Abstract
In recent years, considerable progress has been made in the molecular mechanisms of epigenetics in disease development and progression, the reversible characteristics of epigenetic modification provide new insights for the treatment of such diseases. The pathogenesis of diabetic retinopathy (DR) has not yet been fully understood, treatment of refractory and recurrent diabetic macular edema remains a big change in clinical practice. This review emphasizes that reversibility of epigenetic modification could provide a new strategy for the prevention and treatment of diseases.
Keywords: Diabetic retinopathy, Gene, Epigenetic modifications, DNA methylation, Non-coding RNA, Chromatin remodeling, Histone modification, Therapeutic target
Background
The Human Genome Project found that the protein-coding DNA accounted for only 2% of the entire genome, and thus, proposed that some non-coding regions, such as capable of producing non-coding RNA, repeat fragments, and transposons, may also exercise certain functions. Moreover, the development of some diseases cannot be explained solely by the change in DNA sequence; other factors, including living environment, mental conditions, and stress may also play a vital role in the onset of diseases. Thus, the term epigenetics refers to heritable modifications that are not involved in the changes in the DNA sequence.
Epigenetics is also known as “prefix genetics,” “external genetics,” or “post-genetics,” indicating that in the absence of changes in the DNA sequence, the function of genes could undergo changes that are reversible and heritable. Occasionally, it may also refer to the studies on the process of physiological development [1]. Vital mechanisms of epigenetic modification include DNA methylation, histone acetylation, non-coding RNA regulation, and chromatin remodeling. These mechanisms are intrinsically involved in the development of various diseases including cancer, autoimmune diseases, neurodegenerative diseases, psychiatric diseases, and addictive diseases [2]. In addition to systemic diseases, other ophthalmic diseases such as dry eye, keratitis, uveitis, glaucoma, myopia, and fundus neovascularization may also be associated with epigenetic modifications. Recently, DR, the leading cause of blindness among the working population, has attracted particular attention, due to its high rate of causing blindness, increasing incidence, and significance in public health.
The four mechanisms of epigenetic modification are discussed below.
DNA methylation
DNA methylation is the earliest discovered mechanism in epigenetic modification. It refers to the addition of methyl groups of S-adenosylmethionine (SAM) to DNA molecules catalyzed by DNA methyl transferase (DNMT). In mammalian cells, CpG exists mainly in two forms: one evenly distributed throughout the DNA sequences (60–90% are methylated), and the other grouped in clusters known as the CpG islands (generally protected and remain unmethylated). In the eukaryotic cells, CpG islands are often found in the regulatory regions of coding genes and are involved in gene expression and chromatin structure modification [2]. Hitherto, five enzymes have been identified in the DNMT family, namely DNMT1, DNMT2, DNMT3a, DNMT3b, and DNMT3L; however, among them, only DNMT1, DNMT3a, and DNMT3b are active for DNA methylation. DNA methylation deactivates the target genes or causes conformational changes in DNA, thereby affecting the protein-DNA interaction. In a human body, there are three states of DNA methylation: persistent hypomethylation, induced demethylation, and hypermethylation. The states of methylation are closely related to disease development; persistent hypomethylation and hypermethylation are found in some cancer cells.
Histone modification
Histone modifications refer to methylation, acetylation, phosphorylation, adenylation, ubiquitination, ADP-ribosylation, and other modifications of histones in the presence of related enzymes. Histones are an integral part of epigenetics. The core histones (H2A, H2B, H3, and H4) could form a nucleosome with two H2A–H2B dimers and one H3–H4 tetramer. The histone core is globular, and only the N-terminal protein tails are unstructured [2]. Histone H1 is the linker histone but does not make up the nucleosome beads. It rather binds to the linker DNA (DNA that distinguishes the two histone complexes), and fixes the position of the nucleosome for the entry and exit of DNA [3]. Histone modification affects the transcriptional activity of genes, thereby playing a significant role in regulating DNA transcription, repair, and replication, as well as, alternative splicing and chromosome condensation [2].
