Abstract
Background
Given the uncertainties inherent in clinical measures of prostate cancer aggressiveness, clinically validated tissue biomarkers are needed. We tested whether Alpha-2-Glycoprotein 1, Zinc-Binding (AZGP1) protein levels, measured by immunohistochemistry, and RNA expression, by RNA in situ hybridization (RISH), predict recurrence after radical prostatectomy independent of clinical and pathological parameters.
Methods
AZGP1 IHC and RISH were performed on a large multi-institutional tissue microarray resource including 1275 men with 5 year median follow-up. The relationship between IHC and RISH expression levels was assessed using the Kappa analysis. Associations with clinical and pathological parameters were tested by the Chi-square test and the Wilcoxon rank sum test. Relationships with outcome were assessed with univariable and multivariable Cox proportional hazards models and the Log-rank test.
Results
Absent or weak expression of AZGP1 protein was associated with worse recurrence free survival (RFS), disease specific survival and overall survival after radical prostatectomy in univariable analysis. AZGP1 protein expression, along with pre-operative serum PSA levels, surgical margin status, seminal vesicle invasion, extracapsular extension and Gleason score predicted RFS on multivariable analysis. Similarly, absent or low AZGP1 RNA expression by RISH predicted worse RFS after prostatectomy in univariable and multivariable analysis.
Conclusions
In our large, rigorously designed validation cohort, loss of AZGP1 expression predicts RFS after radical prostatectomy independent of clinical and pathological variables.
Keywords: Prostate cancer, AZGP1, Immunohistochemistry, Prognosis
Introduction
Despite decreasing prostate cancer death rates over the past decade, systematic screening with serum prostate specific antigen (PSA) testing has been heavily criticized (1,2). Two large randomized trials (PLCO and ERSPC) have noted little or no survival benefit derived from PSA testing and suggested that prostate cancer is over treated (3,4). Paralleling these trials has been a growing realization that low risk prostate cancers left untreated can often show an indolent clinical course, giving rise to the concept of active surveillance for low risk lesions (5). Long-term follow-up from several active surveillance cohorts suggests that this is a safe approach, although not entirely without risk (6–8). Virtually all of the active surveillance programs involve relatively intense testing with PSA, digital rectal examinations, repeat biopsies, and, more recently, MRI examinations (9–11). This follow-up is necessitated by the inability to characterize the biological potential of low risk prostate cancers, as well as by sampling errors in biopsy and the poor performance of clinical measures of tumor aggressiveness. This follow-up also incurs significant financial and human costs due to repeated testing. Complicating matters further, some localized prostate cancers treated with surgery or radiation therapy alone appear to be more aggressive than clinical and pathological features suggest, and these might benefit from adjuvant therapy. Given the risks of over-treatment and under treatment in localized prostate cancer, new biomarkers to help characterize tumor aggressiveness are needed.
To address this need, our group has assembled a retrospective cohort of patients who have undergone radical prostatectomies and who have long-term follow-up (12). Using a case-control design with a quota-sampling plan, we have constructed a multi-institutional tissue microarray (TMA) resource for validation of candidate biomarkers of clinical outcome, with both pathological and clinical outcomes, such as recurrence free survival, recorded for nearly all patients (13,14).
We have previously demonstrated that loss of expression of zinc-alpha 2-glycoprotein (AZGP1 or ZAG) protein expression by immunohistochemistry is associated with an increased risk of recurrence after radical prostatectomy (15). This finding has been validated in several later studies, and loss of AZGP1 expression has also been shown to predict subsequent development of metastatic disease and death from prostate cancer (16–19). In addition, loss of transcriptional expression of AZGP1 has been associated with prostate cancer recurrence and death and is one of 12 prognostic transcripts measured in a commercially available tissue-based test called OncotypeDX from Genomic Health (20). OncotypeDX scores have been shown to correlate with adverse pathology on low and intermediate risk patients undergoing radical prostatectomy (20).
Our objective is to validate candidate biomarkers of prognosis to aid in treatment selection for men with localized prostate cancer. Based on strong preliminary data implicating loss of AZGP1 expression as a marker of adverse outcome in prostate cancer, we tested whether loss of RNA expression, using chromogenic RNA in situ hybridization (RISH), and loss of protein expression, by immunohistochemistry, were associated with recurrence free survival after radical prostatectomy.
Materials and Methods
TMA cases and construction
The study was carried out under IRB-approved protocols at each participating site (Stanford University, University of California San Francisco, University of Washington, University of British Colombia, University of Texas Health Sciences Center at San Antonio, Eastern Virginia Medical Center) and a materials transfer agreement that allowed sharing of tissue microarrays, clinical information and tissue samples. Cases included in the TMA cohort were selected randomly by the study statistician (ZF) using de-identified clinical data from each site such that recurrent and non-recurrent cases were balanced. Constraints were placed on selection such that recurrent cases in patients with Gleason score 3+3=6 and non-recurrent cases in those with Gleason score 4+4=8 were oversampled. Details of case selection, tissue microarray construction and statistical considerations have been detailed elsewhere (12).
TMAs were constructed at 6 participating centers using agreed upon standard operating procedures and TMA layouts (12). Briefly, 3 cores of the highest grade cancer from the largest cancer area were harvested as 1 mm cores and transferred to the recipient block. In addition, one core of histologically normal prostate tissue was included from each case. A common set of tissue cores (colon, tonsil, kidney, healthy prostate, liver) from a single study site were placed in each TMA block as a staining control and for normalization. Once constructed, the TMAs were baked and stored under nitrogen gas at each site.
Immunohistochemistry
Freshly cut 5 micron sections from each site were shipped to Stanford University for immunohistochemical staining. AZGP1 immunohistochemistry was performed using a commercial antibody (1:1500 dilution; HPA012582, Sigma Aldrich). All stained slides were digitalized using the Leica SCN400 scanning system with the SL801 autoloader (Leica Microsystems; Concord, Ontario, Canada) at magnification equivalent to 40×. The images were exported and stored in the SlidePath digital imaging hub (DIH; Leica Microsystems). Separate TMA sections were stained with hematoxylin and eosin (H & E) and high molecular weight keratins (HMWK, 34bE12, Dako) and scored for the presence of cancer in each core on the TMA as described previously (13). AZGP1 protein and RNA staining were scored by a single pathologist (LF) only in cores in which cancer was present as determined using the H & E and HMWK stains.
