Abstract
Claudin proteins are components of epithelial tight junctions; a subtype of breast cancer has been defined by the reduced expression of mRNA for claudins and other genes. Here, we characterize the expression of glycoproteins in breast cell lines for the claudin-low subtype using liquid chromatography/tandem mass spectrometry. Unsupervised clustering techniques reveal a group of claudin-low cell lines that is distinct from nonmalignant, basal, and luminal lines. The claudin-low cell lines express F11R, EPCAM, and other proteins at very low levels, whereas CD44 is expressed at a high level. Comparison of mRNA expression to glycoprotein expression shows modest correlation; the best agreement occurs when the mRNA expression level is lowest and little or no protein is detected. These findings from cell lines are compared to those for tumor samples by the Clinical Proteomic Tumor Analysis Consortium (CPTAC). The CPTAC samples contain a group low in CLDN3. The samples low in CLDN3 proteins share many differentially expressed glycoproteins with the claudin-low cell lines. In contrast to the situation for cell lines or patient samples classified as claudin-low by RNA expression, however, most of the tumor samples low in CLDN3 protein express the estrogen receptor or HER2. These tumor samples express CD44 protein at low rather than high levels. There is no correlation between CLDN3 gene expression and protein expression in these CPTAC samples; hence, the claudin-low subtype defined by gene expression is not the same group of tumors as that defined by low expression of CLDN3 protein.
Keywords: breast cancer, glycoprotein, claudin, mass spectrometry, supervised classification, cell lines
Graphical abstract
INTRODUCTION
Analysis of microarray data has led to a classification system for breast tumors that is based on gene expression rather than on clinical features. A pioneering analysis of mRNA expression data described four intrinsic subtypes of breast tumor: HER2-enriched, a normal-like class, and tumors thought to derive from either the basal or luminal cells of the normal epithelium.1 Luminal tumors often express estrogen receptor (ER). Subsequent research subdivided luminal tumors into two groups, A and B, and demonstrated that the five different tumor subtypes are associated with different outcomes for patients.2,3 Others have expanded these subtypes by adding genomic data to the RNA expression data4 or have proposed alternative classification schemes based on gene expression data.5
An additional breast tumor class, based on the low expression of mRNA for claudin proteins, has subsequently been recognized.6–10 The claudin-low subtype of breast cancer comprises tumors with low expression of mRNA for claudins 3, 4, and 7. Claudin proteins are components of the tight junctions between epithelial cells. Tight junctions in the lateral surfaces of epithelial cells join neighboring cells, thereby restricting epithelial transport via the paracellular pathway.11 Tight junctions also maintain the polarity of epithelial cells by reducing the diffusion of membrane proteins between their apical and basal surfaces. Abnormal expression of claudin proteins is associated with tumor grade and invasiveness in breast and colorectal cancers.12–15
Claudin-low tumors identified by RNA expression are usually negative for ER, progesterone receptor (PR), and HER2 (ERBB2), thus constituting an important group that in earlier studies was considered to be a subclass of the basal group. Claudin-low tumor cells display evidence of the epithelial-to-mesenchymal transition in their gene expression patterns and share several features with metaplastic breast tumors.16,17
It has also been reported that putative breast tumor stem cells, CD44+/CD24-low cells, are of the claudin-low type.16,17 Under the cancer stem cell hypothesis, tumors replenish their cells continuously from a population of stem cells. If this view is correct, therapeutics that specifically target claudin-low carcinoma cells may be especially effective on tumors.
Secreted proteins and transmembrane proteins with extracellular domains are frequently glycosylated; this group of proteins includes those that participate in the various intercellular junctions and signaling pathways of an epithelium. In this study, we characterized the differences in glycoprotein expression between claudin-low and other breast cell lines using a data set of 26 breast cell lines in which the glycoproteins were identified and quantitated by liquid chromatography/tandem mass spectrometry. Prat et al.9 identified nine breast cancer cell lines as claudin-low: MDAMB157, MDAMB231, MDAMB435, MDAMB436, SUM159PT, SUM1315, BT549, HS578T, and HBL100 (although the origin of one of these lines has been questioned18). There have been previous studies on the N-linked glycoproteome of breast cancer cell lines,19–21 including studies focused on breast cancer progression,22,23 but not on the claudin-low subgroup. Our goals are to characterize the glycoproteome of a set of claudin-low lines, compare them to basal, luminal, and nonmalignant cells, and identify drugs that may be especially effective against these cell lines using a publicly available data set.24 In addition, we evaluate the extent to which findings on cell lines apply to tumor samples, using a breast tumor protein data set also obtained using mass spectrometry (Clinical Proteomic Tumor Analysis Consortium, CPTAC; https://cptac-data-portal.georgetown.edu/cptac/public).25
EXPERIMENTAL SECTION
Twenty-one breast cancer cell lines (BT474, BT549, HCC1143, HCC1395, HCC1428, HCC1954, HCC1937, HCC38, HCC70, HS578T, MB157, MB231, MB436, MB468, MCF7, SKBR3, SUM149PT, SUM185, SUM229, T47D, and ZR751) obtained from Dr. Susan Fisher of UCSF or ATCC (Manassas, VA), two benign breast tumor cell lines (MCF10A, MCF12A), and three normal human mammary epithelial cell lines (HMEC1, 2, 3) obtained from Cell Applications, Inc. (San Diego, CA), totaling twenty-six cell lines, were grown in 10 cm culture dishes with 10 mL of culture medium based on methods published previously.21,26 Cells were grown at 37 °C with 5% CO2, except for MB468, which was grown without CO2. Each sample [12 mL of lysate for LTQ analysis (Thermo Scientific, San Jose, USA) and 2–4 mL of lysate for Q Exactive analysis (Thermo Scientific, Bremen, Germany)] for analysis was prepared from 5 to 15 culture dishes (10 cm). Biological replicates were prepared and analyzed for all 26 cell lines. Four technical replicates of five cell lines, HCC1395, HCC1428, HCC38, MB468 and HMEC3, were analyzed using Q Exactive MS. For the other 21 cell lines, six technical replicates were analyzed as described below. BT549, HS578T, MB157, MB231, and MB436 are classified here as claudin-low (see Results and Discussion).
Glycoproteins were enriched using a method previously described.21 Briefly, periodate-treated cells on the dish were lysed with buffer that contains 1% octyl-β-D-1-thioglucopyranoside and 1% protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO). Cell lysate was homogenized and glycoproteins were enriched using hydrazide magnetic beads (Bioclone, San Diego, CA). (Oxidized ovalbumin was spiked into the samples as an internal standard to estimate hydrazide bead binding.) Glycoproteins bound to the magnetic beads were denatured, reduced, and alkylated prior to overnight tryptic digestion. After digestion, the tryptic peptide fraction was collected and the N-linked glycopeptides bound to the hydrazide magnetic beads were released from the beads with N-glycosidase F. The eluates were processed by solid-phase extraction to concentrate peptides.
Protein Identification by LC–MS/MS Analysis
Tryptic peptides and deglycosylated N-linked peptides derived from each cell lysate were separately analyzed using an extensive set of LC–MS/MS analyses using a LTQ with a Surveyor HPLC system (Thermo Scientific, San Jose, USA) or a Q Exactive MS with a Dionex UtiMate3000 HPLC system (Thermo Scientific, Germering, Germany) to maximize glycoprotein identification and to improve the reproducible detection of glycoproteins.
