Abstract
To synthesize multi-component nanochains, we developed a simple ‘one-pot’ synthesis, which exhibited high yield and consistency. The nanochains particles consist of parent nanospheres chemically linked into a higher-order, chain-like assembly. The one-pot synthesis is based on the addition of two types of parent nanospheres in terms of their surface chemical functionality (e.g., decorated with PEG-NH2 or PEG-COOH). By reacting the two types of parent nanospheres at a specific ratio (~2:1) for a short period of time (~30 min) under rigorous stirring, nanochains were formed. For example, we show the synthesis of iron oxide nanochains with lengths of about 125 nm consisting of 3–5 constituting nanospheres. The chain-like shaped nanoparticle possessed a unique ability to target and rapidly deposit on the endothelium of glioma sites via vascular targeting. To target and image invasive brain tumors, we used iron oxide nanochains with the targeting ligand being the fibronectin-targeting peptide CREKA. Overexpression of fibronectin is strongly associated with the perivascular regions of glioblastoma multiforme and plays a critical role in migrating and invasive glioma cells. In mice with invasive glioma tumors, 3.7% of the injected CREKA-targeted nanochains was found in gliomas within 1 h. Notably, the intratumoral deposition of the nanochain was ~2.6-fold higher than its spherical variant. Using MR imaging, the precise targeting of nanochains to gliomas provided images with the exact topology of the disease including their margin of infiltrating edges and distant invasive sites.
1. Introduction
Nanoparticle technology provides great flexibility in designing nanoparticles for biomedical applications with various sizes, shapes and composition, resulting in highly controllable structure-function relationships. Considering the near-limitless nanoparticle designs and their adjustable in vivo ability to seek and destroy disease, nanoparticles can be optimally ‘tweaked’ in terms of biophysical and biochemical interactions for site-specific targeting of very distinct microenvironments of diseased tissues. In particular, shaping the geometry of nanoparticles offers additional control over the navigation of nanoparticles through different biological processes, such as interactions with cell surfaces and cell-surface receptors.1, 2 Here, we report a new synthetic concept for making multicomponent chain-like nanoparticles (termed nanochains) using a one-pot approach. Using standard nanoparticles (e.g., iron oxide, gold) or combinations of them as starting particles, each nanochain particle is comprised of at least three nanoparticles chemically linked into a linear, chain-like assembly. The one-pot synthesis utilizes two types of parent nanoparticles based on the functional group on the particle’s surface (Fig. 1). For example, particle A is decorated with amine groups (NP-NH2), whereas particle B is decorated with carboxyl groups (NP-COOH). Upon activation of the carboxyl groups on the NP-COOH particles, particles A and B will react with each other forming larger agglomerates. By performing the reaction in an organic solvent (i.e., absence of water), hydrolysis of the activated COOH intermediate is constrained. Thus, the reaction rate and growth of the agglomerate are dictated by mixing (e.g., stirring rate) and concentration of the starting particles. Let’s assume that NP-NH2 and NP-COOH particles are mixed at a ratio of ~2:1. At early time points, two NP-NH2 particles will react with one ‘activated’ NP-COOH forming a short linear nanochain (Fig. 1b). If the reaction is allowed to continue, the particles will continue growing into large agglomerates (Fig. 1c). We have optimized the reaction time and conditions that result in well-defined short nanochains. To illustrate the flexibility of this one-pot synthetic approach to be applied to different classes of nanoparticles (e.g., iron oxide, gold, silica, etc.) and sizes, as first examples, we synthesized two nanochain particles consisting of iron oxide or gold nanoparticles.
Figure 1.
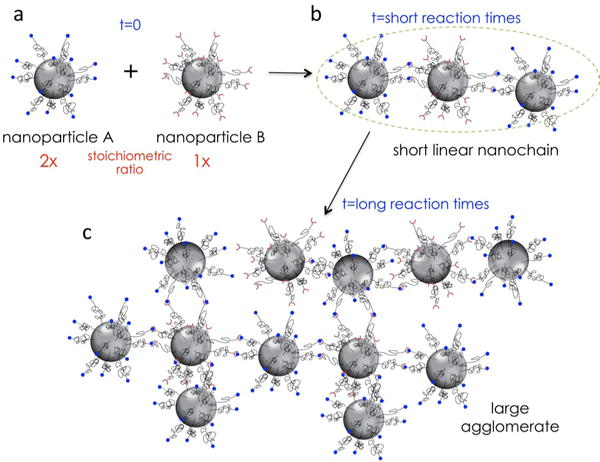
Illustration shows the synthetic concept of short linear nanochain particles.
In particular, we focused on iron oxide nanochains to illustrate the precise targeting of nanochains to hard-to-reach cancers. As a case study, we selected glioblastoma multiforme (GBM), an invasive brain cancer. GBMs are characteristically diffuse and invasive with infiltrating edges and distant glioma cell dispersion.3–5 In addition to the non-localized topology of glioma cells, targeting molecules and nanoparticles into brain tumors is challenging due to the blood-brain barrier (BBB). To evaluate the targeting efficiency of nanochains, we decorated the nanoparticle with a ligand that targets an overexpressed biomarker found on the remodeled endothelium of brain tumors. The remodeled endothelium associated with brain tumors and its surrounding microenvironment offers a diverse set of targetable biomarkers, which differs from that of healthy vascular beds. Overall, our targeting strategy was based on two important design criteria: 1) the size and the multivalent avidity, due to formation of multiple receptor-ligand bonds, makes nanoparticles ideal for targeting of vascular-associated pathologies, and 2) the oblong-shaped nanochain possesses a unique ability to seek and rapidly deposit on the blood vessels of glioma sites via vascular targeting. Using mouse models of GBM and histological analyses, we show that nanochains achieved superior deposition at primary and invasive sites of GBM when compared to their spherical nanoparticle counterparts. Considering the hard-to-reach nature of gliomas, the very high intratumoral levels of iron oxide facilitated MR imaging with precise demarcation of glioma sites including locations with dispersing glioma cells.