Non-coding RNA
Non-coding RNA refers to RNA molecules that do not encode proteins. These RNAs include those with known functions, such as rRNA, tRNA, snRNA, snoRNA, microRNA (miRNA), and circular RNA and others with unknown functions. miRNA is a single strand RNA containing 21–25 nucleotides that is involved in post-transcriptional regulation. It can bind to the 3′-UTR of target sequences and modulate the activity of mRNA, mainly by degrading the RNA or suppressing the protein translation [4, 5]. In humans, this process affects 1/3rd of the entire genome, and in mammals, it regulates the expression of >60% proteins. Furthermore, miRNA also participates in various pathological processes, including the inhibition of translation initiation, inhibition of translation, as well as, the regulation of mRNA deadenylation and degradation. Each miRNA has multiple target genes, and the same gene can be regulated by different miRNAs. Since the inhibition of protein translation is a critical regulation pathway, the complex mechanism of non-coding RNA realizes the fine regulation of gene expression.
Chromatin remodeling
Chromatin remodeling refers to epigenetic mechanisms based on altered chromatin conformation. Since the nucleosome is the basic structural unit of chromatin, the remodeling of chromatin primarily involves structural or molecular changes in nucleosomes, histones and DNAs during the expression, replication, and recombination of genes. Chromatin remodeling can affect DNA methylation, DNA replication, recombination, and repair as well as gene expression. A previous study showed that, during the embryonic and fetal period, chromatin remodeling could encode “molecular memories” of diseases that a susceptible individual may later suffer in adulthood. A typical example is a cross-sectional study of a large population, which found that prenatal malnutrition and low body weight were associated with the metabolic X syndrome (type II diabetes, cardiovascular disease, and hypertension). The underlying mechanism could be elucidated as such: instead of DNA changes, chromatin remodeling during the embryonic or fetal period may affect the normal development and metabolism in organs [6].
Although different modification mechanisms affect specific epigenetic phenomena independently, they interact with each other and together determine the complex physiological processes. For example, the regulation of DNA methylation and miRNA modification requires the participation of DNTMs; the function of DNTMs, in turn, relies on histone modification, such as H3K9 methylation or histone acetylation. Thus, the inhibition of DNMTS would lead to H3K9 demethylation, and subsequently, the release of miRNA [7].
Advancements in DR epigenetics
Previous studies suggested that the four mechanisms of epigenetic modification play significant roles in DR development. The most typical example was hyperglycemia, considered as a major determinant of the progression of DR. However, despite the well-controlled blood glucose in advanced DM patients, the development or progression of DR has been shown to not be alleviated or improved. This indicated that the exposure to hyperglycemia resulted in a “metabolic memory phenomenon”, wherein the epigenetic modifications played essential roles [8]. In the following section, the significance of the four epigenetic mechanisms in DR development will be elucidated further.
DNA methylation
Previous studies suggest a positive association between DNA methylation and DR development; however, different forms of diabetes may involve various CpG islands to be methylated during the development of DR. Maghbooli et al. studied the content of 5-methylcytosine in leukocytes sampled from the peripheral blood of type II diabetes patients and found that patients with DR showed a significantly higher level of DNA methylation as compared to those without DR. This phenomenon indicated that higher DNA methylation level could be a potential risk factor for DR development. In addition, the study found that with the progression of DR, the level of systemic DNA methylation did not continue to increase in DR patients, suggesting that it occurred only in the early stages of diabetes [9]. Agardh et al. performed a genome-wide analysis of blood DNA methylation in 28 patients with proliferative DR (PDR) and 30 healthy controls. The study identified a total of 349 methylated CpG sites within 233 genes, including TNF, CHI3L1 (YKL-40), CHN2, GIRP, GLRA1, GPX1, AHRR, and BCOR. Most of the CpG islands in PDR patients showed a decreased level of DNA methylation, whereas 28 CpG sites within 17 genes (including AHRR, GIRP, GLRA1, and BCOR) showed increased methylation (P < 0.05). Notably, the genes involved in the natural killer (NK) cell-mediated toxicity pathway showed a significantly higher level of methylation (P = 0.006) [10], indicating that methylation in peripheral blood cells could be used as a predictor for type I diabetes-complicated PDR.