LTQ MS Analysis
Samples of tryptic and PNGase F released fractions were analyzed using a data-dependent scan procedure (triple playtop 4 CID, CID = 30%) at an MS scan range of m/z 400–1800 and with MS/MS scans of the precursor with an isolation width of 3 m/z and a minimum ion intensity of 800. Four individual aliquots (5 μL/injection) were injected for the LC–MS/MS analyses. The dynamic exclusion (DE) duration for these LC–MS/MS analyses was variably set to 30, 45, 60, or 90s. A maximum of two MS/MS spectra/precursor ion were collected. Gas-phase fractionation was repeated three times for each sample. The full MS scan range was set at either m/z 400–900, m/z 700–1200, or m/z 1000–1800. Analysis via gas-phase fractionation used the data-dependent MS/MS scan procedure as described above except that the DE rule was set for a 60 s duration and only a single MS/MS scan was acquired for each precursor ion. A self-packed C18 column (75 μm × 130 mm, Nucleosil 5 μm C18; Phenomenex, Torrance, USA) was used for the LC–MS/MS analyses. Mobile phase A was 0.1% formic acid/water, and mobile phase B was 0.1% formic acid in acetonitrile. A gradient was used for separation (5–35% B in 65 min, 35–80% B in 10 min, holding at 80% B for 5 min, and back to hold at 5% B). Additionally, a portion of each tryptic digest solution (25 μL, 5× greater injection volume than that used for one-dimensional LC–MS/MS analyses) was injected for the online 2D-LC–MS/MS analyses. The first dimension of the LC used an SCX column (320 μm × 100 mm, 5 μm BioBasic; ThermoFisher) to chromatographically separate the tryptic peptide mixtures into six fractions. The tryptic peptides were eluted from the SCX column using a series of NH4Cl solutions (0, 20, 40, 60, 80, 200 to 400 mM). Each fraction was then desalted in-line with a C18 trap column (300 μm × 5 mm, 5 μm Zorbax C18; Agilent, Germany). After desalting with 95% water/5% ACN, the trapped peptides were analyzed using the same LC–MS/MS method and setup described for the 1D analysis except that the elution time increased from 90 to 120 min.
Q Exactive MS Analysis
The nanoLC setup used for LTQ was used for Q Exactive MS analysis with the following settings: data-dependent acquisition of DE = 10 s at an MS scan range of m/z 400–1800 and MS/MS (HCD = 27 eV) analyses for the 10 most abundant ions. The precursor ion width was set to 3 m/z, and the minimum ion intensity was set to 5 × 104. The charge state screen of precursor ions for MS/MS scans was set to exclude singly charged ions and ions with charge states greater than 7. Resolving power for the Q Exactive was set as 70 000 for the MS scan and 17 500 for the MS/MS scan at m/z 200. The tryptic digest and PNGase released fractions were analyzed separately (5 μL/injection) by LC–MS/MS. The elution methods described for LTQ analyses were used for Q Exactive analyses.
Data Processing Protocol
The same data processing protocol was used for the LTQ and Q Exactive analyses. The Mascot (v2.3) algorithm was used to identify peptides from the resulting MS/MS spectra by searching against the combined human protein database (a total of 22 670 proteins) extracted from SwissProt (v57.14; 2010 February) using taxonomy “homo sapiens”. The sequences for bovine serum albumin and fetuin were included in the search to provide an estimate of the level of protein contamination resulting from fetal bovine serum proteins contained in the cell culture medium. The sequence for chicken ovalbumin was included in the search to estimate glycoprotein recovery. In all analyses, the level of bovine serum proteins was found to be low; thus, the cell washing procedure was determined to be effective. The ovalbumin level for each sample verified that the hydrazide bead binding was similar for all samples. Searching parameters for parent-/fragment-ion mass tolerances were set as 1.6/0.8 Da for LTQ MS and 20 ppm/0.1 Da for Q Exactive MS. Other parameters used were a fixed modification of carbamidomethyl-Cys and variable modifications of deamidation-Asn (or/and Gln), and oxidation-Met. Trypsin was set as the protease with a maximum of two missed cleavages. The Scaffold program (Proteome Software) was used to merge a total of 20 LC–MS/MS files acquired from each LTQ MS analysis (7 runs for the PNGFase F released fraction (deglycosylated peptides) and 13 runs for the tryptic fraction, including 6 runs of 2D-LC–MS/MS) into a single biosample MudPIT file. For the LC–MS/MS data sets acquired using Q Exactive MS, a total of 8 LC–MS/MS files (4 runs for the tryptic fraction and 4 runs for the PNGase F released fraction) were merged into a single biosample MudPIT file. Protein identifications were based on a minimum detection of two peptides with 99% protein identification probability using the algorithm ProteinProphet. Each peptide identified had a minimum peptide identification probability of 95% using the algorithm PeptideProphet. The false positive rate for the peptide identification in this study was less than 1% based on results obtained with decoy database searching. Each biosample MudPIT file derived from an individual cell line generated an Excel file that contained a list of identified proteins and the corresponding total spectrum count. Each Excel file was converted into a two-column text file. The first column of the file contained the UniProt accession number, and the second column contained the total spectrum count. Each text file was input into the ProteinID Finder program (Proteome Solutions). The ProteinID Finder output file contained all of the data derived from the 26 breast cell lines. Thus, the output file contained two sets of data for each cell line, which represent the analyses of biological replicates for each cell line. The ProteinID Finder program output file contains the following information for each protein: UniProt accession, protein name, N-linked site(s), O-linked site(s), and spectral counts. Comparison of mass spectrometric results from biological replicates demonstrates that the cell lines were stable for the duration of cell culture (Figure S1). The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE27 partner repository with the data set identifiers PXD005390, PXD005292, PXD005295, and PXD005339.
To combine samples from the two LTQ and Q Exactive data sets, the data from a cell line studied on both instruments were plotted in a quantile–quantile plot and a line was fit. Using the slope and intercept, an inverse transform was applied to the Q Exactive data, forcing it to have the same center and dispersion as the LTQ data. For five of the cell lines (HCC1395, HCC1428, HCC38, HMEC3, and MB468), there were only Q Exactive data. Spectral counts for these cell lines were normalized to the LTQ data using household proteins. The seven proteins in the LTQ data set with the lowest coefficient of variation were P20645, Q9BT09, P62937, Q16563, Q9BVK6, Q08722, and P07602. The Euclidean length of the means of the spectral counts for these seven glycoproteins over all the LTQ samples was calculated. For each sample with only Q Exactive data, the Euclidean length was calculated for the seven proteins and the ratio of the Q Exactive length to the LTQ length was used to normalize the Q Exactive spectral count data.28 Glycoproteins with fewer than 10 spectral counts when summed over all cell lines were eliminated from the data set analyzed here.
CPTAC Data
Data used in this publication were generated by the Clinical Proteomic Tumor Analysis Consortium (NCI/NIH). The data set TCGA Breast BI Proteome CDAP.r2.itraq was downloaded and processed as follows: (1) duplicate samples from the same patient were removed, (2) six patient samples with no entry for CLDN3 were removed, (3) genes/proteins with empty cells in any of the remaining patients were removed, and (4) shared log ratio data was retained for analysis. The resulting data set has 99 patients and 6990 genes/proteins.