2. Experimental section
2.1 Synthesis of parent iron oxide nanospheres
Iron oxide nanoparticles were synthesized by the co-precipitation of Fe(II) and Fe(III) ions in the presence of sodium hydroxide solution.6 The surface of the iron oxide nanoparticles was modified with PEG (2 kDa) using silane-PEG-COOH or silane-PEG-NH2. For more details about the synthesis of the iron oxide nanoparticles, see additional methods in ESI.
2.2 Synthesis of nanochains
First, each mono-functionalized nanoparticles (IONP-COOH and IONP-NH2) were transferred from water to organic phase. The concentrated samples of functionalized nanoparticles were added to dimethylformamide (DMF) and heated to evaporate all water. The concentrations of nanoparticles were then adjusted to 1mg/mL by adding more DMF. To activate carboxyl groups on the IONP-COOH nanoparticles, catalytic amount of pyridine and 50 molar excess N,N′-Dicyclohexylcarbodiimide (DCC) relative to the available COOH groups were added and allowed to react for 30 minutes. To this reaction mixture 2.5 excess IONP-NH2 nanoparticles over the number of IONP-COOH nanoparticles was added and shaken for another 30 minutes. At this time point, the reaction was arrested by ‘deactivating’ the carboxyl groups. To deactivate carboxyl groups, 10 molar excess ethylenediamine (relative to the number of carboxyl groups) was added and shaken for another 30 minutes. Finally, 10 times excess distilled water (relative to the volume of DMF) was added to the above reaction mixture. Nanochain particles were separated by centrifugation with Amicon® Ultra-15 centrifugal filters. To further clean nanochains from any unreacted parent nanoparticles, a strong magnet was used. Briefly, synthesized nanochains at a concentration of 10 mg/mL were transferred to a small test tube and were exposed to a powerful magnetic field for 90 minutes. Nanochain particles attached at the walls of the test tube were then collected and stored at 4 °C.
2.3 Functionalization of nanochains with targeting ligand
The CREKA peptide was conjugated onto the particles via maleimide chemistry. First, amine-functionalized nanochains in PBS were vortexed with 10 molar excess sulfosuccinimidyl 4-(N-maleimidomethyl)cyclohexane-1-carboxylate (sulfo-SMCC) for 15 minutes. Next, CREKA was added at a 5 molar excess over the number of amines on nanochains and allowed to react for 2 hours. The product was dialyzed against PBS using a 100,000 Da MW cut-off membrane to remove unbound peptide.
The number of CREKA per nanochain particle was determined by the measurement of collected free CREKA solution after separation using a desalination column. The concentration of CREKA was determined using a high-performance liquid chromatography (LC) system with an analytical column (250 mm× 4.6 mm, pore size 5 micron, Luna-C18). An isocratic elution technique was used where the mobile phase contained a mixture of solvent A (0.1% trifluoroacetic acid in water) and solvent B (50% acetonitrile solution and 50 % methanol) (A:B 20:80, v/v). The flow rate was set at 1.0 ml/min. The sample injection volume was 50 μL, and the detector wavelength was 220 nm.
2.4 Tumor models
All animal procedures were conducted under a protocol approved by the IACUCs of Case Western Reserve University and Cleveland Clinic. For the CNS-1 rodent glioma tumor model, 5–8-week-old athymic nude mice (~25 g) were housed in the Athymic Animal Core Facility at Case Western Reserve University according to institutional policies. CNS-1 cells were infected with green fluorescent protein (GFP) encoding lentivirus, harvested for intracranial implantation by trypsinization, and concentrated to 1×105 cells/mL in PBS. Mice were anesthetized by intraperitoneal administration of ketamine and xylazine and fitted into a stereotaxic rodent frame. Cells were implanted at AP= +0.5 and ML= −2.0 from bregma at a rate of 1 mL/min in the right striatum at a depth of −3 mm from dura. A total of 200,000 cells were implanted per mouse. Similar procedures were employed for the human T4121 glioma model. For more details about the human GBM specimen and derivative glioma stem cells, see additional methods in ESI.
2.5 Histological evaluation
Immunohistochemistry was performed to evaluate the topology of fibronectin expression with respect to glioma cells and blood vessels. The mice were anesthetized with an IP injection of ketamine/xylazine and transcardially perfused with heparinized PBS followed by 4% paraformaldehyde in PBS. Brains were explanted and post-fixed overnight in 4% paraformaldehyde in PBS. The tissues were soaked in 30% sucrose (w/v) in PBS at 4 °C for cryosectioning. Serial tissue sections of 12 μm in thickness were obtained. Direct fluorescence of GFP (green) imaging was performed for imaging the location of glioma cells. To visualize the tumor microvasculature and fibronectin, the tissue slices were immunohistochemically stained for the endothelial antigen CD31 or anti-fibronectin primary antibody (BD Biosciences, Pharmingen). The tissues were also stained with the nuclear stain DAPI. The tissue sections were imaged at 5, 10 or 40× on the Zeiss Axio Observer Z1 motorized FL inverted microscope. To obtain an image of the entire large tissue section (i.e., entire brain section), a montage of each section was made using the automated tiling function of the microscope.
2.6 MR Imaging
MR images were acquired on a 7 T Bruker MRI system. A volume coil (3.5 cm inner diameter) was employed. The sequence used was a Rapid Acquisition with Relaxation Enhancement (RARE). High-resolution images were obtained before and 60 min after IV injection of the nanochains (at a dose of 10–20 mg Fe/kg b.w.) using a T2-weighted RARE sequence with the following parameters: TR/TE = 3,646.6/31 ms, matrix = 256 × 256, FOV = 3 × 3 cm, and 5 averages. The acquisition time was 9 minutes 43 seconds. This resulted in an in-plane spatial resolution of 111.7 μm and a slice thickness of 0.5 mm.