Studies on animal models also suggested that retinal cells under hyperglycemia exhibited an altered level of CpG methylation as compared to normal cells. Tewari et al. analyzed DNA methylation in the retinal tissue of streptozotocin (STZ)-induced diabetic rats with stringently regulated (glycosylated blood glucose 6%) and less controlled blood glucose (glycosylated blood glucose 11%). The study found that when a rat was administered less controlled blood glucose for 3 months followed by another 3 months of strictly controlled blood glucose, the level of polymerase γ-1 (POLG1), an enzyme that participates in the process of prokaryotic DNA replication. continued to decrease, the D-loop was damaged, and the CpG islands located within the regulatory region of POLG were hypermethylated. Further studies also found that the hypermethylated CpG islands in POLG affected its binding with mitochondrial RNA and inhibited the transcriptional activity [11]. This phenomenon was also observed in retinal endovascular cells that were exposed to hyperglycemia, indicating that DNA methylation plays a major role in metabolic memory [12]. An in vitro study revealed that methylation and activation of the matrix metalloproteinase 9 (MMP 9) gene are significantly associated with DR [12]; activated MMP-9 could cause mitochondrial damage and accelerate the apoptosis of retinal vascular endothelia, thereby inducing DR development and progression. Studies performed in human cadaver eyes confirmed that hyperglycemia caused H3K9 acetylation in MMP-9 (Ac-H3K9), and p65 activation resulted in CpG methylation within MMP-9. Both the changes increased the MMP-9 expression and further aggregated the mitochondrial damage. Therefore, a potential therapeutic target may be to regulate methylation and demethylation of MMP-9, thus reducing mitochondrial damage, and DR could be prevented and controlled [13] (Table 1).
Table 1.
The role of DNA methylation in the pathogenesis of diabetic retinopathy
| Refs | Study type | DNA methylation |
|---|---|---|
| Diabetes Metab [9] | Clinical study (serum) | Global DNA methylation↑ |
| BMC Med [10] | Clinical study (blood) | DNA methylation↓ in PDR, differential DNA methylation of 28 CpG sites in 17 genes (AHRR, GIPR, GLRA1, and BCOR) |
| Invest Ophthalmol Vis Sci [11] | Invitro/animal (STZ diabetic mice) Retina |
DNA methylation of polymerase gamma (POLG1) gene ↑ |
| Lab Invest [13] Diabetes [12] |
Invitro/animal (STZ diabetic mice) Retina |
Dnmt1 binding to Matrix metalloproteinase -9 promoter→MMP↑→5mC↓ |
| Invest Ophthalmol Vis Sci [14] | Invitro/animal study (Bovine retinal endothelial) | mtDNA methylation↑ |
Histone modification
Diabetic retinopathy is a neurovascular disease caused by hyperglycemia. Histone modification participates in neuronal apoptosis and vascular leakage, thereby facilitating DR development and progression [15, 16]. Histone methylation and acetylation are the two major mechanisms by which histone modification affects DR development.
Histone methylation
Histone lysine methylation (HLM) plays a vital role in various physiological processes such as gene expression, gene transcription (activation and inhibition), genomic stability [17], stem cell pluripotency [18], tumorigenesis [19] and inflammatory reaction [20]. The methyl transferase SUV39H1 methylates the lysine of histone H3 at the N-terminus, resulting in H3K9me3 [21]. The methyl transferase encoded by SUV39H2 has been demonstrated to lead to H3K9 methylation, resulting in the onset of DR. A recent cohort study comprising of 3000 DM patients also demonstrated that a polymorphism in the SUV39H2 gene was significantly associated with microvascular complications [22]. Since the EZH2- and G9A-mediated epigenetic regulation of HLM is speculated to play a key role in delaying retinal neuronal degradation, it might represent a promising therapeutic target in the future treatment and prevention of DR.
Histone acetylation
The level of histone acetylation was significantly increased in the retina of rats with diabetes. Hyperglycemia caused H3 hyperacetylation, and the inflammatory cytokines, such as TNF and Cox, were significantly increased in cultured Muller cells. However, these changes could be reversed by the inhibitors of histone acetyltransferase [23]. Histone acetylation is associated with the activation of the NF-κB-dependent inflammation signaling pathway. Also, an imbalanced histone acetylation would result in the “metabolic memory phenomenon”, which mainly manifests as hypoacetylation in the promoter region of protective genes and hyperacetylation in the promoter region of pathogenic genes. Zhong et al. found that in STZ-induced diabetic rats, the transcription activity of HDAC1/2/8 (histone deacetylase) was increased in retinal endovascular cells, whereas the activity of HAT (histone acetyltransferase) and the expression of acetylated histone H3 were both decreased. After the blood glucose of the rats was restored to normal level, the above changes were irreversible, indicating that DR development is significantly associated with H3 acetylation, and the latter may participate in the formation of “metabolic memory” [24]. Based on the above results, we speculate that in retinal cells, the H3 deacetylation may downregulate the genes maintaining the redox state and inhibiting cell apoptosis, thereby resulting in the progression of DR.