RNA Data
RNA seq data for the glycoproteins are from Daemen et al.24 (Gene Expression Omnibus accession number GSE48216). RNA scores for the nine cell line claudin-low predictor are from Supplemental Data of Prat et al.9 Gene expression microarray data for the CPTAC samples were downloaded from the TCGA portal.
Statistical calculations were carried out in R. For hierarchical clustering analysis, one-half spectral count was added to each value and the base 2 logarithm was taken. Pairwise Euclidean distances and the average linkage method were used to create the dendrogram, with the hclust function (Figure 1). The prcomp function was used for principal components analysis. One-half spectral count was added to the data, and the base 2 logarithm was taken. The glycoprotein variables were centered but not scaled (Figure 2). In Figure 3, decision boundaries were drawn perpendicular to the linear discriminants. The t.test function (var.equal = F) was used to calculate t statistics. Lasso logistic regression and random forest classification were conducted using glmnet and randomForest, respectively.
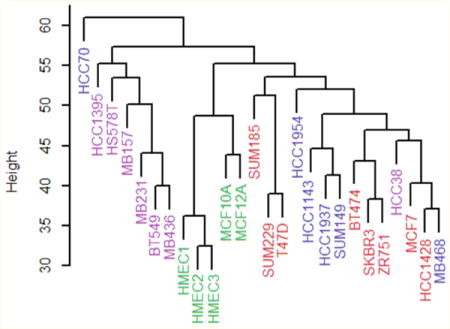
Figure 1.
Hierarchical clustering analysis of the 26 breast cancer cell lines. Color key: nonmalignant, green; claudin-low, violet; basal, blue; luminal, red.
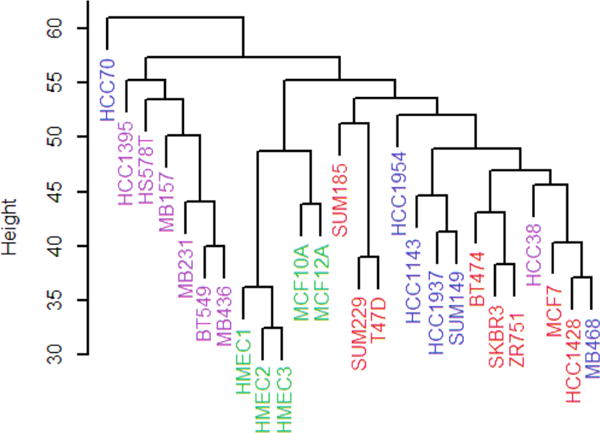
Figure 2.
Principal components analysis of the cell lines. (a) Analysis using all 399 glycoproteins. The decision boundary separates the claudin-low cell lines from the others. (b) Principal components analysis using a random set of 50 glycoproteins. Color key: nonmalignant, green; basal, blue (HCC38 is the triangle, HCC1395 the diamond); claudin-low, violet; luminal, red. The asterisks correspond to cell lines that over-express HER2.
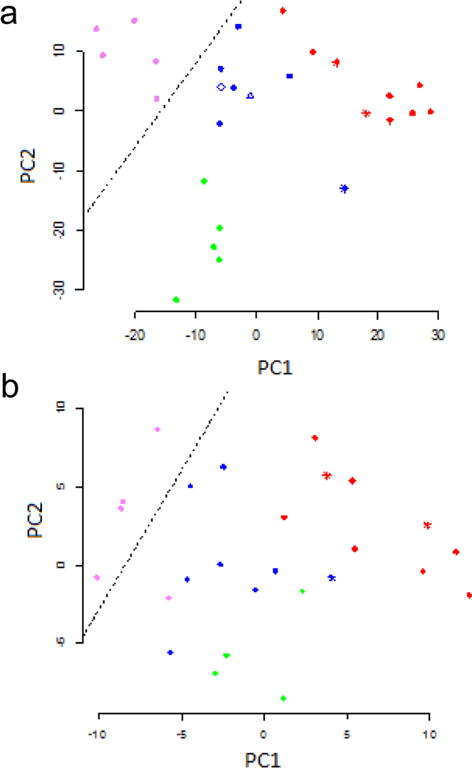
Figure 3.
Frequency distribution for two-sample t statistics. The curve is the sum of two normal distributions with means of −1.63 and 0.82 and standard deviations of 0.77 and 0.61.
RESULTS AND DISCUSSION
LC–MS/MS analysis of tryptic digests prepared from 26 breast cell lines generated a data set that includes 399 glycoproteins (Tables S1 and S2). The cell lines studied included claudin-low, basal, and luminal groups based on array RNA evidence.8,9,29 There were also three samples of normal human mammary epithelial cells (HMEC) grown in primary culture and two from cell lines established from benign tumors (MCF10A, MCF12A). Claudin proteins are not N-linked glycoproteins and are not present in the data set.30
Unsupervised Classification
Hierarchical clustering of the glycoprotein data largely confirms the existence of nonmalignant and claudin-low groups of cell lines that are distinct from the others (Figure 1). The figure shows cell line subtypes as assigned by other laboratories using cluster analysis of mRNA data.8,9,29 All but one (HCC38) of the claudin-low cell lines (violet) cluster together. Similarly, the nonmalignant cell lines form a group (green). The basal (blue) and luminal (red) cell lines, however, do not form clearly defined groups.
Principal components analysis of the glycoprotein expression data also sorts the cell lines into groups similar to those expected from clustering by RNA expression8,9,29 (Figure 2a). Two cell lines, HCC38 and HCC1395, classified as claudin-low by Prat et al.,8 fall clearly within the basal cluster (blue in Figure 2a). Hence, HCC38 and HCC1395 are grouped here with the basal cell lines, leaving BT549, HS578T, MB157, MB231, and MB436 as the claudin-low group. In the principal component analysis, each of the four subtypes, including the basal and luminal ones, can be separated from the others into nonoverlapping subtypes by a line (for these cell lines, basal and luminal are distinct groups), which is not the case with hierarchical clustering.
Hierarchical clustering and principal component analysis both support the existence of a distinct claudin-low class of breast cell lines. This evidence, based on protein expression, does not depend on the measurement of RNA expression levels. There are only a few discrepancies with the earlier classification by gene expression.9,29
In both the hierarchical clustering and the principal component analysis, the HER2-overexpressing cell lines are found in the basal and luminal classes rather than forming a separate class by themselves. This result contrasts with that of Perou et al., who find that HER2-overexpressing tumors form one of the intrinsic subtypes,9 but is in agreement with the classification system proposed by Guedj et al.5
Along the first principal component, the cell lines appear in the following order: claudin-low, basal/nonmalignant, luminal. Assuming that the claudin-low cell lines have stem cell-like characteristics, this sequence is similar to the order of differentiation proposed by Lim et al.31 and Prat et al.8 The second principal component distinguishes the nonmalignant (cells originating from mammoplasties or benign tumors) from malignant cell lines. The first and second principal components form a coordinate system in which the cell lines have the largest variance (projected onto the first principal component), followed by the next largest variance (second principal component).32 Hence, these cell lines differ more in their state of differentiation from claudin-low to luminal than they do between nonmalignant and malignant.
The information required to group the cell lines is distributed widely among the glycoproteins, as can be seen when the principal component analysis is performed with 50 randomly selected glycoproteins (Figure 2b). The same four subtypes are present, although the clusters corresponding to the subtypes are more diffuse and there is some overlap among subtypes.