2.7 3D Cryoimaging
After the entire brain was harvested, they were preserved in 4% paraformaldehyde, soaked in 30% sucrose in PBS, and frozen in OCT. The BioInVision CryoViz was used to section through each brain in slices of 25-μm thickness, while bright-field and fluorescence images of each tissue section were collected at 1.25× magnification. Using CryoViz software, a 3D visualization was created for the bright-field organ view as well as a corresponding 3D visualization of the distribution of GFP glioma cells in the brain. Subsampling in 3D reconstruction created the final images with a voxel size of 10.418 × 10.418 × 50 μm. To reflect visually distinguishable signal, a maximum intensity projection of the green signal in the fluorescent acquisition image was obtained and thresholded. This image was overlaid with the 3D bright-field reconstruction of the brain.
2.8 Statistical analysis
Means were determined for each variable in this study and the resulting values from each experiment were subjected to one-way analysis of variance with post hoc Bonferroni test (SPSS 15, Chicago, IL). A P value of less than 0.05 was used to confirm significant differences. Normality of each data set was confirmed using the Anderson-Darling test.
3. Results and discussion
3.1 Fabrication of nanochains
The synthetic process is shown in Fig. 2 using iron oxide nanoparticles (IONP) as the parent particles. First, the IONP-COOH and IONP-NH2 were transferred from water to organic phase. The particle concentration for both IONP suspensions was then adjusted to 1 mg/mL. In the nanochain formation reaction, DCC acted as the activating agent of the carboxylic acid groups on the surface of IONP-COOH. The IONP suspension was added to dimethylformamide (DMF) and heated to evaporate water before adding DCC, because the activation efficiency of DCC is very high in anhydrous solutions. After 30 minutes, IONP-NH2 particles were added at a particle ratio of 2.5:1 (IONP-NH2:IONP-COOH) under rigorous stirring. During the nanochain formation reaction, the activated leaving groups on IONP-COOH are displaced by terminal amines of IONP-NH2 nanoparticles. The reaction was allowed for 30 min. To arrest the reaction, the rest of the leaving groups are displaced by ethylenediamine. Following cleaning steps with water, the nanochains were separated from unreacted parent IONP using magnetic separation for 90 min.
Figure 2.
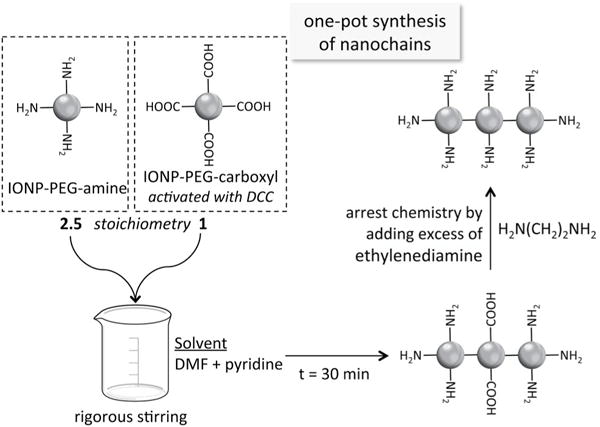
Reaction scheme of the synthesis of iron oxide nanochains
As shown in Fig. 3a, DLS measurements show the hydrodynamic sizes of the parent IONP and the final nanochain. It can be seen that the distributions of the parent particles disappear and a new peak appears, representing the population of nanochain particles. Considering the size distributions of the starting particles, the nanochain’s size indicates chain-like particles consisting of 3–5 IONP members. TEM images confirm the structure and number of constituting IONP in the nanochains (Fig. 3b). Further, the yield of the synthetic process is very high and consistent. Analysis of TEM images indicates that the majority of the parent IONP particles are incorporated in chain-like structures. Notably, the size of the nanochains remained unchanged over 30 weeks (Fig. 3c). Finally, T2 relaxometry measured the T2 relaxivity of the nanochains to be 103.2 s−1 mM−1. Table 1 summarizes some of the characteristics of the particles.
Figure 3.
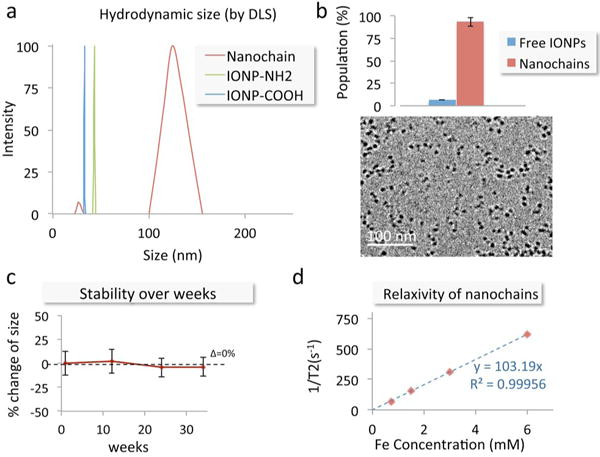
Characterization of the nanochain particles. (a) The size of the parent iron oxide nanoparticles and nanochains was measured using dynamic light scattering (DLS). (b) TEM images of nanochain particles are shown. The number of parent IONP being free or incorporated in nanochain was obtained from visual analysis of TEM images (minimum count was 400 particles; data presented as mean ± standard deviation). (c) The size of the nanochains was measured for many weeks using DLS. The suspension was kept in PBS at room temperature. (d) The transverse (R2) relaxation rate of the nanochains was measured at 1.4 Tesla using a relaxometer.
Table 1.