Oxidative stress also plays a major role in regulating hyperglycemia-dependent histone deacetylation, which is speculated to be a pathogenic factor for the development of diabetic microvascular complications [25, 26]. The reactive oxygen species (ROS) could increase the activity of HDAC while decreasing that of HAT, thereby inhibiting histone acetylation. The mitochondrial dysfunction in the retina could cause apoptosis of endothelial cells and further lead to the development of DR [8]. The lysine-specific demethylase 1 (LSD1) is ascribed as the underlying factor, primarily functioning by decreasing the level of histone H3 dimethyl lysine 9 (H3K9me2) in the promoter region of MMP-9 and increasing the levels of acetyl H3K9 (Ac-H3K9) and p65 [27]. SOD catalyzes the disproportionation of superoxide radicals into oxygen or hydrogen peroxide, thereby aiding the cells in the removal of superoxide molecules. Since hyperglycemia inhibits the protective effect of Mn-SOD on retinal mitochondria, it also accelerates the apoptosis of endovascular cells and induces the development of DR in STZ-induced diabetic rats [28, 29]. In addition, histone modification within the SOD2 gene is another crucial factor causing DR; significantly increased H3K9 methylation and H4K20 acetylation was observed in the promoter and enhancer regions of SOD2, respectively [30]. The lysine in different histones could be modified, and these modifications would result in various alterations. Under hyperglycemia, LSD1 could be activated, which reduces H3K4 acetylation in the SOD2 gene, and ultimately induces the development and progression of DR.
miRNA and DR
The blood retinal barrier (BRB) breakdown and neovascularization are the basic pathogenic characteristics of DR [31]; non-coding RNA, especially miRNA, could regulate the expression of genes involved in DR development, thereby complicating the sophisticated epigenetic modifications of DR.
The blood retinal barrier breakdown is the hallmark of DR [32]. Several miRNAs have been shown to be involved in the pathogenesis of DR. miR-23b-3p regulates high-glucose-induced cellular metabolic memory in cultured human endothelium cells through an SIRT1-dependent signaling pathway [33]. In a diabetic rat model, miR-126 was found to play a potential role in the pathogenesis of DR, which might regulate the expressions of VEGF, Ang-1, and VCAM-1 [34].
Previous studies have shown that miRNA plays an important role in regulating retinal neovascularization [35]. In mice with ischemic retinopathy, microarray analysis identified seven miRNAs with increased expression (miRNA-106a, -146, -181, -199a, -214, -424, and -451) and three miRNAs with decreased expression (miRNA-31, -150, and -184) [36]. miRNA-31 and miRNA-184 are specifically expressed in non-vascular cornea and lens and are found in the retina at a relatively low level [37], suggesting that these miRNAs may inhibit neovascularization in the cornea, lens, and retinal tissues. In rats with ischemic retinopathy, injection of pre-miRNA-31, -150, and -184 downregulate the expression of vascular endothelial growth factor (VEGF), platelet-derived growth factor B (PDGF-B), and hypoxia inducible factor-1α (HIF-1α) [36]; however, the potential targets of these miRNAs have not yet been identified. Nevertheless, the study suggested that elevating the level of miRNA through intraocular injection may become a potential choice for the treatment of DR [38]. An in vitro experiment on human retinal endothelial cells (HRECs) found that high-glucose induction resulted in decreased level of miR-18b, which facilitated cell proliferation and VEGF expression. Notably, miR-18 mediates signal transduction via the activation of the IGF-1/IGFR-1 signaling pathway [39] (Table 2).