Representative cores (clearly positive, clearly negative and mixed positive/negative) were manually identified and values on a four-point scale were assigned to each immunostain and RISH. Immunohistochemical staining for AZGP1 was defined as absent, weak (faint cytoplasmic staining of scattered cells), moderate (intermediate or heterogeneous cytoplasmic staining in tumor cells), and strong (dense cytoplasmic staining of nearly all tumor cells) as defined previously (15). Similarly, AZGP1 RISH staining was scored as absent, weak, moderate and strong.
RNA in situ hybridization (RISH)
AZGP1 RNA expression was performed on 5 micron sections using the RNAscope® 2.0 HD Detection Kit (Red) assay (Cat. No. 310034, Advanced Cell Diagnostics) using probes for AZGP1 (Advanced Cell Diagnostics). Sections were deparaffinized in a series of xylene and ethanol and allowed to dry before incubation with “pretreatment 1” for 10 minutes at room temperature, boiled in “pretreatment 2” for 15 minutes, and protease-digested in “pretreatment 3” at 40°C for 30 minutes. Slides were then processed according to the manufacturer’s instructions. The bacterial gene DapB was used as a negative control and the housekeeping gene POLR2A served as a positive control. Sections were counterstained with Gill’s hematoxylin (Sigma-Aldrich) and mounted with Ecomount (Biocare Medical).
Statistical methods
A total of 1326 subjects are represented on the TMA and had their clinical data collected. The clinical and pathological characteristics included in the analysis were age, pre-surgery PSA, post-surgical Gleason score, seminal vesicle invasion (SVI), extra-capsular invasion (ECE), and surgical margin status. Subjects with 25% or more of their clinical or pathological characteristics missing were excluded from this analysis (N= 51). A total of 1275 patients with evaluable AZGP1 staining data and the clinical and pathological data were included in the analysis.
The primary endpoint of this analysis was post-surgical recurrence-free survival (RFS) defined as the absence of PSA (biochemical) recurrence, local recurrence, prostate cancer metastases, or death from prostate cancer, with events scored at the earliest date noted after surgery. Disease-specific survival (DSS), defined as death from prostate cancer or development of advanced metastatic disease, and overall survival (OS) were secondary endpoints. For all endpoints the baseline was set at the date of surgery. AZGP1 IHC and RISH score for each patient was the maximum score of all the cores from that patient as defined above. Based on previous work, AZGP1 stained cases were grouped as negative/weak staining and compared to moderate/strong staining (15).
Summary statistics of patients’ AZGP1 protein and RNA scores were provided in frequencies and percentages. The association between AZGP1 expression levels by IHC and RISH was assessed by Kappa analysis and the Chi-square test. The association between AZGP1 expression levels and categorical values (seminal vesicle invasion, extracapsular extension, and positive surgical margins) was assessed by Chi-square test. The association between AZGP1 expression and continuous variables (pre-operative serum PSA levels and age) was assessed by the Wilcoxon rank sum test. The Kaplan-Meier (KM) method was used to estimate survival endpoints by AZGP1 expression group. Cox proportional hazard model was used to estimate effects AZGP1 expression on each survival endpoint. Unweighted and weighted analyses were performed, with the latter accounting for the oversampling of patients with recurrence less than 5 years after surgery. All tests were two-sided and p-values of 0.05 or less were considered statistically significant. Statistical analysis was carried out using SAS version 9 (SAS Institute, Cary, NC). Kaplan Meier plots were generated using Spotfire S+ 8.2 (TIBCO Inc., Palo Alto, CA).
Results
Patient population and staining results
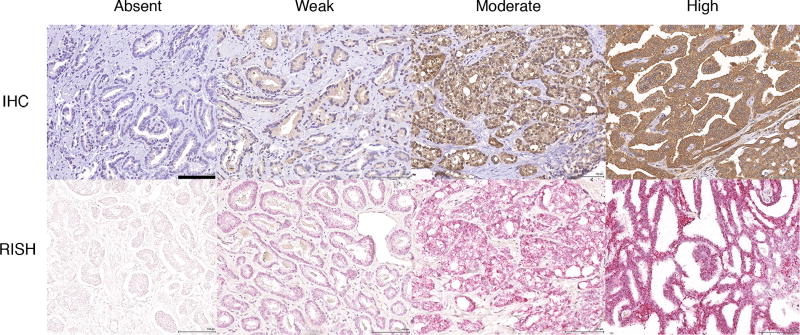
A total of 1275 patients had available clinical and pathological data, as well as evaluable AZGP1 expression status by either IHC or RISH (representative images shown in Figure 1). For AZGP1 IHC, a total of 139 cases (11%) did not have evaluable staining data either because of core loss or because lack of cancer in the core samples. Of the remaining tumors, 22% (252/1136) showed absent expression, 21% (240/1136) showed weak expression, 33% (372/1136) showed moderate expression, and 24% (272/1136) showed strong expression. The distribution of AZGP1 RISH staining was very similar - absent expression: 23%; weak: 32%; moderate: 24%; and strong: 22%. For RISH, 186 cases did not have evaluable AZGP1 staining. AZGP1 expression levels, clinical and pathological data are summarized in Table 1.
Figure 1. Immunohistochemical and RISH AZGP1 staining in representative prostate cancer samples showing absent, weak, moderate and strong staining.
Black bar in the upper left micrograph corresponds to 100 microns. All images scaled the same.
Table 1.