Differential Expression of Glycoproteins in Cell Lines
To compare the differences in glycoprotein expression between claudin-low cell lines and the non-claudin-low cell lines, we calculated a two-sample t statistic for each of the 399 glycoproteins (Table S3). One group was claudin-low and the other contained the remaining cell lines, including the normal and nonmalignant ones. The frequency distribution of these statistics shows two large peaks, corresponding to proteins expressed at lower or higher levels in claudin-low cell lines compared to that in the other cell lines (Figure 3). A mixture of two normal distributions fits the data well. It is clear that the expression of many proteins is different, either higher or lower, in the claudin-low cell lines compared to that in the other cell lines. Glycoproteins with reduced expression (t < 0) outnumber those with increased expression.
The widespread changes in glycoprotein expression illustrated in Figure 3 suggest an explanation for the modest success in identifying separate subgroups of cell lines with randomly chosen proteins (Figure 2b). A sample of 50 proteins chosen randomly will include ones that are differentially expressed to various degrees, allowing the claudin-low lines to be separated along the first principal component.
Although two normal components describe most of the observed data well, there are excess glycoproteins with t < −3 or t > 3 (Figure 3). These glycoproteins at the extremes of the distribution are the ones with the greatest differential expression. The dozen proteins with the greatest reduction in expression and the three proteins with the greatest increase are given in Table 1. Several of these are known to have low (e.g., EPCAM) or high (e.g., CD44) RNA expression in claudin-low tumors compared to that in luminal tumors.9 They constitute a glycoprotein expression signature for claudin-low cell lines.
Table 1.
Differentially Expressed Glycoproteinsa
| UniProt accession | gene name | common name | cell line data
|
CPTAC data
|
||
|---|---|---|---|---|---|---|
| t | q | t | q | |||
| Q9Y624 | F11R | junctional adhesion molecule A | −6.7 | 2.0 × 10−4 | −9.8 | 7.8 × 10−11 |
| P16422 | EPCAM | epithelial cell adhesion molecule | −5.2 | 1.8 × 10−3 | −11.1 | 3.5 × 10−13 |
| O95274 | LYPD3 | Ly6/PLAUR domain-containing protein | −5.2 | 1.8 × 10−3 | −1.6 | 0.03 |
| P15529 | CD46 | membrane cofactor protein | −5.1 | 1.8 × 10−3 | −12.8 | 9.2 × 10−12 |
| O43278 | SPINT1 | Kunitz-type protease inhibitor 1 | −4.9 | 1.8 × 10−3 | −4.8 | 4.2 × 10−5 |
| Q92896 | GLG1 | Golgi apparatus protein 1 | −4.8 | 1.8 × 10−3 | −9.4 | 6.9 × 10−11 |
| Q8IWA5 | SLC44A2 | choline transporter-like protein 2 | −4.3 | 5.5 × 10−3 | −1.7 | 0.11 |
| O14672 | ADAM10 | disintegrin and metalloproteinase domain-containing protein 10 | −4.2 | 5.6 × 10−3 | −4.1 | 2.9 × 10−4 |
| O43291 | SPINT2 | Kunitz-type protease inhibitor 2 | −4.2 | 6.3 × 10−3 | −11.9 | 7.0 × 10−12 |
| Q9UHL4 | DPP7 | dipeptidyl peptidase 2 | −4.2 | 6.1 × 10−3 | 2.7 | 3.7 × 10−3 |
| P09758 | TACSTD2 | tumor-associated calcium signal transducer 2 | −4.0 | 7.6 × 10−3 | −4.5 | 9.4 × 10−5 |
| Q92542 | NCSTN | nicastrin | −3.8 | 1.0 × 10−3 | 5.2 | 1.2 × 10−5 |
| P50454 | SERPINH1 | serine-type endopeptidase inhibitor | 2.7 | 0.07 | 7.6 | 3.4 × 10−8 |
| P27797 | CALR | calreticulin | 2.8 | 0.07 | 5.0 | 2.6 × 10−5 |
| P16070 | CD44 | hyaluronic acid receptor | 4.1 | 0.01 | −13.7 | 4.6 × 10−15 |
For each protein, a t statistic (Welch’s two-sample test) was calculated for the difference in expression between the claudin-low cells or tumor samples and the rest. The dozen proteins with the greatest reduction in expression in the cell line data and the three proteins with the greatest increase are shown. q values were calculated using qvalue() by John D. Storey with contributions from Andrew J. Bass, Alan Dabney and David Robinson (2015). qvalue: Q-value estimation for false discovery rate control. R package, version 2.2.2 (http://github.com/jdstorey/qvalue).
Similar glycoprotein signatures were calculated for the nonmalignant, basal, and luminal groups (Table S3). In general, the proteins with the greatest differential expression differ for the four subtypes of cell lines. However, there are some proteins that appear in more than one signature; for example, EPCAM and SLC44A2 are expressed at very low levels in both the claudin-low and nonmalignant cell lines.
Glycoprotein Expression Compared to mRNA Expression
To what extent do changes in glycoprotein expression follow mRNA expression? RNA seq data for the cell lines studied here is available from Daemen et al.24 We calculated t statistics from the mRNA data as described above for the glycoprotein data. There is a positive association between the variables, i.e., the mRNA and protein expression levels tend to change in the same direction, but the scatterplot displays a noisy relationship (Figure 4a). For the lower values of t, the mRNA and glycoprotein statistics are correlated. Using the first quartile of t values for the change in mRNA, in which the glycoproteins are expressed at a lower level in claudin-low cell lines, the coefficient of correlation is 0.31 and is significantly different from 0 (p value < 0.01). In contrast for the fourth quartile, in which the glycoproteins are expressed at a higher level, the coefficient of correlation is 0.005 and is not significantly different from 0.
Figure 4.
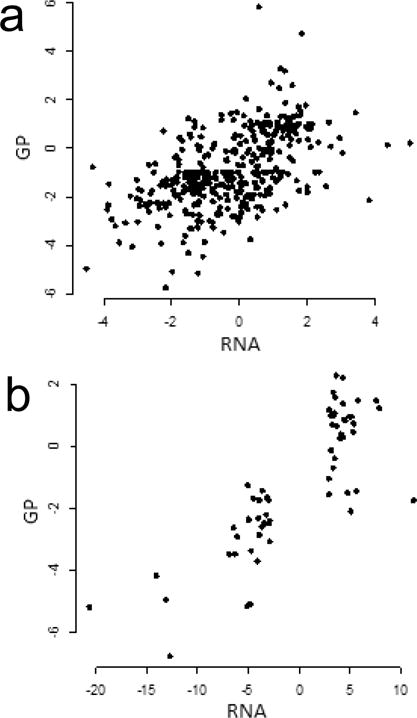
Relation between changes in expression of mRNA and glycoproteins. (a) Scatterplot of t statistics for mRNA and the claudin-low glycoproteins identified in this study. There are 391 proteins for which both mRNA and glycoprotein data are available. (b) Scatterplot of scores (logarithms of fold change) for differentially expressed mRNA from the nine cell line claudin-low predictor data set (Supplemental Data from Prat et al.9) and t statistics for glycoproteins. There are 61 genes for which both mRNA and glycoprotein data are available.