Summary of nanoparticle characteristics
| Hydrodynamic size (mean ± s.d.) | Number of functional groups | Fe content per 1 mg | |
|---|---|---|---|
| IONP-NH2 | 43.4 ± 2.3 nm | 200 (amines) | 52 % |
| IONP-COOH | 33.2 ± 3.3 nm | 500 (carboxyls) | 43 % |
| Nanochain | 121.1 ± 33.4 nm | 700 (amines) | 51 % |
In addition to iron oxide nanochains, our synthetic method offers great flexibility to employ other types of starting nanoparticles composed of different material. As an example, we describe the synthesis of gold nanochains in the Supporting Information (see ESI Fig. S1).
In conclusion, two mono-functionalized nanoparticles (i.e., amine and carboxyl) were mixed at a specific stoichiometry and allowed to react for a well-defined finite time. The formed nanochains were then separated by centrifugation and magnetic separation. Other synthetic approaches have resulted in nanochain particles.7–9 For example, we previously reported the synthesis and biomedical applications of chain-like nanoparticles using an elegant but multifaceted method.10–15 To assemble nanochains, the previous method was based on a stepwise solid-phase chemistry approach. Briefly, nanospheres were introduced in the resin in a step-by-step manner followed by multiple washing cycles for each step. The new one-pot method is significantly simpler and highly efficient (yield > 90%).
3.2 Histological evaluation of the vascular target
Nanoparticles are capable of targeting brain tumors via highly specific vascular targeting of the vascular bed associated with the primary tumor mass and its invasive sites.11, 16 Notably, the remodeled endothelium associated with brain tumors and its surrounding microenvironment offers a diverse set of targetable biomarkers. For example, vascular targets overexpressed in gliomas include αvβ3 integrin,17–20 platelet-selectin (P-selectin),21 vascular endothelial growth factor receptors (VEGFR) and fibronectin.22–25 In particular, we were interested in fibronectin due to its role in migration of glioma cells. Most importantly, overexpression of fibronectin is strongly associated with the perivascular regions of GMB tumors.26 In addition to its selective perivascular expression, fibronectin is abundant and plays a critical role in migrating and invasive glioma cells. Considering its insignificant expression on the endothelium of normal tissues, fibronectin is an ideal fit to our vascular targeting scheme.
To confirm the availability of fibronectin as a vascular target, we performed histological analysis using the orthotopic CNS-1 glioma model in mice. In terms of pathological and genetic similarities to the human disease,27, 28 the CNS-1 model is one of the few rodent models that recapitulate the microenvironment of the human disease and displays several histological features and diffuse growth and invasive pattern similar to human GBM. In addition to their ability to express several glioma markers, a three-dimensional cryo-imaging technique showed that the rodent CNS-1 glioma cells is a valid system to study the highly dispersive nature of glioma tumor cells along blood vessels and white matter tracts in vivo.29
Animals were euthanized 8 days after orthotopic inoculation of CNS-1 cells, which were infected with GFP encoding lentivirus. The brains were collected for histological analysis of brain tumor location (CNS-1-GFP cells). Using fluorescence microscopy, images of entire histological sections of the organs were obtained at a low magnification (5×) using the automated tiling function of the microscope. Fig. 4 shows a representative image of the entire left hemisphere of a brain displaying the location of the primary tumor and presence of clusters of invasive glioma cells dispersed in the brain parenchyma. Notably, abundance of fibronectin was seen in the primary tumor (Fig. 4b) as well as invasive sites (Fig. 4c). Further, we assessed the location of glioma cells with respect to blood vessels and the associated presence of fibronectin. Blood vessels associated with the primary and invasive glioma sites presented a remodeled endothelium as indicated by the abundance of fibronectin (Fig. 4c). It should be noted that no fibronectin can be seen in healthy brain tissues, which indicates that it is a highly selective vascular target associated with GBM sites.
Figure 4.
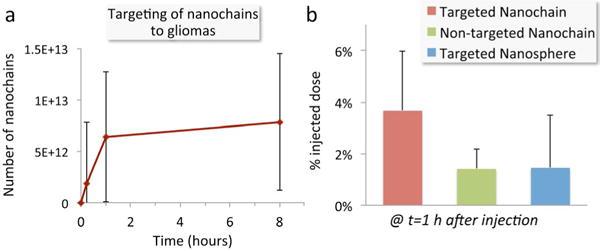
Evaluation of the ability of the fibronectin-targeting nanochains to deposit in brain tumors. (a) Quantification of the time course of accumulation of CREKA-targeted nanochains in the brain of mice bearing orthotopic glioma CNS-1 tumors. The animals were euthanized at 0, 1 and 8 hours after injection (n=5 mice per time point). Brains were collected and the concentration of the nanochains in tissues was quantified by direct measurement of iron using ICP-OES. Tumor-bearing mice injected with saline were used for correction of the background levels of iron in the tumor tissue. (b) The deposition of CREKA-targeted nanochains was compared to their non-targeted variant and CREKA-targeted nanospheres based on the parent IONP (n=5 mice per condition). All formulations were administered at a dose containing an equal number of particles.
3.3 Quantitative evaluation of targeting brain tumors
To target the glioma-associated endothelium, we employed the fibronectin-targeting peptide CREKA, which has been shown to specifically bind fibrin–fibronectin complexes in tumors with very high specificity.9, 30–33 The CREKA peptide was conjugated onto the available amines on the surface of the nanochains using the heterobifunctional crosslinker sulfo-SMCC. An HPLC assay was used to quantify the number of CREKA peptides per nanochain particle, confirming that all the 700 amines available for conjugation were consumed.