Table 2.
miRNA and diabetic retinopathy
| Refs | Study type | Identified miRNA |
|---|---|---|
| Graefes Arch Clin Exp Ophthalmol [40] | Clinical study (serum & vitreous) | miR-15a, miR-320b, miR-93, miR-423-5p, miR-29a, miR-320a |
| Diabetes [23] | Clinical study (serum) | miR-27b, miR-454, miR-28-3p, miR-122, miR-320a, miR-125b, miR-221 |
| Sci Rep [60] | Clinical study (serum) | miR-125b, miR-221, miR-132, miR-100, miR-376a |
| IOVS Nuria [41] | Clinical study (E/PCs) | miR-221, miR-126, miR-222 |
| Cell Physiol Biochem [42] | Clinical study (serum) | miR-21, miR-181c, miR-1179 |
| FEBS Lett [43] | Invitro/animal OIR mice study Retina | miR-184 |
| BRBC [44] | Invitro/animal study Retina | miR-192-5P, miR-335 |
| Diabetologia [34] | Invitro/animal diabetic rats (E/PCs) | miR-195 |
| Circ Res [45] | Invitro/animal (STZ diabetic rat/db/db mice) Retina/(E/PCs) | IncRNA-myocardial infarction-associated transcript (MIAT)and miR—150 |
| Int J Med Sci [46] | Invitro/animal STZ diabetic rat study Retina | miR-126 |
| Mol Vis [47] | Invitro/animal STZ diabetic rat study Retina | miR-29b |
| IOVS [48] | Invitro/animal Akita mouse/(E/PCs) | miR-146,miR-200b |
| IOVS [49] | Invitro/animal Retina/(E/PCs) | miR-200b |
| IOVS [50] | Invitro/animal Akita and wild-type (WT) mice study Retina | miR-200b |
| Diabetes [51] | Invitro/animal (STZ diabetic rat) study Retina | miR-200b |
| Molecular Therapy [52] | Animal study (mice with oxygen-induced ischemic retinopathy) retina | miR-106a,-146,-181,-199a,-214,-424, -451 miR-31, miR-150, miR-184 |
| The International Journal of Biochemistry & Cell Biology [32] | Invitro study In vitro cell proliferation analyses |
MiR-18b |
Circular RNA and DR
Contrasting to linear RNA, the 3′ and 5′ in circular RNA, are joined and renamed circRNA. The circular RNA has been predominantly found in the cytoplasm and is usually composed of 1–5 exons, approximately 100 nucleotides in length with thousands of members in mammalian cells. Although these circular RNAs are more stable than the linear RNAs [53], their functions are yet to be clarified. Some of the potential roles might include: (1) evolutionary conservation of circularization mechanisms and signals and interactions with miR-7 [54–56], miRNA 26-28 due to its sponge capacity, as well as binding to RNA-binding proteins and initiating the in vivo protein production from the start codon [57]. (2) Regulation of mRNA in the cell through limited base pairing. (3) The physiological role of circular RNAs may be related to senescence, aging, and cell death [58]. (4) Circular RNAs have been shown to have a protective effect on colorectal cancer through downregulation of cir-ITCH expression [59]. (5) A study has shown that the circular RNA, Cdr1, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells in vitro. Nonetheless, the functions of circular RNAs remain uncertain in the pathogenesis of DR. The role of genetics and epigenetics in the pathogenesis of diabetic retinopathy is summarized in Fig. 1.
Fig. 1.
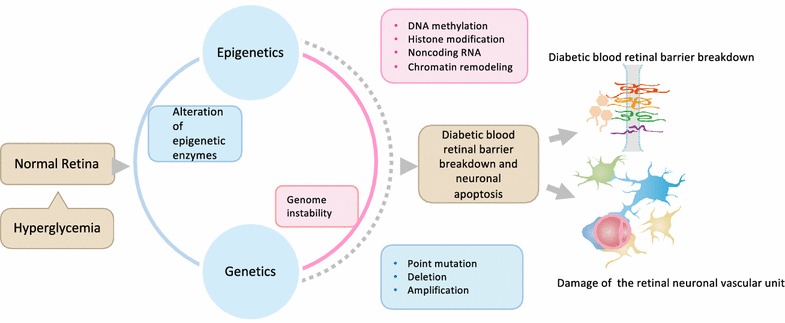
The role of genetics and epigenetics in the pathogenesis of diabetic retinopathy. Hyperglycemia induced breakdown of the blood retinal barrier (BRB) damage of the retinal neuronal vascular unit are the early pathogenic characteristics of DR. This figure illustrates the role of genetics (point mutation, deletion, amplification) and epigenetics (DNA methylation, histone modification, noncoding RNA, Chromatin remodeling) downstream of hyperglycemia in leading to diabetic retinopathy
Reversibility of epigenetic modifications and development of new molecular targets
The epigenetic modifications can precede disease pathology and serve as biomarkers to predict the development or progression of diseases. Furthermore, the main characteristics of epigenetic modification are also reversible, rendering them as promising therapeutic candidates.