Summary of pathological characteristics
| Variable | Status | Number | Percentage |
|---|---|---|---|
| Gleason Score | Missing | 10 | 0.78 |
| ≤ 6 | 549 | 43.06 | |
| 3+4=7 | 458 | 35.92 | |
| 4+3=7 | 143 | 11.22 | |
| 8–10 | 115 | 9.02 | |
| Extracapsular extension | Missing | 17 | 1.33 |
| No | 877 | 68.78 | |
| Yes | 381 | 29.88 | |
| Seminal vesicle Invasion | Missing | 17 | 1.33 |
| No | 1177 | 92.31 | |
| Yes | 81 | 6.35 | |
| Surgical Margins | Missing | 179 | 14.04 |
| Positive | 385 | 30.20 | |
| Negative | 711 | 55.76 | |
| AZGP1 protein IHC | Missing | 139 | 10.90 |
| Absent | 252 | 19.76 | |
| Weak | 240 | 18.82 | |
| Moderate | 372 | 29.18 | |
| Strong | 272 | 21.33 | |
| AZGP1 RISH | Missing | 186 | 14.59 |
| Absent | 252 | 19.76 | |
| Weak | 344 | 26.98 | |
| Moderate | 257 | 20.16 | |
| Strong | 236 | 18.51 |
Expression levels of AZGP1 protein measured by IHC and RNA by RISH were associated with each other, although the correlation was modest. When compared on a per core basis, the Kappa value for the 4 staining groups between IHC and RISH was only 0.15 (95% CI: 0.12 – 0.18), although the correlation by Chi-square test was highly significant (p < 0.0001). When IHC and RISH were assessed for each patient by grouping results on the 3 cores for each patient tumor sample the Kappa improved to 0.34 (95% CI: 0.30–0.38) and the Chi-square test remained highly significant (p < 0.0001). Grouping patient samples into absent/weak compared to moderate/strong expression did not improve the correlation between RISH and IHC (Kappa: 0.34; 95% CI: 0.29–0.40; p < 0.0001 by Chi-square test).
AZGP1 expression and RFS after radical prostatectomy
Previous reports have demonstrated that AZGP1 expression assessed by IHC is prognostic in prostate cancer when cases are split categorically into absent/weak expression compared to moderate/high level expression (15,17). Our objective was to test whether splitting samples in this fashion could be validated in our carefully selected cases of patients who had undergone radial prostatectomy and had associated detailed clinical data including long-term follow-up. Kaplan-Meier analysis demonstrated that absent and weak expression of AZGP1 protein showed significantly worse RFS compared to moderate and high expressing tumors (Supplementary Figure 1A). Moreover, RFS for the absent and weak staining were virtually identical as was RFS for the moderate and high level expression categories. These findings validated previous groupings of staining into 2 categories for IHC and therefore samples were divided into absent weak vs. moderate/strong for the remaining analyses.
AZGP1 expression levels measured by RISH showed a similar pattern to those seen with IHC, although with some differences (Supplementary Figure 1B). RISH expression level was a weaker predictor of RFS after prostatectomy (P = 0.011, log-rank test) compared to IHC (P < 0.0001, log-rank test). In addition, AZGP1 expression level assessed by RISH did not segregate into 2 discrete groups. While RFS appeared to be similar between absent and weak staining by RISH, moderate staining appeared to have intermediate outcomes compared to these groups and those that expressed high levels of AZGP1 RNA. However, the differences in RFS between moderate expressing cases and high and low expressing cases was small. Therefore, to allow for comparison of IHC and RISH results, we grouped RISH cases into absent/weak and moderate/strong staining.
AZGP1 protein and RNA expression and clinicopathological features
AZGP1 levels by IHC and RISH were tested for their association with clinical and pathologic features (Table 2). For both IHC and RISH, absent/weak expression of AZGP1 was associated with adverse clinical features including positive surgical margins (PSM), extracapsular extension (ECE), and higher Gleason score (GS). However, absent/weak expression was not associated with seminal vesicle invasion (SVI), pre-operative serum PSA levels, or patient age (all P > 0.05). Lymph node status was not available for approximately half of the cases and therefore was not included in the analysis. Taken together, AZGP1 expression status is associated with many, but not all clinical features important in prostate cancer prognosis after surgery.
Table 2.
AZGP1 staining status and pathological parameters
| Feature | Status | IHC low | IHC high | P value | RISH low | RISH high | P value |
|---|---|---|---|---|---|---|---|
| Surgical margins | Yes | 166 | 181 | 0.004 | 199 | 131 | 0.006 |
| No | 242 | 389 | 306 | 297 | |||
| SV invasion | Yes | 38 | 35 | 0.14 | 43 | 27 | 0.27 |
| No | 448 | 599 | 547 | 457 | |||
| Extracaps exten | Yes | 169 | 173 | 0.004 | 213 | 116 | <0.0001 |
| No | 313 | 467 | 375 | 372 | |||
| Gleason score | ≤ 6 | 189 | 281 | 0.003 | 272 | 181 | 0.009 |
| 3+4=7 | 177 | 243 | 209 | 192 | |||
| 4+3=7 | 54 | 74 | 56 | 70 | |||
| 8–10 | 66 | 45 | 56 | 46 |
P values by Chi-square test
AZGP1 expression and clinical outcomes
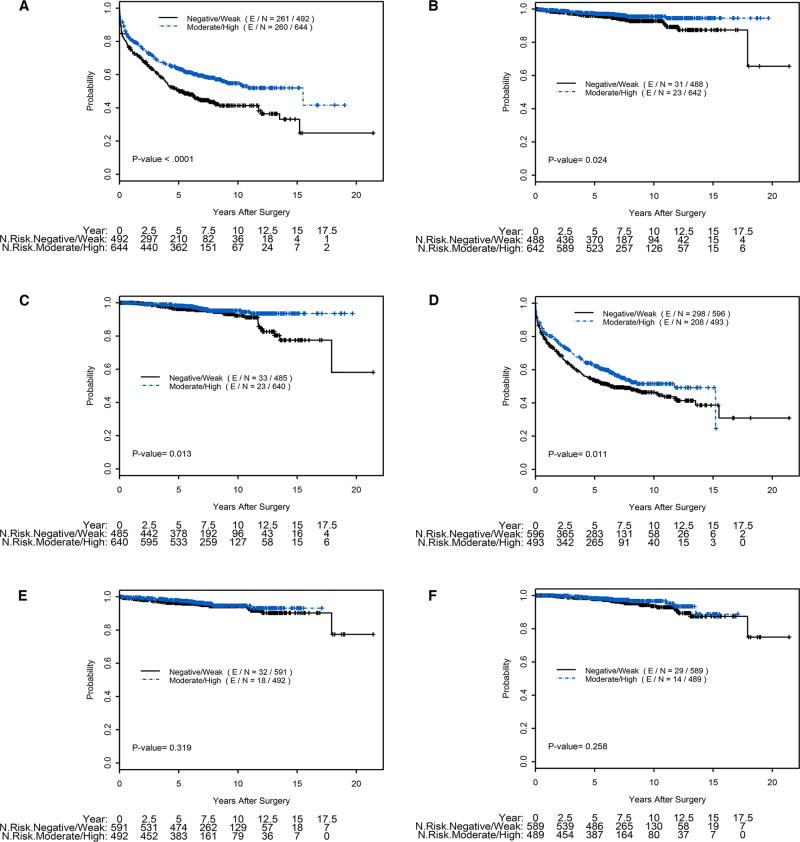
In univariable Cox proportional hazards analysis, absent or weak staining for AZGP1 by IHC was associated with significantly worse RFS (HR=1.49; 95% CI 1.26, 1.77; P<0.0001). Absent/weak AZGP1 expression was also associated with worse DSS (HR=1.84; P=0.03) and OS (HR=1.94;p=0.01). Likewise, absent/weak expression of AZGP1 by RISH was associated with worse RFS, although to a lesser degree (HR=1.26; 95% CI 1.05, 1.50; P=0.01). However, AZGP1 expression by RISH was not associated with DSS or OS (P=0.32 and P=0.26, respectively). Univariable Cox proportional hazards analysis for AZGP1 expression levels and clinical and pathological data are summarized in Table 3. Kaplan-Meier analysis demonstrated that absent/weak expression AZGP1 IHC expression compared to moderate/high expression was significantly associated with RFS (P < 0.0001, log-rank test), overall survival (P = 0.013, log-rank test), and disease specific survival (P = 0.024, log-rank test) (Figure 2 A–C). Absent/weak expression AZGP1 RNA by RISH was also associated with worse RFS (P = 0.011, log-rank test), but not with OS or DSS (Figure 2 D–F).