Animals were systemically injected with nanochains 8 days after orthotopic inoculation of the CNS-1 cells. Each dose contained 4–10 mg of iron per kg of body weight, which corresponded to 1.6 × 1014 nanochain particles being administered to each animal. The concentration of the nanochains in tissues was quantified by direct measurement of iron ex vivo. To determine the time course of the intratumoral deposition of CREKA-targeted nanochains, animals were euthanized at 0, 1 and 8 hours post-injection (n=5 mice per time point). Whole brains were perfused and tissues were homogenized. The iron concentration in the homogenate was directly measured ex vivo using ICP-OES. Tumor-bearing mice injected with saline were used for correction of the background levels of iron in the tumor tissue. Fig. 5a shows the intratumoral deposition of nanochains reaches very high levels quickly within 1 h after injection. In fact, vascular targeting of the nanochains is rapid as the later time point indicates (t=8 h). The number of nanoparticles that successfully targeted glioma sites corresponded to a significant portion of the injected dose (3.7% as shown in Fig. 5b). Using the same methodology, we quantitatively compared the CREKA-targeted nanochains to their non-targeted variant and a CREKA-targeted nanosphere based on the parent IONP. All formulations were administered at a dose containing an equal number of particles per kilogram of body weight. At t = 1 h after injection, the tumor deposition of the targeted nanochain was ~2.6-fold higher than its nontargeted variant and the targeted nanosphere. This indicates that the oblong shape and flexibility of the chain-like nanoparticle enhanced its vascular targeting capabilities.
Figure 5.
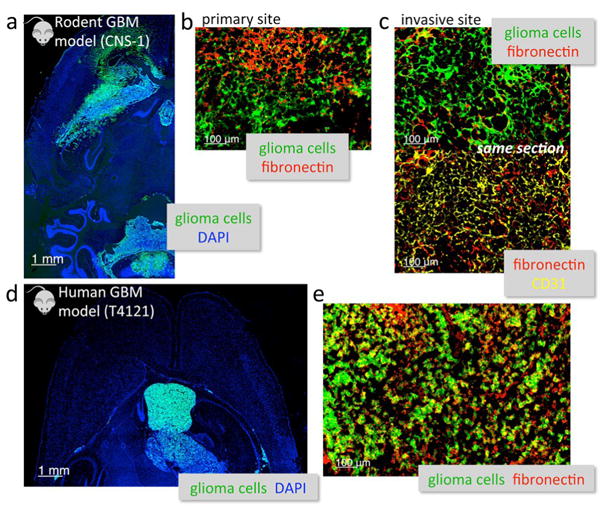
Histological analysis of the degree and topology of fibronectin expression in two glioma models in mice. (a) Fluorescence imaging of the entire left hemisphere of the brain shows the primary tumor and its invasive sites (5× magnification; green: CNS-1 glioma cells (GFP); blue: nuclear stain (DAPI)). (b) The abundance of fibronectin (red) is shown in the primary CNS-1 tumor (20× magnification). (c) High magnification imaging (20×) of an invasive site shows the location of fibronectin (red) with respect to the location of endothelial cells (yellow: CD31) and brain tumor cells in the same histologic section. (d) Fluorescence imaging of an entire histologic section of the brain shows the primary tumor and its invasive sites (5× magnification; green: T4121 glioma cells (GFP)). (e) High magnification imaging (20×) shows the overexpression of fibronectin (red) in the T4121 GBM.
Typically, brain tumors are nearly inaccessible to most molecules due to poor penetration across the blood-tumor barrier (BTB).34–36 Notably, nanoparticles have shown promise, because they can “squeeze” into intracranial tumors through their leaky vasculature due to the enhanced permeation and retention (EPR) effect.37, 38 For example, it was demonstrated in patients with GBM that long circulating liposomal nanoparticles could penetrate the BTB.37 Furthermore, contrast enhancement of GBMs in CT and MR imaging has generated controversy as to whether the BTB barrier presents significant limitations to drug delivery. Many studies have shown that while the blood-brain barrier (BBB) is partially breached resulting in an increased penetration of drugs into brain tumors compared to normal brain, BBB is still present in GBMs, which consist of blood vessels that are not as leaky as the angiogenic vessels observed in other cancer types.11, 16, 39–42 This results in low penetration of nanoparticles into the brain tumor interstitium with a patchy, near-perivascular distribution, resulting in failure to reach the majority of the primary tumor mass and especially its invasive sites.38, 43 Furthermore, the EPR effect is typically noticeable at the primary regions of GBM, while it is attenuated at the invasive sites of brain tumors with dispersing cancer cells. This is due to the fact that the BBB of invasive sites has a very high likelihood to remain intact. Most importantly, it is not uncommon to find dispersing brain tumor cells as far away as 4 cm from the primary site.44
Contrary to EPR-driven passive targeting or deep-tissue active targeting, vascular targeting of nanoparticles to GBMs is a very attractive strategy. Fibronectin and fibrinogen are not innate brain matrix proteins. However, the endothelium and perivascular regions of tumors contain high levels of products of blood clotting. Fibrin based proteins typically arise due to systemic intervention upon injury to the endothelium. A fibrin meshwork is initially formed by conversion of fibrinogen, which leaks into tumor via its leaky endothelium. Plasma fibronectin becomes covalently linked or otherwise bound to the fibrin meshwork. These fibrin–fibronectin complexes on the tumor endothelium are targetable by CREKA-decorated nanoparticles.30, 33 Further, fibronectin plays a key role in the migration of glioma cells.26 Our histological analysis confirmed the abundance of fibronectin in two GBM models in mice. Thus, the remodelled endothelium associated with glioma cells served as an ideal targetable site of the disease, which accurately reflected the changes that occurred behind the vascular bed in the interstitium.