As discussed above, histone acetylation resulted in the acceleration of DR. Garcinol, an inhibitor of the histone acetyltransferases [60], can prevent the epigenetic alterations that are implicated in the metabolic memory in DR. Histone deacetylase/DNA methylation inhibitors have been approved by the USA FDA and have been successfully applied clinically (ref). MiRNAs have also gained attention as potential therapeutic targets.
A novel gene therapeutic technique that efficiently inhibits the early pathologies in DR has been established: the epigenetic silencing of thioredoxin-interacting protein (TXNIP) in the diabetic retina [61]. TXNIP is induced early by hyperglycemia (in vitro) and diabetes (in vivo diabetic retina). TXNIP plays a central role in promoting neurovascular dysfunction in DR, and its epigenetic silencing prevents the progression of DR [61, 62]. This technique consists of injecting the retina of STZ diabetic rats with small interfering RNA (siRNA). These siRNAs are targeted to the TXNIP promoter and coupled via electrostatic bonds (not covalent) with cell penetrating peptides (CPP) containing a nuclear localization signal, in order to target the delivery into the cell nucleus and the TXNIP gene regulatory region [61]. Recent data show that miRNA plays a role in DR progression; thus, this technique may be used to restore the expression of specific miRNA in late DR by coupling with CPP.
Several possibilities for epigenetic treatment of DR have been studied. One approach is to inhibit the methylation of SOD2 and MMP-9. The DNMT inhibitors, 5-azacytidine, and 5-aza-20-deoxycytidine, have both been approved by the FDA for the treatment of other conditions. Further studies have examined the effects of those compounds on DR; wherein they seem to inhibit the methylation patterns with some success at reducing the symptoms. The DNA methylation inhibitor, Zebularine, has also been studied, although results are currently inconclusive. A second approach is to reduce the miRNAs observed at elevated levels in patients with DR, although the exact role of those miRNAs is yet unclear. The histone acetyltransferase (HAT) inhibitors, epigallocatechin-3-gallate, vorinostat, and romidepsin, have also been extensively studied albeit with limited success [63]. The possibility of using siRNAs to target the miRNAs mentioned above has also been speculated; however, no methods are currently known. The method described in the present study is partially impeded by the difficulty involved in delivering the siRNAs to the affected tissues [63].
In summary, the impact of epigenetics in DR is an emerging area, reversibility of epigenetic modification could provide a new strategy for the prevention and treatment of DR.
Authors’ contributions
All authors have contributed to manuscript writing. All authors read and approved the final manuscript.
Acknowledgements
The study is partly supported by the grant from “The National Natural Science Foundation of China (Grant No: 81170859 and 81570850)”, “Beijing health system high-level talent project (Grant No: 2013-3-056)” the Grant 2016YFC1305600, 2016YFC1305601 from the Major Chronic Non-communicable Disease Prevention and Control Research Key Project of the Ministry of Science and Technology of the People’s Republic of China.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Consent for publication has been made online.
Ethics approval and consent to participate
This is a review article, there are no issues relevant to ethics approval and consent to participant.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- BRB
the blood retinal barrier
- CPP
cell penetrating peptides
- DEM
diabetic macular edema
- DM
diabetes mellitus
- DNMT
methyltransferase
- DR
diabetic retinopathy
- FDA
food and drug administration
- HAT
histone acetyltransferase
- HDAC
histone deacetylases
- H3K9me2
H3 dimethyl lysine 9
- HIF-1α
hypoxia inducible factor-1α
- HLM
histone lysine methylation
- HRECs
human retinal endothelial cells
- LSD1
lysine-specific demethylase 1
- miRNA
microRNA
- MMP9
matrix metalloproteinase 9
- NK
natural killer
- PDGF-B
platelet-derived growth factor B
- POLG1
polymerase γ-1
- ROS
reactive oxygen species
- SAM
S-adenosylmethionine
- siRNA
small interfering RNA
- SOD
superoxide dismutase
- STZ
streptozotocin
- TXNIP
thioredoxin
- VEGF
vascular endothelial growth factor
Contributor Information
Xinyuan Zhang, Phone: +86 10 58269911, Email: mmzxy2010@163.com.