Table 3.
Univariable Cox proportional hazards model for recurrence free survival
| Factor | Comparison | Hazard Ratio | 95% LCL |
95% UCL |
P-value | #Event | #Censored | Total #Pts |
|---|---|---|---|---|---|---|---|---|
| AZGP1 IHC | Negative/Weak vs. Moderate/Strong | 1.49 | 1.26 | 1.77 | <0.0001 | 521 | 615 | 1136 |
| AZGP1 CISH | Negative/Weak vs. Moderate/Strong | 1.26 | 1.05 | 1.50 | 0.01 | 506 | 583 | 1089 |
| Margin | Pos vs. Neg | 2.08 | 1.74 | 2.48 | <.0001 | 495 | 601 | 1096 |
| SVI | Pos vs. Neg | 3.38 | 2.61 | 4.38 | <.0001 | 568 | 690 | 1258 |
| ECE | Pos vs. Neg | 1.92 | 1.62 | 2.27 | <.0001 | 571 | 687 | 1258 |
| Gleason | 3+4 vs. ≤6 | 1.43 | 1.18 | 1.74 | 0.0003 | 570 | 695 | 1265 |
| 4+3 vs. ≤6 | 2.39 | 1.87 | 3.06 | <.0001 | ||||
| 8–10 vs. ≤6 | 2.39 | 1.82 | 3.13 | <.0001 | ||||
| Age | 1 unit increase | 1.00 | 0.99 | 1.01 | 0.65 | 559 | 610 | 1169 |
| Log(PSA) | 1 unit increase | 1.87 | 1.63 | 2.15 | <.0001 | 531 | 616 | 1147 |
Figure 2. Kaplan-Meier plots of clinical outcome with staining categorized as absent/weak vs. moderate/high AZGP1 expression levels.
A) AZGP1 IHC and Recurrence Free Survival; B) AZGP1 IHC and Disease Specific Survival; C) AZGP1 IHC and Overall Survival; D) AZGP1 RISH and Recurrence Free Survival; E) AZGP1 RISH and Disease Specific Survival; F) AZGP1 RISH and Overall Survival.
To evaluate whether AZGP1 IHC or RISH expression levels provided prognostic information independent of clinical variables, we performed multivariable Cox proportional hazards analysis using a backwards elimination procedure to identify the final model for each endpoint (Table 4). For RFS, absent/weak AZGP1 expression levels assessed by either IHC or RISH were independently associated with worse clinical outcome (HR=1.39; P=0.002 and HR 1.28; P=0.02, respectively), as were presence of positive surgical margins, extracapsular extension, seminal vesicle invasion, higher pre-operative PSA and increasing Gleason score. The concordance index (C-index) for the model including margins, SVI, GS and log[PSA] was 0.656 and improved to 0.659 with the addition of ECE, or to 0.661 with the addition AZGP1 RISH. A model including margins, SVI, ECE, GS, log[PSA] improved the C-index from 0.659 to 0.665 with addition of AZGP1 IHC. However, AZGP1 expression assessed either by IHC or RISH was not associated with DSS or OS on multivariable analysis. DSS was associated only with Gleason score and pre-operative PSA and OS survival was associated only with Gleason score and age as we have reported previously (13). The relatively small number of prostate cancer deaths or metastases (n=54) or deaths from all causes (n=71) limited our ability to test the association of the biomarkers with these endpoints.
Table 4.