Prat et al.9 identified genes that were differentially expressed, comparing claudin-low cell lines to other malignant cell lines. They established a nine cell line claudin-low predictor list, for use in classifying tumor samples. The four genes for glycoproteins with the most reduced RNA expression in that list, EPCAM, SPINT1, SPINT2, and F11R, are among the glycoproteins with the most reduced expression (Table 1). CD46 and LYPD3 likewise have reduced expression of both mRNA and protein. Yet, several of the glycoproteins with reduced expression, including GLG1, SLCC44A2, DPP7, TACSTD2, and NCSTN, do not have sufficiently large changes in mRNA expression to appear in the nine cell line claudin-low predictor list. None of the three overexpressed glycoproteins (CD44, CALR, SERPINH1) have mRNA changes large enough to appear in the upregulated genes of the nine cell line claudin-low predictor list.
Figure 4b shows the positive association between gene expression from Prat et al.9 and changes in glycoprotein expression. Overall, the correlation between the RNA expression score and t statistics for glycoproteins is 0.82 (Figure 4b). The correlation is stronger than that seen for all of the gene/glycoprotein data, as expected given that Prat et al. selected for differential expression. For the 28 glycoproteins with low expression in the claudin-low cell lines (t < 0), the correlation was 0.66 (p < 0.01), whereas for the 33 high expression proteins, the correlation was −0.17.
Very low mRNA expression must limit glycoprotein expression, but for higher levels of mRNA, other factors, such as processing in the endomembrane system, may be more important in determining glycoprotein levels. Except at very low levels of mRNA, the level of mRNA gives little quantitative information about the amount of glycoprotein expressed.
Supervised Classification
After classifying the 26 cell lines using principal component analysis, how might it be determined whether a new cell line is claudin-low? The task should be straightforward since the claudin-low cell lines form a distinct group that does not overlap with the others (Figure 2a) and since there are many glycoproteins that are differentially expressed (Figure 3).
Linear discriminant analysis can be used to find a linear decision boundary to distinguish, without error, between the 5 claudin-low cell lines and the 21 other lines (Figure 2a). The decision boundary can be used to classify other cell lines; any new cell lines above the line would be categorized as claudin-low, whereas those falling below the line would be categorized as not claudin-low. Linear discriminant analysis was carried out using the coordinates of the cell lines on the two principle component axes, which depend on all 399 glycoproteins.
The distribution of two-sample t statistics (Figure 3) shows that many glycoproteins are expressed differently in the claudin-low cell lines compared to the others, raising the possibility that a random sample of proteins may also serve for classification. If so, the success of the random predictor classifier would provide additional evidence for the widespread distribution of information relevant for classification in the data.
Fifty of the 399 proteins were selected randomly and used for principal component analysis followed by linear discriminate analysis. Figure 2b shows an example of a principal component analysis and the corresponding decision boundary from a random sample of 50 glycoproteins. All non-claudin-low cell lines are below the decision boundary, giving 100% specificity (the probability that it is correctly classified as not claudin-low when the cell line is not claudin-low). Four of 5 claudin-low cell lines are above the decision boundary, giving 80% sensitivity (the probability that it is correctly classified as claudin-low when the cell line is claudin-low). When the same analysis was performed 1000 times with different random samples, the sensitivity and specificity were found to be 0.71 and 0.97, respectively.
The performance of the 50 random variable classifier may be understood as follows. There are 5 claudin-low cell lines and 21 others. A single misclassification of the claudin-low lines would give a sensitivity of 0.8, whereas a single misclassification of the other lines would yield a specificity of 0.95. These values of sensitivity and specificity are close to the values of 0.71 and 0.97 in 1000 trials. Hence, the 50 random variable classifier misclassifies 1–2 claudin-low and ~1 other cell line per trial on average. Even with no attempt to identify the most useful markers, it is possible to classify the cell lines fairly well. The performance of this random variable classifier varies with the size of the sample, with more variables giving better performance.
The phenomenon in which very different sets of predictor variables can give similar classification performance is well-known in the gene expression analysis of breast tumors, where prognostic classifiers may give similar results even though they are based on different sets of genes.33–35
For these glycoprotein data, a classifier may also be constructed using fewer than 50 predictor proteins. Lasso logistic regression performs variable selection in parallel with constructing a logistic regression model.32 On the glycoprotein data set, lasso logistic regression returned a model that correctly classified all cell lines using three glycoproteins: Q9Y624 (F11R, junctional adhesion protein), P50897 (PPT1, palmitoyl-protein thioesterase 1), and P02452 (COL1A1, collagen alpha1(I)).
A very different method, Random Forests, classifies the 26 cell lines into claudin-low vs others with a sensitivity of 0.20 and a specificity of 1.00. The developers of Random Forests note that the method may perform poorly if the two groups are unbalanced in size, which is the case here (Breiman and Cutler, https://www.stat.berkeley.edu/~breiman/RandomForests/). Even so, the single most important predictor variable is junctional adhesion molecule A (F11R). This protein is also one of the three predictors in the logistic regression model and is the protein in the data set that differs most in expression between the claudin-low and other cell lines (Table 1).
Drug Sensitivity of the Claudin Low Cell Lines
Are the claudin-low cell lines particularly sensitive to one drug or class of drugs? Daemen et al.24 provide the growth inhibition 50% (GI50) values for 90 drugs on 70 breast cancer cell lines. Using the Daemen et al. data, we calculated for each drug the difference in GI50 between the claudin-low cell lines and the others. The 18 drugs most active on claudin-low cell lines are presented in Table 2. Most of these compounds interfere directly with the cell cycle, e.g., with DNA integrity, mitotic spindle function, or chromosome segregation.
Table 2.
Drugs Most Active against Claudin-Low Cell Linesa
| p value | drug | target |
|---|---|---|
| 5.7 × 10−5 | GSK461364 | polo-like kinase, active in M phase |
| 0.00031 | docetaxel | mitotic spindle |
| 0.00065 | vinorelbine | mitotic spindle |
| 0.0025 | paclitaxel | mitotic spindle |
| 0.0036 | etoposide | topoisomerase |
| 0.0039 | ibandronate | inhibits bone resorption |
| 0.0051 | bosutinib | tyrosine kinases |
| 0.0052 | ixabepilone | mitotic spindle |
| 0.019 | GSK923295 | centromere-associated protein |
| 0.022 | PF_3814735 | Aurora kinase, chromatid segregation |
| 0.023 | lestaurtinib | tyrosine kinases |
| 0.027 | TCS2312 | checkpoint kinase inhibitor |
| 0.028 | irinotecan | topoisomerase |
| 0.031 | XRP44X | ras-net (ELK3) |
| 0.033 | sunitinib | receptor tyrosine kinases |
| 0.035 | cisplatin | DNA |
| 0.043 | GSK1070916 | Aurora kinase, chromatid segregation |
| 0.049 | IKK_16 | IkB kinase |
For each drug, two-sample t tests were performed comparing drug sensitivity of the claudin-low cell lines to the others. The results were sorted on the p values.