In this context, due to its size, one nanoparticle can be decorated with a high number of targeting ligands, which results in the formation of multiple receptor-ligand bonds. This geometrically enhanced multivalent avidity makes nanoparticles ideal for targeting vascular biomarkers.29, 41, 42 Besides blood components, the endothelium is the closest point-of-contact for blood circulating nanoparticles. Thus, by having direct access to the vascular bed, nanoparticles continuously scavenge the endothelium for the vascular biomarkers associated to brain tumors. An additional benefit of nanoparticle technology is that adjusting the shape can significantly dictate targeting avidity. Not surprisingly, the oblong shape and flexibility of the nanochains resulted in superior deposition at glioma sites when compared to their spherical counterparts. We have previously seen that chain-like nanoparticles exhibit similar organ distribution to their spherical counterparts and end up in organs of the reticuloendothelial system.15 The safety profile of the nanochain particles is part of ongoing short and long-term toxicity studies.
3.4 Imaging of invasive brain tumors using MRI
To explore the in vivo performance of the new nanochains, we explored the ability of iron oxide nanochains to target the hard-to-reach invasive brain tumors for imaging and diagnostic applications. Treatment options for GBM typically involve surgery and radiation, with the addition of Temozolomide based chemotherapy for glioma patients. Despite advancements, the recurrence of glioma is >90%. This stems from the invasive nature and blood-brain barrier (BBB) limiting the effectiveness of surgery and systemic chemotherapy, respectively.4, 5 Thus, radiation is an essential part of the tumor treatment protocol. However, a major limitation for successful radiotherapy is the lack of accurate image guidance and exact topology of the disease.
Fig. 6 shows representative coronal T2-weighted images of mice with orthotopic CNS-1 tumors (n=5) obtained using a 7 T MRI before and after administration of the CREKA-targeting nanochains (at a dose of 20 mg Fe/kg b.w.). This is a typical dose of IO nanoparticles used in imaging small animals with MRI (e.g., 10 mg Fe/kg).45 MR images were acquired a few minutes prior to injection of the agent and 60 min after injection. The MR parameters in the pre- and post-injection images were identical. Fig. 6a compares the pre-injection and 1 h post-injection images of the brain of the same animal. Due to effective vascular targeting of the nanochains, a significant negative contrast was observed in the post-injection image highlighting the periphery and the core of the primary site. Most importantly, infiltrating edges and invasive sites were also clearly marked (Fig. 6b, left panel). To confirm the accuracy of the in vivo MR imaging, terminal analysis was employed. At the end of the last in vivo imaging session, the animal was euthanized and the brain was perfused, collected and prepared for 3D cryo-imaging. Serial cryo-sections of entire brains were obtained using 3D reconstructions, providing ultra-high-resolution fluorescence images and the location of primary and invasive glioma sites (i.e., CNS-1-GFP). The in vivo MR image and the ex vivo fluorescence image (Fig. 6b, right panel) confirm the targeting accuracy of the nanochain as indicated by the colocalization of MR signal and glioma cells. The three-dimensional volume of the entire brain from 3D cryo-imaging is shown in ESI Fig. S2.
Figure 6.
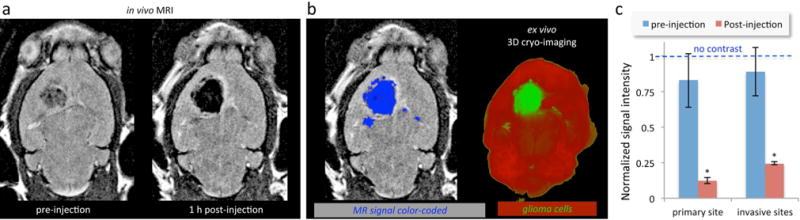
Representative in vivo MR images of the brain of mice bearing orthotopic glioma CNS-1 tumors using a 7 T MRI. (a) Coronal T2-weighted images of the brain of a mouse before and 1 h after injection of CREKA-targeted nanochains. (b) In the 1 h post-injection MR image, the signal enhancement was thresholded and color-coded in blue (left). In the end of MR imaging, the brains of the animals were perfused, excised, and imaged ex vivo using 3D cryo-imaging. 3D cryo-imaging provided an ultra-high-resolution fluorescence volume of the brain showing the topology of CNS-1-GFP cells. The in vivo MR image (left) and the ex vivo fluorescence image (right) show the colocalization of MR signal and glioma cells. (c) The absolute MR signal intensity in gliomas and the healthy brain was measured in manually drawn ROIs. The signal intensity in glioma sites was normalized to the signal of the corresponding healthy brain region (scale: 0–1). Since lower values indicate greater contrast in T2 images, normalized intensity values of 0 and 1 correspond to maximum and minimum contrast, respectively (data presented as mean ± standard deviation; n = 5; *P < 0.05).
The ability of the nanochains to target invasive GBM was evaluating by quantitatively analyzing the MR signal intensity (Fig. 6c). The absolute MR signal intensity in primary and invasive glioma sites and the corresponding healthy brain tissues was measured using manually drawn regions of interest. By normalizing the tumor signal to that of corresponding healthy tissues, the normalized signal intensity had a scale of 0–1 with lower values indicating greater contrast in T2 images. A normalized intensity value of 1 corresponds to no contrast compared to healthy tissue. The pre-injection image exhibited values for both the primary and invasive glioma sites that were fairly similar and statistically close to 1. However, precise targeting of the nanochain to glioma sites generated a significantly high negative contrast. In the post-injection images, the primary and invasive glioma sites exhibited a normalized intensity value of 0.12 and 0.24 respectively, indicating significantly higher contrast compared to the post-injection background signal of the healthy or uninvolved regions of the brain.
4. Conclusions
To synthesize multi-component nanochains, we developed a new and simple ‘one-pot’ synthesis, which exhibited high yield, consistency and scalability. The shape and size of the nanochains facilitated the precise targeting of nanochains to GBM sites, which can provide MR images with the exact topology of the disease including their margin of infiltrating edges and distant invasive sites. By accurately defining the tumor borders and distant glioma cell dispersion, a more effective image guidance can lead to successful radiotherapy and improved patient outcomes.