Multivariable Cox proportional hazards model for recurrence free survival
| Factor | Comparison | Hazard Ratio | 95% LCL | 95% UCL | P-value |
|---|---|---|---|---|---|
| AZGP1 IHC N=835; E=382 | Negative/Weak vs. Moderate/Strong | 1.39 | 1.13 | 1.71 | 0.002 |
| Log(PSA) | 1 unit increase | 1.43 | 1.21 | 1.68 | <.0001 |
| margin | Pos vs. Neg | 1.62 | 1.31 | 2.02 | <.0001 |
| SVI | Pos vs. Neg | 2.20 | 1.58 | 3.06 | <.0001 |
| ECE | Pos vs. Neg | 1.26 | 1.01 | 1.58 | 0.04 |
| Gleason | 3+4 vs. ≤6 | 1.19 | 0.93 | 1.52 | 0.16 |
| 4+3 vs. ≤6 | 1.99 | 1.47 | 2.69 | <.0001 | |
| 8–10 vs. ≤6 | 1.43 | 1.02 | 1.99 | 0.04 | |
| AZGP1 CISH N=811; E=377 | Negative/Weak vs. Moderate/Strong | 1.28 | 1.04 | 1.58 | 0.02 |
| Log(PSA) | 1 unit increase | 1.46 | 1.24 | 1.73 | <.0001 |
| margin | Pos vs. Neg | 1.71 | 1.39 | 2.12 | <.0001 |
| SVI | Pos vs. Neg | 2.26 | 1.62 | 3.15 | <.0001 |
| Gleason | 3+4 vs. <=6 | 1.22 | 0.96 | 1.57 | 0.11 |
| 4+3 vs. <=6 | 2.12 | 1.57 | 2.86 | <.0001 | |
| 8–10 vs. <=6 | 1.60 | 1.15 | 2.23 | 0.006 |
Given our interest in identifying prognostic biomarkers for selection of patients for active surveillance, we evaluated whether AZGP1 expression could predict outcome in patients with GS ≤ 3+3=6. In univariable Cox proportional hazards analysis, AZGP1 IHC (HR 1.8) and RISH (HR 1.9) remained significant predictors of outcome. In a multivariable model including PSA, SVI, ECE and SM, both remained significant (HR 1.7 for both) (Supplemental Table 1). Kaplan Meier analysis confirmed strong association of AZGP1 RISH and IHC with RFS (Supplemental Figure 2). The C-index improved from 0.618 to 0.662 for IHC and 0.658 for RISH over the clinical model that included PSA, SVI, ECE and SM for patients with GS ≤ 3+3=6.
Discussion
There are no immunohistochemical markers of prognosis in clinical use to aid in the management of prostate cancer, despite clear clinical needs, and despite the number of candidates reported in the literature. One recurring issue has been the lack of meaningful validation for many biomarkers (21). To address this need, we developed the Canary multi-institutional TMA with the explicit design to validate candidate biomarkers of prognosis in clinically localized low and intermediate risk disease (12). Using this platform, we have validated PTEN copy alterations using FISH and PTEN protein expression using a clinical grade assay, and Ki67 staining as providing prognostic information independent of clinical and pathological variables in our tumor set (14,22,23). Furthermore, we have shown that ERG and SPINK1 protein expression are not predictive of clinical outcome (13). Here we demonstrate that AZGP1 protein and RNA expression provide independent prediction of RFS after prostatectomy. Furthermore, AZGP1 protein expression by itself correlates with OS as well as DSS and metastases. Since AZGP1 protein expression shows greater hazard ratios, more significant p-values and is readily measured in nearly all pathology laboratories, AZGP1 IHC should be further developed for use in clinical practice, possibly as part of a panel of IHC prognostic biomarkers.
Several previous studies have nominated AZGP1 as a candidate biomarker of prognosis. The role of AZGP1 in prostate cancer prognosis was first identified by our group based on the observation that AZGP1 expression was highly correlated with a gene-expression subtype of prostate cancer comprised of low risk tumors with favorable outcome (15). In addition, we showed that moderate/high AZGP1 protein expression was associated with improved RFS on an independent dataset of prostate tumors that did not overlap with cases included in the current study. Subsequently several groups have demonstrated that AZGP1 protein expression assessed by IHC is correlated with recurrence-free survival in univariable and multivariable analyses (16,18,19). Furthermore, Henshall et al. showed that loss of expression of AZGP1 by IHC was associated with the development of metastatic disease in 228 men after prostatectomy, in agreement with our findings (17). Recently Burdelski et al. have shown that AZGP1 protein expression is a strong independent predictor of clinical outcome in a set of 8510 patients operated on in Germany (24). Given the relative strengths of their and our studies, AZGP1 appears to be a highly validated biomarker of prognosis in prostate cancer.
Decreased levels of AZGP1 RNA expression have also been associated with adverse clinical outcome in prostate cancer. AZGP1 transcript levels have been shown in meta-analyses and cross-validation studies to correlate with RFS after radical prostatectomy (15,20,25). In a recent tiered approach to biomarker identification and validation, AZGP1 was identified as one of a set of 12 transcripts that predicts outcome in radical prostatectomy patients, including metastases (20). Analysis of pre-treatment biopsies using a commercial test using these 12 transcripts and 5 control genes has been shown to predict upgrading and upstaging in men undergoing radical prostatectomy. Ours is the first study to assess AZGP1 RNA expression levels by RISH where we found it remains an independent biomarker of prognosis. While the correlation between RNA and protein levels was modest, they were highly significant, suggesting that AZGP1 expression is regulated largely at the transcript level. However, whereas protein expression appears to show a threshold between weak and moderate staining in influencing outcome, the data from RISH and RNA expression studies do not disclose such a threshold, and RNA expression levels appear to be a continuous predictor of risk. Furthermore, RNA expression measured by RISH was a weaker predictor of outcome compared to AZGP1 IHC. However, RISH is a relatively non-quantitative measure of RNA expression, and it is unclear whether measurement of AZGP1 RNA expression using a more quantitative assay by itself or in the context of other genes will better predict outcome compared to AZGP1 IHC. Direct comparison on identical samples will be necessary to evaluate the relative performance of quantitative measures of AZGP1 RNA and AZGP1 IHC.
Our study, coupled with previous work, strongly suggests that AZGP1 IHC could have value as a prognostic biomarker in prostate cancer and could find use in several clinical settings including selection of patients for active surveillance and identification of patients at risk for recurrence after radical prostatectomy that would benefit from adjuvant radiation therapy (26). AZGP1 expression provides independent, albeit modest improvement in predicting recurrence after radical prostatectomy, likely because of its association with adverse pathological features. However, this association with adverse pathology and the strong association with outcome in Gleason score ≤3+3=6 patients suggest its greatest utility could be selection of patients for active surveillance. One significant challenge will be developing clinical grade IHC assays for AZGP1 with well-characterized antibodies. Available antibodies against AZGP1 are polyclonal and ongoing availability for a clinical assay could be a significant issue. It is noteworthy, however, that loss of AZGP1 expression by IHC has been shown to be highly prognostic in several tumor types, including breast, gastric, and liver cancer (27–29). Therefore, development of clinical grade assays with a well-characterized monoclonal antibody could find applications beyond prostate cancer.