Cell lines defined as claudin-low by RNA expression often do not express ER or overexpress HER2.9 The five claudin-low cell lines in our glycoprotein data set are all triple negative. Hence, the claudin-low cell lines may be expected to have low sensitivity to many drugs in the Daemen et al. data set, which includes a large number of agents aimed at specific targets in the growth factor pathways. The rate of proliferation of the cell lines is another factor that may influence drug sensitivity. Prat et al.8 showed that claudin-low cell lines (along with basal cell lines) have higher expression of proliferation-related genes than do other cell lines. Perhaps these cells are also more sensitive to drugs that block the cell cycle.
Tumors with Low Claudin Protein Expression
To what extent are the results for glycoprotein expression in breast cell lines also true for tumors? A data set of proteins from 105 breast cancer tumors measured by ITRAQ (isobaric tags for relative and absolute quantitation) is publicly available from the Clinical Proteomic Tumor Analysis Consortium (CPTAC) and was used for the comparison. These data are a subset of the larger The Cancer Genome Atlas Project (TCGA) data set.25
In the original TCGA publication on breast cancer, only 8 of 525 tumors (from which the CPTAC tumor samples were drawn) were claudin-low, as assessed by gene expression.25 Claudin-low was viewed as a minor subtype and was not used in the classification system based on gene expression.
Nevertheless, there seems to be a claudin-low breast tumor subtype based on protein expression in the CPTAC data. Claudin-low tumors were defined by Herschkowitz et al.7 as having low mRNA expression of claudins (CLDN) 3, 4, and 7. Claudins 3 and 7, but not 4, are present in the CPTAC data set. The frequency distribution of CLDN3 protein expression shows a peak and also a significant tail of tumors with low CLDN3 expression (Figure 5a). The distribution of CLDN7 expression is similar, but it lacks the tail of samples with low expression. In the principal component plot of the CPTAC data, the 18 tumor samples with the lowest CLDN3 expression cluster in one region, consistent with their identification as a distinct group (Figure 5b). Samples low in CLDN7, in contrast, are distributed uniformly across the plot (not shown). On the basis of this data from the CPTAC proteomic data set, we treat the 18 tumor samples with the lowest CLDN3 expression as claudin-low.
Figure 5.
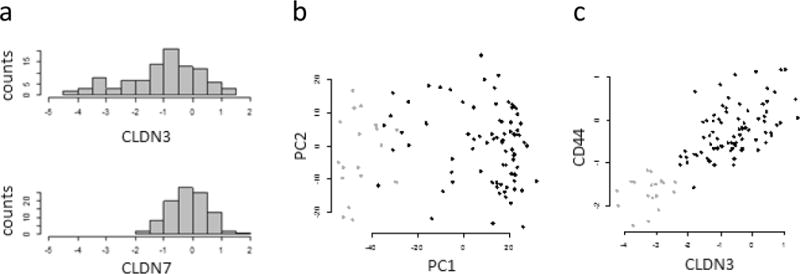
Claudins and CD44 in a breast tumor data set (CPTAC). (a) Frequency distribution of CLDN3 and CLDN7 expression. The tail of 18 samples on the left of the CLDN3 data was used to identify the claudin-low samples. (b) Principal components plot. The claudin-low samples are identified by the violet symbols. (c) Association between CLDN3 and CD44.
An alternative to using CLDN3 to define claudin-low for these tumors would be to add the expression values for CLDN3 and CLDN7 together for each patient sample. When this is done, 14 of 18 with lowest summed CLDN expression are the same samples as in the CLDN3-low group. Thus, the main conclusions described in the following paragraphs are the same regardless of which definition of claudin-low is used for the tumors.
Using low expression of CLDN3 protein in the CPTAC data set to define claudin-low gives a prevalence of ~18%, roughly consistent with other reports (Prat et al.,9 7–14%; Sabatier et al.,10 12.4%), although it is not consistent with mRNA expression in the TCGA data set (<2%).
The claudin-low tumors in the CPTAC data set differ in significant ways from tumors or cell lines defined as claudin-low by gene expression. Most tumors defined as claudin-low by gene expression are triple negative (not expressing the estrogen or progesterone receptors or overexpressing HER2). Prat et al.9 report that 61–70% of claudin-low tumors are triple-negative based on immunohistochemistry; Sabatier et al.10 report this to be 52% based on gene expression. Yet, of the 18 tumor samples low in CLDN3 protein, only 5 were derived from triple negative tumors, according to the TCGA patient annotation. Ten of the 18 tumors also expressed estrogen receptor, and five overexpressed HER2.
Glycoprotein Expression in Tumors Compared to That in Cell Lines
Are the glycoproteins with the greatest differential expression in the breast cancer cell lines also expressed at extreme levels in tumor samples? We calculated t statistics for the difference in glycoprotein expression between the samples low in CLDN3 protein and the rest in the CPTAC data. Table 1 shows the results for the CPTAC proteins that are in the claudin-low signature. The majority (10 of 15) of the differentially expressed glycoproteins observed in the cell line data set are also differentially expressed in the CPTAC data set. These include F11R, EPCAM, CD46, SPINT1, SPINT2, GLG1, ADAM10, and TACSTD2, which are expressed at lower levels in claudin-low cell lines and the 18 CLDN3-low CPTAC tumors. CALR and SERPINH1 are expressed at higher levels in both the cell lines and tumors.
There are exceptions, however. For LYPD3 and SLC44A2, there is little evidence for differential expression in the CPTAC data. For DPP7 and NCSTN, the sign of the expression difference has changed. CD44 is the protein for which the disparity in results is most dramatic: In the cell line data, CD44 is found at higher levels in the claudin-low lines, compared to very low levels in the CPTAC data. In the CPTAC data, CLDN3 and CD44 are positively correlated, with r = 0.77 (Figure 5c).
Is the discrepancy between CD44 protein expression in cell lines and tumor samples due to differences in CD44 gene expression? RNA seq data for most of the cell lines studied here is available from Daemen et al.24 The difference in CD44 mRNA expression between the claudin-low and other cell lines is not significant (p = 0.27). Gene expression data for the tumor samples in the CPTAC data set is also available, through The Cancer Genome Atlas project. Again, the difference in CD44 gene expression among the 18 samples low in CLDN3 expression and the remainder is not significant (p = 0.59). The differences in CD44 protein expression, higher in claudin-low cell lines and lower in tumor samples, must be due to post-transcriptional processing differences between the cell lines and tumors.36
CD44 expression is used as a marker for breast cancer stem cells,37 and high CD44 expression has been cited as evidence that claudin-low tumors have stem cell-like properties.9 The finding that CLDN3-low tumor samples have low CD44 expression seems to contradict this interpretation. However, the CPTAC data describes tumor samples rather than cell lines, and it is possible that the CLDN3-low or other CPTAC samples contain a small proportion of claudin-low/CD44+ cells that function as cancer stem cells.
For CLDN3, as well as CD44, the conclusions drawn from measuring proteins differ from those using gene expression. The relationship between mRNA and protein levels for CLDN3 is very weak, with a correlation coefficient (0.11) that is not significantly different from 0 (Figure 6). It can be seen that the samples with the lowest CLDN3 protein expression have a large range of CLDN3 mRNA levels. The CPTAC samples with low CLDN3 protein expression are not the same as those with low CLDN3 mRNA expression; hence, it is not surprising that they differ in traits such as CD44 protein, estrogen receptor, progesterone receptor, or HER2 expression. The poor correlation between CLDN3 mRNA and protein in breast tumors has also been noted by Tokes et al.38 In an immunohistochemical study, Soini39 reported that the expression of claudins, including CLDN3, is not associated with ER status.