Supplementary Material
Acknowledgments
This work was partially supported by grants from the National Cancer Institute (R01CA177716, U01CA198892), the Prayers from Maria Children’s Glioma Foundation and the Angie Fowler AYA Cancer Research Initiative of the Case Comprehensive Cancer Center (E.K.). G.C. was supported by a fellowship from the NIH Interdisciplinary Biomedical Imaging Training Program (T32EB007509) administered by the Department of Biomedical Engineering, Case Western Reserve University.
Footnotes
Electronic Supplementary Information (ESI) available. See DOI: 10.1039/x0xx00000x
References
- 1.Toy R, Hayden E, Shoup C, Baskaran H, Karathanasis E. Nanotechnology. 2011;22:115101. doi: 10.1088/0957-4484/22/11/115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Toy R, Peiris PM, Ghaghada KB, Karathanasis E. Nanomedicine (Lond) 2014;9:121–134. doi: 10.2217/nnm.13.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black KL, Pikul BK. Clinical neurosurgery. 1999;45:160–163. [PubMed] [Google Scholar]
- 4.Juratli TA, Schackert G, Krex D. Pharmacol Ther. 2013;139:341–358. doi: 10.1016/j.pharmthera.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 5.Adamson C, Kanu OO, Mehta AI, Di C, Lin N, Mattox AK, Bigner DD. Expert opinion on investigational drugs. 2009;18:1061–1083. doi: 10.1517/13543780903052764. [DOI] [PubMed] [Google Scholar]
- 6.Kang YS, Risbud S, Rabolt JF, Stroeve P. Chem Mater. 1996;8:2209–2211. [Google Scholar]
- 7.Huang X, El-Sayed IH, Qian W, El-Sayed MA. JACS. 2006;128:2215–2120. [Google Scholar]
- 8.Sardar R, Shumaker-Parry JS. Nano Lett. 2008;8:731–736. doi: 10.1021/nl073154m. [DOI] [PubMed] [Google Scholar]
- 9.Park JH, von Maltzahn G, Zhang L, Derfus AM, Simberg D, Harris TJ, Ruoslahti E, Bhatia SN, Sailor MJ. Small. 2009;5:694–700. doi: 10.1002/smll.200801789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peiris PM, Schmidt E, Calabrese M, Karathanasis E. PLoS One. 2011;6:e15927. doi: 10.1371/journal.pone.0015927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peiris PM, Abramowski A, Mcginnity J, Doolittle E, Toy R, Gopalakrishnan R, Shah S, Bauer L, Ghaghada KB, Hoimes C, Brady-Kalnay SM, Basilion JP, Griswold MA, Karathanasis E. Cancer Research. 2015;75:1356–1365. doi: 10.1158/0008-5472.CAN-14-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peiris PM, Bauer L, Toy R, Tran E, Pansky J, Doolittle E, Schmidt E, Hayden E, Mayer A, Keri RA, Griswold MA, Karathanasis E. ACS Nano. 2012;6:4157–4168. doi: 10.1021/nn300652p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peiris PM, Tam M, Vicente P, Abramowski A, Toy R, Bauer L, Mayer A, Pansky J, Doolittle E, Tucci S, Schmidt E, Shoup C, Rao S, Murray K, Gopalakrishnan R, Keri RA, Basilion JP, Griswold MA, Karathanasis E. Pharm Res. 2014;31:1460–1468. doi: 10.1007/s11095-013-1102-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peiris PM, Toy R, Abramowski A, Vicente P, Tucci S, Bauer L, Mayer A, Tam M, Doolittle E, Pansky J, Tran E, Lin D, Schiemann WP, Ghaghada KB, Griswold MA, Karathanasis E. J Control Release. 2014;173:51–58. doi: 10.1016/j.jconrel.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peiris PM, Toy R, Doolittle E, Pansky J, Abramowski A, Tam M, Vicente P, Tran E, Hayden E, Camann A, Mayer A, Erokwu BO, Berman Z, Wilson D, Baskaran H, Flask CA, Keri RA, Karathanasis E. ACS Nano. 2012;6:8783–8795. doi: 10.1021/nn303833p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karathanasis E, Ghaghada KB. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2016;8:678–695. doi: 10.1002/wnan.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schnell O, Krebs B, Carlsen J, Miederer I, Goetz C, Goldbrunner RH, Wester HJ, Haubner R, Popperl G, Holtmannspotter M, Kretzschmar HA, Kessler H, Tonn JC, Schwaiger M, Beer AJ. Neuro-oncology. 2009;11:861–870. doi: 10.1215/15228517-2009-024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reardon DA, Nabors LB, Stupp R, Mikkelsen T. Expert opinion on investigational drugs. 2008;17:1225–1235. doi: 10.1517/13543784.17.8.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Danhier F, Vroman B, Lecouturier N, Crokart N, Pourcelle V, Freichels H, Jerome C, Marchand-Brynaert J, Feron O, Preat V. J Control Release. 2009;140:166–173. doi: 10.1016/j.jconrel.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 20.Reddy GR, Bhojani MS, McConville P, Moody J, Moffat BA, Hall DE, Kim G, Koo YE, Woolliscroft MJ, Sugai JV, Johnson TD, Philbert MA, Kopelman R, Rehemtulla A, Ross BD. Clin Cancer Res. 2006;12:6677–6686. doi: 10.1158/1078-0432.CCR-06-0946. [DOI] [PubMed] [Google Scholar]