The mechanisms by which loss of AZGP1 affects cancer aggressiveness are currently unknown. AZGP1 is an androgen-regulated gene, and AZGP1 protein is secreted at high levels in the prostatic fluid (30,31). Gene-expression profiling suggests that androgens regulate pathways associated with terminal differentiation in the prostate including secretory proteins (32,33). It is therefore possible that loss of AZGP1 merely reflects loss of terminal differentiation in more aggressive cancers. However, it is equally possible that AZGP1 plays an active role in suppressing carcinogenesis, particularly since it is prognostic across several tumor types. AZGP1 was originally described as a secreted member of the MHC1 family and it is possible it modulates immune response to the tumor (34,35). Furthermore, in colon cancer cell lines, forced over-expression of AZGP1 results in down-regulation of the mTOR signaling pathway, decreased proliferation and invasion, increased apoptosis and mitotic arrest (36). In pancreatic cancer cell lines, AZGP1 has been shown to act as a tumor suppressor and loss of expression induces epithelial to mesenchymal transition, increases invasion in activates cell survival programs (37). Clearly, additional work will be necessary to discover the role of AZGP1 in cancer progression.
Our study has some limitations. First, the relative age of some of the samples could affect RNA stability and influence RISH results, which might account for its lower predictive performance compared to IHC. Second, patient samples were collected retrospectively and, although we tried to limit biases by using a case control design, potential confounders are possible including changes in practice patterns or patient populations over time. Finally, rather than select cases that reflect the distribution of GS and RFS typical of the population of patients undergoing radical prostatectomy, we over-sampled recurrent low grade (GS 3+3=6), balanced recurrent and non-recurrent cases with GS 3+4=7 and 4+3=7 and oversampled non-recurrent GS≥8 cancers. While this design has advantages in identifying biomarkers independent of GS, it will affect the weight of GS in univariable and multivariable models in predictions of clinical outcome.
Conclusions
Loss of expression of AZGP1 protein and RNA are associated with adverse pathological features at radical prostatectomy and are independently associated with RFS after surgery. Loss of expression is associated with OS and DSS in our cohort. Together these findings identify AZGP1 IHC as an independent prognostic marker in prostate cancer and provide the basis for development of a clinical grade assay. Further work will be necessary to define the relative performance of AZGP1 protein and RNA based assays in assessing clinical outcome and to define the role of AZGP1 in suppressing cancer progression.
Supplementary Material
Supplementary Figure 1: Kaplan-Meier plots of RFS after radical prostatectomy for absent, weak, moderate and strong staining for AZGP1: A) AZGP1 levels by IHC; B) AZGP1 levels by RISH.
Acknowledgments
This work was supported by the Canary Foundation, the Burroughs Wellcome Fund (#1007519 to JRP), the Department of Defense (W81XWH-11-1-0380 to JDB and ZF), the NCI (R01 CA122246 to JRP and U01CA196387 to JDB), and the NCI Early Detection Research Network (CA152737 to JDB and CA08636815 to ZF).
Footnotes
Conflict of Interest: The authors declare no conflicts of interest related to the work described in this manuscript.
References
- 1.USPSTF. Screening for prostate cancer: U.S. Preventive Services Task Force recommendation statement. Ann Internal Med. 2008;149(3):185–191. doi: 10.7326/0003-4819-149-3-200808050-00008. [DOI] [PubMed] [Google Scholar]
- 2.Saquib N, Saquib J, Ioannidis JP. Does screening for disease save lives in asymptomatic adults? Systematic review of meta-analyses and randomized trials. Intl J Epidemiol. 2015;44(1):264–277. doi: 10.1093/ije/dyu140. [DOI] [PubMed] [Google Scholar]
- 3.Andriole GL, Crawford ED, Grubb RL, 3rd, Buys SS, Chia D, Church TR, Fouad MN, Gelmann EP, Kvale PA, Reding DJ, Weissfeld JL, Yokochi LA, O'Brien B, Clapp JD, Rathmell JM, Riley TL, Hayes RB, Kramer BS, Izmirlian G, Miller AB, Pinsky PF, Prorok PC, Gohagan JK, Berg CD. Mortality results from a randomized prostate-cancer screening trial. N Engl J Med. 2009;360(13):1310–1319. doi: 10.1056/NEJMoa0810696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schroder FH, Hugosson J, Roobol MJ, Tammela TL, Ciatto S, Nelen V, Kwiatkowski M, Lujan M, Lilja H, Zappa M, Denis LJ, Recker F, Paez A, Maattanen L, Bangma CH, Aus G, Carlsson S, Villers A, Rebillard X, van der Kwast T, Kujala PM, Blijenberg BG, Stenman UH, Huber A, Taari K, Hakama M, Moss SM, de Koning HJ, Auvinen A. Prostate-cancer mortality at 11 years of follow-up. N Engl J Med. 2012;366(11):981–990. doi: 10.1056/NEJMoa1113135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brooks JD. Managing localized prostate cancer in the era of prostate-specific antigen screening. Cancer. 2013;119(22):3906–3909. doi: 10.1002/cncr.28301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter HB. Active surveillance for favorable risk prostate cancer. Curr Opin Urol. 2015;25(3):230–231. doi: 10.1097/MOU.0000000000000167. [DOI] [PubMed] [Google Scholar]
- 7.Klotz L, Vesprini D, Sethukavalan P, Jethava V, Zhang L, Jain S, Yamamoto T, Mamedov A, Loblaw A. Long-term follow-up of a large active surveillance cohort of patients with prostate cancer. J Clin Onc. 2015;33(3):272–277. doi: 10.1200/JCO.2014.55.1192. [DOI] [PubMed] [Google Scholar]