Figure 6.
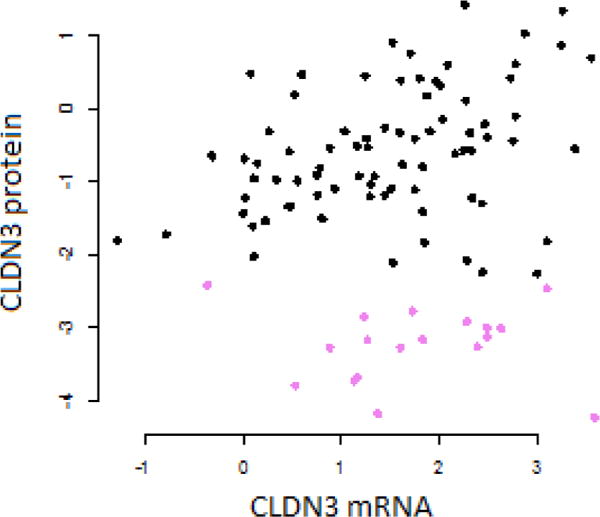
Association between mRNA expression and protein expression for CLDN3 in the CPTAC data. The samples low in CLDN3 protein are identified by violet symbols.
CONCLUSIONS
The claudin-low subtype of breast cancer was studied in two contexts, cell lines and tumor samples from the CPTAC. There are findings that are in common between the cell lines and tumor samples, but there are also important differences. For the breast cell lines, analysis of glycoprotein expression is quite consistent with the claudin-low subtype as defined by RNA expression: (1) hierarchical clustering and principal components analysis support claudin-low as a subtype distinct from basal and luminal, (2) the five claudin-low cell lines that form a distinct group are negative for ER, PR, and HER2 overexpression, (3) many of the same genes that are differentially expressed at the RNA level are also differentially expressed as proteins, and (4) CD44 expression is high, consistent with the claudin-low subtype having stem cell-like properties.
The breast tumor samples in the CPTAC data set were drawn from the larger TCGA data set, in which the claudin-low subtype was reported to have a prevalence <2%. Yet, in the CPTAC data set, there is clearly a set of tumors with very low CLDN3 expression. We categorized 18 of 99 tumor samples as claudin-low. In these samples, there was a fair amount of agreement with the breast cell line data on differential protein expression. For example junctional adhesion molecule A (F11R), EPCAM, SPINT1, and SPINT2 proteins and others are expressed at very low levels in both tumors and cell lines. This finding is also in agreement with results from RNA expression measurements.
It is clear that the CPTAC tumors include a group low in claudin protein, one that resembles the claudin-low subtype defined elsewhere. However, these tumor samples do not share all of the properties of the claudin-low subtype as originally described. Many of the samples low in CLDN3 express ER or overexpress HER2. Furthermore, the tumors low in CLDN3 protein also express CD44 at a low level. These data indicate that there is a group of breast tumors with low claudin protein expression, that these tumors are not mostly triple negative, and, judging from CD44 expression, that they do not have stem cell-like properties.
Supplementary Material
Acknowledgments
Support for this work was provided by grants from the National Institutes of Health, R15 CA164929 and P20 MD000544, and the National Science Foundation, grants CHEM-0619163 and CHE-1228656.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.6b00470.
Stability of glycoprotein expression profiles (Figure S1; PDF)
Cell line subtype classifications (Table S1; XLSX)
Glycoprotein spectral count data (Table S2; XLSX)
Complete list of p values comparing cell line subtypes (Table S3; XLSX)
ORCID
Leslie C. Timpe: 0000-0002-2398-4765
Notes
The authors declare no competing financial interest.
References
- 1.Perou CM, Sorlie T, Eisen MB, de Rijn van M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours Nature. 2000;406(6797):747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, de Rijn van M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lonning PE, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, Lonning PE, Brown PO, Borresen-Dale AL, Botstein D. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100(14):8418–23. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dawson SJ, Rueda OM, Aparicio S, Caldas C. A new genome-driven integrated classification of breast cancer and its implications. EMBO J. 2013;32(5):617–28. doi: 10.1038/emboj.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guedj M, Marisa L, de Reynies A, Orsetti B, Schiappa R, Bibeau F, MacGrogan G, Lerebours F, Finetti P, Longy M, Bertheau P, Bertrand F, Bonnet F, Martin AL, Feugeas JP, Bieche I, Lehmann-Che J, Lidereau R, Birnbaum D, Bertucci F, de The H, Theillet C. A refined molecular taxonomy of breast cancer. Oncogene. 2012;31(9):1196–206. doi: 10.1038/onc.2011.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grigoriadis A, Mackay A, Noel E, Wu PJ, Natrajan R, Frankum J, Reis-Filho JS, Tutt A. Molecular characterisation of cell line models for triple-negative breast cancers. BMC Genomics. 2012;13(1):619. doi: 10.1186/1471-2164-13-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, Rasmussen KE, Jones LP, Assefnia S, Chandrasekharan S, Backlund MG, Yin Y, Khramtsov AI, Bastein R, Quackenbush J, Glazer RI, Brown PH, Green JE, Kopelovich L, Furth PA, Palazzo JP, Olopade OI, Bernard PS, Churchill GA, Van Dyke T, Perou CM. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8(5):R76–R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prat A, Karginova O, Parker JS, Fan C, He X, Bixby L, Harrell JC, Roman E, Adamo B, Troester M, Perou CM. Characterization of cell lines derived from breast cancers and normal mammary tissues for the study of the intrinsic molecular subtypes. Breast Cancer Res Treat. 2013;142(2):237–55. doi: 10.1007/s10549-013-2743-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, He X, Perou CM. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12(5):R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sabatier R, Finetti P, Guille A, Adelaide J, Chaffanet M, Viens P, Birnbaum D, Bertucci F. Claudin-low breast cancers: clinical, pathological, molecular and prognostic characterization. Mol Cancer. 2014;13:228. doi: 10.1186/1476-4598-13-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gunzel D, Yu AS. Claudins and the modulation of tight junction permeability. Physiol Rev. 2013;93(2):525–69. doi: 10.1152/physrev.00019.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwon M. Emerging Roles of Claudins in Human Cancer. Int J Mol Sci. 2013;14(9):18148. doi: 10.3390/ijms140918148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morin PJ. Claudin proteins in human cancer: promising new targets for diagnosis and therapy. Cancer Res. 2005;65(21):9603–6. doi: 10.1158/0008-5472.CAN-05-2782. [DOI] [PubMed] [Google Scholar]
