presenting Cells and Antigen Presentation in Tertiary Lymphoid Organs. Front Immunol. 2016;7:481. doi: 10.3389/fimmu.2016.00481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding T, Yan F, Cao S, Ren X. Regulatory B cell: New member of immunosuppressive cell club. Hum Immunol. 2015;76(9):615–21. doi: 10.1016/j.humimm.2015.09.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flow cytometry analysis of liver metastasis immune cell populations