- 8.Newcomb LF, Thompson IM, Jr, Boyer HD, Brooks JD, Carroll PR, Cooperberg MR, Dash A, Ellis WJ, Fazli L, Feng Z, Gleave ME, Kunju P, Lance RS, McKenney JK, Meng MV, Nicolas MM, Sanda MG, Simko J, So A, Tretiakova MS, Troyer DA, True LD, Vakar-Lopez F, Virgin J, Wagner AA, Wei JT, Zheng Y, Nelson PS, Lin DW, Canary PI. Outcomes of Active Surveillance for Clinically Localized Prostate Cancer in the Prospective, Multi-Institutional Canary PASS Cohort. J Urol. 2016;195(2):313–320. doi: 10.1016/j.juro.2015.08.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dianat SS, Carter HB, Pienta KJ, Schaeffer EM, Landis PK, Epstein JI, Trock BJ, Macura KJ. Magnetic resonance-invisible versus magnetic resonance-visible prostate cancer in active surveillance: a preliminary report on disease outcomes. Urology. 2015;85(1):147–153. doi: 10.1016/j.urology.2014.06.085. [DOI] [PubMed] [Google Scholar]
- 10.Newcomb LF, Brooks JD, Carroll PR, Feng Z, Gleave ME, Nelson PS, Thompson IM, Lin DW. Canary Prostate Active Surveillance Study: design of a multi-institutional active surveillance cohort and biorepository. Urology. 2010;75(2):407–413. doi: 10.1016/j.urology.2009.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walton Diaz A, Shakir NA, George AK, Rais-Bahrami S, Turkbey B, Rothwax JT, Stamatakis L, Hong CW, Siddiqui MM, Okoro C, Raskolnikov D, Su D, Shih J, Han H, Parnes HL, Merino MJ, Simon RM, Wood BJ, Choyke PL, Pinto PA. Use of serial multiparametric magnetic resonance imaging in the management of patients with prostate cancer on active surveillance. Urol Oncol. 2015;33(5):202 e201–207. doi: 10.1016/j.urolonc.2015.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hawley S, Fazli L, McKenney JK, Simko J, Troyer D, Nicolas M, Newcomb LF, Cowan JE, Crouch L, Ferrari M, Hernandez J, Hurtado-Coll A, Kuchinsky K, Liew J, Mendez-Meza R, Smith E, Tenggara I, Zhang X, Carroll PR, Chan JM, Gleave M, Lance R, Lin DW, Nelson PS, Thompson IM, Feng Z, True LD, Brooks JD. A model for the design and construction of a resource for the validation of prognostic prostate cancer biomarkers: the Canary Prostate Cancer Tissue Microarray. Adv Anat Pathol. 2013;20(1):39–44. doi: 10.1097/PAP.0b013e31827b665b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brooks JD, Wei W, Hawley S, Auman H, Newcomb L, Boyer H, Fazli L, Simko J, Hurtado-Coll A, Troyer DA, Carroll PR, Gleave M, Lance R, Lin DW, Nelson PS, Thompson IM, True LD, Feng Z, McKenney JK. Evaluation of ERG and SPINK1 by Immunohistochemical Staining and Clinicopathological Outcomes in a Multi-Institutional Radical Prostatectomy Cohort of 1067 Patients. PloS One. 2015;10(7):e0132343. doi: 10.1371/journal.pone.0132343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Troyer DA, Jamaspishvili T, Wei W, Feng Z, Good J, Hawley S, Fazli L, McKenney JK, Simko J, Hurtado-Coll A, Carroll PR, Gleave M, Lance R, Lin DW, Nelson PS, Thompson IM, True LD, Brooks JD, Squire JA. A multicenter study shows PTEN deletion is strongly associated with seminal vesicle involvement and extracapsular extension in localized prostate cancer. Prostate. 2015;75(11):1206–1215. doi: 10.1002/pros.23003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E, Montgomery K, Ferrari M, Egevad L, Rayford W, Bergerheim U, Ekman P, DeMarzo AM, Tibshirani R, Botstein D, Brown PO, Brooks JD, Pollack JR. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci, USA. 2004;101(3):811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Descazeaud A, de la Taille A, Allory Y, Faucon H, Salomon L, Bismar T, Kim R, Hofer MD, Chopin D, Abbou CC, Rubin MA. Characterization of ZAG protein expression in prostate cancer using a semi-automated microscope system. Prostate. 2006;66(10):1037–1043. doi: 10.1002/pros.20405. [DOI] [PubMed] [Google Scholar]
- 17.Henshall SM, Horvath LG, Quinn DI, Eggleton SA, Grygiel JJ, Stricker PD, Biankin AV, Kench JG, Sutherland RL. Zinc-alpha2-glycoprotein expression as a predictor of metastatic prostate cancer following radical prostatectomy. JNCI. 2006;98(19):1420–1424. doi: 10.1093/jnci/djj378. [DOI] [PubMed] [Google Scholar]
- 18.Mills J, Oliver A, Sherwin JC, Frydenberg M, Peters JS, Costello A, Harewood L, Love C, Redgrave N, van Golen KL, Bailey M, Pedersen J. Utility of RhoC and ZAG protein expression as biomarkers for prediction of PSA failure following radical prostatectomy for high grade prostate cancer. Pathol. 2012;44(6):513–518. doi: 10.1097/PAT.0b013e3283581780. [DOI] [PubMed] [Google Scholar]
- 19.Severi G, FitzGerald LM, Muller DC, Pedersen J, Longano A, Southey MC, Hopper JL, English DR, Giles GG, Mills J. A three-protein biomarker panel assessed in diagnostic tissue predicts death from prostate cancer for men with localized disease. Cancer Med. 2014;3(5):1266–1274. doi: 10.1002/cam4.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein EA, Cooperberg MR, Magi-Galluzzi C, Simko JP, Falzarano SM, Maddala T, Chan JM, Li J, Cowan JE, Tsiatis AC, Cherbavaz DB, Pelham RJ, Tenggara-Hunter I, Baehner FL, Knezevic D, Febbo PG, Shak S, Kattan MW, Lee M, Carroll PR. A 17-gene assay to predict prostate cancer aggressiveness in the context of Gleason grade heterogeneity, tumor multifocality, and biopsy undersampling. Eur Urol. 2014;66(3):550–560. doi: 10.1016/j.eururo.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 21.Brooks JD. Translational genomics: the challenge of developing cancer biomarkers. Genome Res. 2012;22(2):183–187. doi: 10.1101/gr.124347.