- 14.Singh AB, Dhawan P. Claudins and cancer: Fall of the soldiers entrusted to protect the gate and keep the barrier intact. Semin Cell Dev Biol. 2015;42:58–65. doi: 10.1016/j.semcdb.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 15.Swisshelm K, Macek R, Kubbies M. Role of claudins in tumorigenesis. Adv Drug Delivery Rev. 2005;57(6):919–28. doi: 10.1016/j.addr.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 16.Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A, Rimm DL, Wong H, Rodriguez A, Herschkowitz JI, Fan C, Zhang X, He X, Pavlick A, Gutierrez MC, Renshaw L, Larionov AA, Faratian D, Hilsenbeck SG, Perou CM, Lewis MT, Rosen JM, Chang JC, Weinberg RA. Residual Breast Cancers after Conventional Therapy Display Mesenchymal as Well as Tumor-Initiating Features. Proc Natl Acad Sci U S A. 2009;106(33):13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hennessy BT, Gonzalez-Angulo A-M, Stemke-Hale K, Gilcrease MZ, Krishnamurthy S, Lee J-S, Fridlyand J, Sahin A, Agarwal R, Joy C, Liu W, Stivers D, Baggerly K, Carey M, Lluch A, Monteagudo C, He X, Weigman V, Fan C, Palazzo J, Hortobagyi GN, Nolden LK, Wang NJ, Valero V, Gray JW, Perou CM, Mills GB. Characterization of a Naturally Occurring Breast Cancer Subset Enriched in Epithelial-to-Mesenchymal Transition and Stem Cell Characteristics. Cancer Res. 2009;69(10):4116–4124. doi: 10.1158/0008-5472.CAN-08-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rae JM, Creighton CJ, Meck JM, Haddad BR, Johnson MD. MDA-MB-435 cells are derived from M14 Melanoma cells––a loss for breast cancer, but a boon for melanoma research. Breast Cancer Res Treat. 2007;104(1):13–19. doi: 10.1007/s10549-006-9392-8. [DOI] [PubMed] [Google Scholar]
- 19.Drake PM, Schilling B, Niles RK, Prakobphol A, Li B, Jung K, Cho W, Braten M, Inerowicz HD, Williams K, Albertolle M, Held JM, Iacovides D, Sorensen DJ, Griffith OL, Johansen E, Zawadzka AM, Cusack MP, Allen S, Gormley M, Hall SC, Witkowska HE, Gray JW, Regnier F, Gibson BW, Fisher SJ. Lectin chromatography/mass spectrometry discovery workflow identifies putative biomarkers of aggressive breast cancers. J Proteome Res. 2012;11(4):2508. doi: 10.1021/pr201206w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Timpe LC, Yen R, Haste NV, Litsakos-Cheung C, Yen TY, Macher BA. Systemic alteration of cell-surface and secreted glycoprotein expression in malignant breast cancer cell lines. Glycobiology. 2013;23(11):1240–9. doi: 10.1093/glycob/cwt060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yen TY, Macher BA, McDonald CA, Alleyne-Chin C, Timpe LC. Glycoprotein profiles of human breast cells demonstrate a clear clustering of normal/benign versus malignant cell lines and basal versus luminal cell lines. J Proteome Res. 2012;11(2):656–67. doi: 10.1021/pr201041j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yen TY, Haste N, Timpe LC, Litsakos-Cheung C, Yen R, Macher BA. Using a cell line breast cancer progression system to identify biomarker candidates. J Proteomics. 2014;96:173–83. doi: 10.1016/j.jprot.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boersema PJ, Geiger T, Wisniewski JR, Mann M. Quantification of the N-glycosylated secretome by super-SILAC during breast cancer progression and in human blood samples. Mol Cell Proteomics. 2013;12(1):158–71. doi: 10.1074/mcp.M112.023614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daemen A, Griffith OL, Heiser LM, Wang NJ, Enache OM, Sanborn Z, Pepin F, Durinck S, Korkola JE, Griffith M, Hur JS, Huh N, Chung J, Cope L, Fackler MJ, Umbricht C, Sukumar S, Seth P, Sukhatme VP, Jakkula LR, Lu Y, Mills GB, Cho RJ, Collisson EA, Veer van’t LJ, Spellman PT, Gray JW. Modeling precision treatment of breast cancer. Genome Biol. 2013;14(10):R110. doi: 10.1186/gb-2013-14-10-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.CancerGenomeAtlasNetwork. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo WL, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10(6):515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vizcaino JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, Xu QW, Wang R, Hermjakob H. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44(D1):D447–56. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Timpe LC, Li D, Yen TY, Wong J, Yen R, Macher BA, Piryatinska A. Mining the Breast Cancer Proteome for Predictors of Drug Sensitivity. J Proteomics Bioinf. 2015;8(9):204–211. doi: 10.4172/jpb.1000370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heiser LM, Sadanandam A, Kuo WL, Benz SC, Goldstein TC, Ng S, Gibb WJ, Wang NJ, Ziyad S, Tong F, Bayani N, Hu Z, Billig JI, Dueregger A, Lewis S, Jakkula L, Korkola JE, Durinck S, Pepin F, Guan Y, Purdom E, Neuvial P, Bengtsson H, Wood KW, Smith PG, Vassilev LT, Hennessy BT, Greshock J, Bachman KE, Hardwicke MA, Park JW, Marton LJ, Wolf DM, Collisson EA, Neve RM, Mills GB, Speed TP, Feiler HS, Wooster RF, Haussler D, Stuart JM, Gray JW, Spellman PT. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc Natl Acad Sci U S A. 2012;109(8):2724–9. doi: 10.1073/pnas.1018854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu F, Koval M, Ranganathan S, Fanayan S, Hancock WS, Lundberg EK, Beavis RC, Lane L, Duek P, McQuade L, Kelleher NL, Baker MS. Systems Proteomics View of the Endogenous Human Claudin Protein Family. J Proteome Res. 2016;15(2):339–59. doi: 10.1021/acs.jproteome.5b00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, Asselin-Labat ML, Gyorki DE, Ward T, Partanen A, Feleppa F, Huschtscha LI, Thorne HJ, Fox SB, Yan M, French JD, Brown MA, Smyth GK, Visvader JE, Lindeman GJ. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15(8):907–13. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 32.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning. Springer; New York: 2009. [Google Scholar]
- 33.Fan C, Oh DS, Wessels L, Weigelt B, Nuyten DSA, Nobel AB, van’t Veer L J, Perou CM. Concordance among Gene-Expression–Based Predictors for Breast Cancer. N Engl J Med. 2006;355(6):560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 34.Haibe-Kains B, Desmedt C, Piette F, Buyse M, Cardoso F, Van’t Veer L, Piccart M, Bontempi G, Sotiriou C. Comparison of prognostic gene expression signatures for breast cancer. BMC Genomics. 2008;9:394. doi: 10.1186/1471-2164-9-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomassen M, Tan Q, Eiriksdottir F, Bak M, Cold S, Kruse TA. Comparison of gene sets for expression profiling: prediction of metastasis from low-malignant breast cancer. Clin Cancer Res. 2007;13(18):5355–5360. doi: 10.1158/1078-0432.CCR-07-0249. [DOI] [PubMed] [Google Scholar]
- 36.Louderbough JM, Schroeder JA. Understanding the dual nature of CD44 in breast cancer progression. Mol Cancer Res. 2011;9(12):1573–86. doi: 10.1158/1541-7786.MCR-11-0156. [DOI] [PubMed] [Google Scholar]
- 37.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tokes AM, Kulka J, Paku S, Szik A, Paska C, Novak PK, Szilak L, Kiss A, Bogi K, Schaff Z. Claudin-1, –3 and –4 proteins and mRNA expression in benign and malignant breast lesions: a research study. Breast Cancer Res. 2005;7(2):R296–305. doi: 10.1186/bcr983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soini Y. Claudins 2, 3, 4, and 5 in Paget’s disease and breast carcinoma. Hum Pathol. 2004;35(12):1531–6. doi: 10.1016/j.humpath.2004.09.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