Lin Zhao, Email: judyzl1991@163.com.
Brett Hambly, Email: brett.hambly@sydney.edu.au.
Shisan Bao, Email: bob.bao@sydney.edu.au.
Kaiyue Wang, Email: kaiyuewang94@163.com.
References
- 1.Moosavi A, Ardekani AM. Role of epigenetics in biology and human diseases. Iran Biomed J. 2016;20(5):246–258. doi: 10.22045/ibj.2016.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28(10):1057. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 3.Daujat S, Zeissler U, Waldmann T, Happel N, Schneider R. Hp1 binds specifically to lys26-methylated histone h1.4, whereas simultaneous ser27 phosphorylation blocks hp1 binding. J Biol Chem. 2005;280(45):38090. doi: 10.1074/jbc.C500229200. [DOI] [PubMed] [Google Scholar]
- 4.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(5):522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teif VB, Rippe K. Predicting nucleosome positions on the DNA: combining intrinsic sequence preferences and remodeler activities. Nucleic Acids Res. 2009;37(17):5641. doi: 10.1093/nar/gkp610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baer C, Claus R, Plass C. Genome-wide epigenetic regulation of miRNAs in cancer. Cancer Res. 2013;73(2):473–477. doi: 10.1158/0008-5472.CAN-12-3731. [DOI] [PubMed] [Google Scholar]
- 8.Frank RF. Diabetic retinopathy. N Engl J Med. 2004;350(1):48–58. doi: 10.1056/NEJMra021678. [DOI] [PubMed] [Google Scholar]
- 9.Maghbooli Z, Hosseinnezhad A, Larijani B, Amini M, Keshtkar A. Global dna methylation as a possible biomarker for diabetic retinopathy. Diabetes/metab Res Rev. 2015;31(2):183. doi: 10.1002/dmrr.2584. [DOI] [PubMed] [Google Scholar]
- 10.Agardh E, Lundstig A, Perfilyev A, Volkov P, Freiburghaus T, Lindholm E, et al. Genome-wide analysis of dna methylation in subjects with type 1 diabetes identifies epigenetic modifications associated with proliferative diabetic retinopathy. BMC Med. 2015;13(1):182. doi: 10.1186/s12916-015-0421-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tewari S, Zhong Q, Santos JM, Kowluru RA. Mitochondria dna replication and dna methylation in the metabolic memory associated with continued progression of diabetic retinopathy. Investig Ophthalmol Visual Sci. 2012;53(8):4881. doi: 10.1167/iovs.12-9732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhong Q, Kowluru RA. Regulation of matrix metalloproteinase-9 by epigenetic modifications and the development of diabetic retinopathy. Diabetes. 2013;62(7):2559. doi: 10.2337/db12-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kowluru RA, Shan Y, Mishra M. Dynamic DNA methylation of matrix metalloproteinase-9 in the development of diabetic retinopathy. Lab Investig J Tech Methods Pathol. 2016;96(10):1040–1049. doi: 10.1038/labinvest.2016.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mishra M, Kowluru RA. Epigenetic modification of mitochondrial DNA in the development of diabetic retinopathy. Investig Ophthalmol Visual Sci. 2015;56(9):5133. doi: 10.1167/iovs.15-16937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, Bao S, Lai D, Rapkins RW, Gillies MC. Intravitreal triamcinolone acetonide inhibits breakdown of the blood-retinal barrier through differential regulation of VEGF-A and its receptors in early diabetic rat retinas. Diabetes. 2008;57(4):1026–1033. doi: 10.2337/db07-0982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, Lai D, Bao S, Hambly BD, Gillies MC. Triamcinolone acetonide inhibits p38MAPK activation and neuronal apoptosis in early diabetic retinopathy. Curr Mol Med. 2013;13(6):946–958. doi: 10.2174/1566524011313060007. [DOI] [PubMed] [Google Scholar]