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lotan TL, Wei W, Morais CL, Hawley ST, Fazli L, Hurtado-Coll A, Troyer D, McKenney JK, Simko J, Carroll PR, Gleave M, Lance R, Lin DW, Nelson PS, Thompson IM, True LD, Feng Z, Brooks JD. PTEN Loss by Clinical-Grade Immunohistochemistry Assay is Associated with Worse Recurrence Free Survival in Prostate Cancer. Eur Urol Focus. 2015 doi: 10.1016/j.euf.2015.07.005. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tretiakova MS, Wei W, Boyer HD, Newcomb LF, Hawley ST, Auman H, Vakar-Lopez F, McKenney JK, Fazli L, Simko J, Troyer D, Hurtado-Coll A, Thompson IM, Carroll PR, Ellis WJ, Gleave ME, Nelson PS, Lin DW, True LD, Feng Z, Brooks JD. Prognostic Value of Ki67 in Localized Prostate Carcinoma: A Multi-Institutional Study of >1,000 Prostatectomies. Prostate Cancer Prostate Dis. 2016 doi: 10.1038/pcan.2016.12. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burdelski C, Kleinhans S, Kluth M, Hube-Magg C, Minner S, Koop C, Graefen M, Heinzer H, Tsourlakis MC, Wilczak W, Marx A, Sauter G, Wittmer C, Huland H, Simon R, Schlomm T, Steurer S. Reduced AZGP1 expression is an independent predictor of early PSA recurrence and associated with ERG-fusion positive and PTEN deleted prostate cancers. Intl J Cancer Journal. 2016;138(5):1199–1206. doi: 10.1002/ijc.29860. [DOI] [PubMed] [Google Scholar]
- 25.Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd C, Tamayo P, Renshaw AA, D'Amico AV, Richie JP, Lander ES, Loda M, Kantoff PW, Golub TR, Sellers WR. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell. 2002;1(2):203–209. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- 26.Yip PY, Kench JG, Rasiah KK, Benito RP, Lee CS, Stricker PD, Henshall SM, Sutherland RL, Horvath LG. Low AZGP1 expression predicts for recurrence in margin-positive, localized prostate cancer. Prostate. 2011;71(15):1638–1645. doi: 10.1002/pros.21381. [DOI] [PubMed] [Google Scholar]
- 27.Huang CY, Zhao JJ, Lv L, Chen YB, Li YF, Jiang SS, Wang W, Pan K, Zheng Y, Zhao BW, Wang DD, Chen YM, Yang L, Zhou ZW, Xia JC. Decreased expression of AZGP1 is associated with poor prognosis in primary gastric cancer. PloS One. 2013;8(7):e69155. doi: 10.1371/journal.pone.0069155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang Y, Li LZ, Zhang CZ, Yi C, Liu LL, Zhou X, Xie GB, Cai MY, Li Y, Yun JP. Decreased expression of zinc-alpha2-glycoprotein in hepatocellular carcinoma associates with poor prognosis. J Translat Med. 2012;10:106. doi: 10.1186/1479-5876-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parris TZ, Kovacs A, Aziz L, Hajizadeh S, Nemes S, Semaan M, Forssell-Aronsson E, Karlsson P, Helou K. Additive effect of the AZGP1, PIP, S100A8 and UBE2C molecular biomarkers improves outcome prediction in breast carcinoma. Intl J Cancer. 2014;134(7):1617–1629. doi: 10.1002/ijc.28497. [DOI] [PubMed] [Google Scholar]
- 30.Bohm M, Locke WJ, Sutherland RL, Kench JG, Henshall SM. A role for GATA-2 in transition to an aggressive phenotype in prostate cancer through modulation of key androgen-regulated genes. Oncogene. 2009;28(43):3847–3856. doi: 10.1038/onc.2009.243. [DOI] [PubMed] [Google Scholar]
- 31.Zhao H, Kim Y, Wang P, Lapointe J, Tibshirani R, Pollack JR, Brooks JD. Genome-wide characterization of gene expression variations and DNA copy number changes in prostate cancer cell lines. Prostate. 2005;63(2):187–197. doi: 10.1002/pros.20158. [DOI] [PubMed] [Google Scholar]
- 32.DePrimo SE, Diehn M, Nelson JB, Reiter RE, Matese J, Fero M, Tibshirani R, Brown PO, Brooks JD. Transcriptional programs activated by exposure of human prostate cancer cells to androgen. Genome Biol. 2002;3(7) doi: 10.1186/gb-2002-3-7-research0032. RESEARCH0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nelson PS, Clegg N, Arnold H, Ferguson C, Bonham M, White J, Hood L, Lin B. The program of androgen-responsive genes in neoplastic prostate epithelium. Proc Natl Acad Sci, USA. 2002;99(18):11890–11895. doi: 10.1073/pnas.182376299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hassan MI, Waheed A, Yadav S, Singh TP, Ahmad F. Zinc alpha 2-glycoprotein: a multidisciplinary protein. Mol Cancer Res. 2008;6(6):892–906. doi: 10.1158/1541-7786.MCR-07-2195. [DOI] [PubMed] [Google Scholar]
- 35.Sanchez LM, Lopez-Otin C, Bjorkman PJ. Biochemical characterization and crystalization of human Zn-alpha2-glycoprotein, a soluble class I major histocompatibility complex homolog. Proc Natl Acad Sci, USA. 1997;94(9):4626–4630. doi: 10.1073/pnas.94.9.4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang L, Tian X, Lu Y, Jia M, Wu P, Huang P. Alpha-2-glycoprotein 1(AZGP1) regulates biological behaviors of LoVo cells by down-regulating mTOR signaling pathway and endogenous fatty acid synthesis. PloS One. 2014;9(6):e99254. doi: 10.1371/journal.pone.0099254. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Kong B, Michalski CW, Hong X, Valkovskaya N, Rieder S, Abiatari I, Streit S, Erkan M, Esposito I, Friess H, Kleeff J. AZGP1 is a tumor suppressor in pancreatic cancer inducing mesenchymal-to-epithelial transdifferentiation by inhibiting TGF-beta-mediated ERK signaling. Oncogene. 2010;29(37):5146–5158. doi: 10.1038/onc.2010.258. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Kaplan-Meier plots of RFS after radical prostatectomy for absent, weak, moderate and strong staining for AZGP1: A) AZGP1 levels by IHC; B) AZGP1 levels by RISH.