- 21.Shamay Y, Elkabets M, Li H, Shah J, Brook S, Wang F, Adler K, Baut E, Scaltriti M, Jena PV, Gardner EE, Poirier JT, Rudin CM, Baselga J, Haimovitz-Friedman A, Heller DA. Science translational medicine. 2016;8:345ra387. doi: 10.1126/scitranslmed.aaf7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Serres E, Debarbieux F, Stanchi F, Maggiorella L, Grall D, Turchi L, Burel-Vandenbos F, Figarella-Branger D, Virolle T, Rougon G, Van Obberghen-Schilling E. Oncogene. 2014;33:3451–3462. doi: 10.1038/onc.2013.305. [DOI] [PubMed] [Google Scholar]
- 23.Ohnishi T, Hiraga S, Izumoto S, Matsumura H, Kanemura Y, Arita N, Hayakawa T. Clin Exp Metastasis. 1998;16:729–741. doi: 10.1023/a:1006532812408. [DOI] [PubMed] [Google Scholar]
- 24.Neri D, Bicknell R. Nat Rev Cancer. 2005;5:436–446. doi: 10.1038/nrc1627. [DOI] [PubMed] [Google Scholar]
- 25.Borsi L, Balza E, Bestagno M, Castellani P, Carnemolla B, Biro A, Leprini A, Sepulveda J, Burrone O, Neri D, Zardi L. Int J Cancer. 2002;102:75–85. doi: 10.1002/ijc.10662. [DOI] [PubMed] [Google Scholar]
- 26.Castellani P, Borsi L, Carnemolla B, Biro A, Dorcaratto A, Viale GL, Neri D, Zardi L. Am J Pathol. 2002;161:1695–1700. doi: 10.1016/S0002-9440(10)64446-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barth RF, Kaur B. J Neurooncol. 2009;94:299–312. doi: 10.1007/s11060-009-9875-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacobs VL, Valdes PA, Hickey WF, De Leo JA. ASN neuro. 2011;3:e00063. doi: 10.1042/AN20110014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burden-Gulley SM, Qutaish MQ, Sullivant KE, Lu H, Wang J, Craig SE, Basilion JP, Wilson DL, Brady-Kalnay SM. Cancer Res. 2011;71:5932–5940. doi: 10.1158/0008-5472.CAN-11-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simberg D, Duza T, Park JH, Essler M, Pilch J, Zhang L, Derfus AM, Yang M, Hoffman RM, Bhatia S, Sailor MJ, Ruoslahti E. Proc Natl Acad Sci U S A. 2007;104:932–936. doi: 10.1073/pnas.0610298104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chung EJ, Cheng Y, Morshed R, Nord K, Han Y, Wegscheid ML, Auffinger B, Wainwright DA, Lesniak MS, Tirrell MV. Biomaterials. 2014;35:1249–1256. doi: 10.1016/j.biomaterials.2013.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Z, Qutaish M, Han Z, Schur RM, Liu Y, Wilson DL, Lu ZR. Nat Commun. 2015;6:7984. doi: 10.1038/ncomms8984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pilch J, Brown DM, Komatsu M, Jarvinen TA, Yang M, Peters D, Hoffman RM, Ruoslahti E. Proc Natl Acad Sci U S A. 2006;103:2800–2804. doi: 10.1073/pnas.0511219103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohnishi T, Tamai I, Sakanaka K, Sakata A, Yamashima T, Yamashita J, Tsuji A. Biochemical pharmacology. 1995;49:1541–1544. doi: 10.1016/0006-2952(95)00082-b. [DOI] [PubMed] [Google Scholar]
- 35.Mankhetkorn S, Dubru F, Hesschenbrouck J, Fiallo M, Garnier-Suillerot A. Molecular pharmacology. 1996;49:532–539. [PubMed] [Google Scholar]
- 36.Rousselle C, Clair P, Lefauconnier JM, Kaczorek M, Scherrmann JM, Temsamani J. Molecular pharmacology. 2000;57:679–686. doi: 10.1124/mol.57.4.679. [DOI] [PubMed] [Google Scholar]
- 37.Koukourakis MI, Koukouraki IFS, Kelekis N, Kyrias SAG, Karkavitsas N. Br J Cancer. 2000;83:1281–1286. doi: 10.1054/bjoc.2000.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karathanasis E, Park J, Agarwal A, Patel V, Zhao F, Annapragada AV, Hu X, Bellamkonda RV. Nanotechnology. 2008;19:315101. doi: 10.1088/0957-4484/19/31/315101. [DOI] [PubMed] [Google Scholar]
- 39.Lockman PR, Mittapalli RK, Taskar KS, Rudraraju V, Gril B, Bohn KA, Adkins CE, Roberts A, Thorsheim HR, Gaasch JA, Huang S, Palmieri D, Steeg PS, Smith QR. Clin Cancer Res. 2010;16:5664–5678. doi: 10.1158/1078-0432.CCR-10-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boyle FM, Eller SL, Grossman SA. Neuro-oncology. 2004;6:300–305. doi: 10.1215/S1152851703000516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blanchette M, Fortin D. Methods in molecular biology. 2011;686:447–463. doi: 10.1007/978-1-60761-938-3_23. [DOI] [PubMed] [Google Scholar]
- 42.Sato S, Kawase T, Harada S, Takayama H, Suga S. Acta neurochirurgica. 1998;140:1135–1141. doi: 10.1007/s007010050227. disc 1141–1132. [DOI] [PubMed] [Google Scholar]
- 43.Baumann BC, Kao GD, Mahmud A, Harada T, Swift J, Chapman C, Xu X, Discher DE, Dorsey JF. Oncotarget. 2013;4:64–79. doi: 10.18632/oncotarget.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iacob G, Dinca EB. Journal of medicine and life. 2009;2:386–393. [PMC free article] [PubMed] [Google Scholar]
- 45.Crayton SH, Tsourkas A. ACS Nano. 2011;5:9592–9601. doi: 10.1021/nn202863x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