- 17.Plath K, Fang J, Mlynarczykevans S, Cao R, Worringer KA, Wang H, Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300(5616):131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 18.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 19.Schneider R, Bannister AJ, Kouzarides T. Unsafe SETs: histone lysine methyltransferases and cancer. Trends Biochem Sci. 2002;27(27):396–402. doi: 10.1016/S0968-0004(02)02141-2. [DOI] [PubMed] [Google Scholar]
- 20.Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating cd4+ T cells. Immunity. 2009;30(1):155. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muramatsu D, Kimura H, Kotoshiba K, Tachibana M, Shinkai Y. Pericentric H3K9me3 formation by HP1 interaction-defective histone methyltransferase suv39h1. Cell Struct Function. 2016;41:145–152. doi: 10.1247/csf.16013. [DOI] [PubMed] [Google Scholar]
- 22.Joglekar MV, Januszewski AS, Jenkins AJ, Hardikar AA. Circulating microRNA biomarkers of diabetic retinopathy. Diabetes. 2016;65(1):22–24. doi: 10.2337/dbi15-0028. [DOI] [PubMed] [Google Scholar]
- 23.Zampetaki A, Willeit P, Burr S, Yin X, Langley SR, Kiechl S, et al. Angiogenic microRNAs linked to incidence and progression of diabetic retinopathy in type 1 diabetes. Diabetes. 2016;65(1):216. doi: 10.2337/db15-0389. [DOI] [PubMed] [Google Scholar]
- 24.Zhong Q, Kowluru RA. Role of histone acetylation in the development of diabetic retinopathy and the metabolic memory phenomenon. J Cell Biochem. 2010;110(6):1306–1313. doi: 10.1002/jcb.22644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baynes JW. Role of oxidative stress in development of complications in diabetes. Diabetes. 1991;40(4):405–412. doi: 10.2337/diab.40.4.405. [DOI] [PubMed] [Google Scholar]
- 26.Brownlee M. The pathobiology of diabetic complications: a unifying mechanism. Diabetes. 2005;54(6):1615. doi: 10.2337/diabetes.54.6.1615. [DOI] [PubMed] [Google Scholar]
- 27.Miao F, Smith DD, Zhang L, Min A, Feng W, Natarajan R. Lymphocytes from patients with type 1 diabetes display a distinct profile of chromatin histone H3 lysine 9 dimethylation: an epigenetic study in diabetes. Diabetes. 2008;57(12):3189. doi: 10.2337/db08-0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kowluru RA, Kowluru V, Xiong Y, Ho YS. Overexpression of mitochondrial superoxide dismutase in mice protects the retina from diabetes-induced oxidative stress. Free Radic Biol Med. 2006;41(8):1191–1196. doi: 10.1016/j.freeradbiomed.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 29.Kanwar M, Chan PS, Kern TS, Kowluru RA. Oxidative damage in the retinal mitochondria of diabetic mice: possible protection by superoxide dismutase. Investig Ophthalmol Visual Sci. 2007;48(8):3805–3811. doi: 10.1167/iovs.06-1280. [DOI] [PubMed] [Google Scholar]
- 30.Zhong Q, Kowluru RA. Epigenetic changes in mitochondrial superoxide dismutase in the retina and the development of diabetic retinopathy. Diabetes. 2011;60(4):1304–1313. doi: 10.2337/db10-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fong DS, Aiello L, Gardner TW, King GL, Blankenship G, Cavallerano JD, et al. Diabetic retinopathy. Diabetes Care. 2003;26(Suppl 1(1)):S99. doi: 10.2337/diacare.26.2007.S99. [DOI] [PubMed] [Google Scholar]
- 32.Wu JH, Wang YH, Wang W, Shen W, Sang YZ, Liu L, et al. Mir-18b suppresses high-glucose-induced proliferation in HRECs by targeting IGF-1/IGF1R signaling pathways. Int J Biochem Cell Biol. 2016;73:41–52. doi: 10.1016/j.biocel.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 33.Zhao S, Li T, Li J, Lu Q, Han C, Wang N, et al. Mir-23b-3p induces the cellular metabolic memory of high glucose in diabetic retinopathy through a SIRT1-dependent signalling pathway. Diabetologia. 2016;59(3):644. doi: 10.1007/s00125-015-3832-0. [DOI] [PubMed] [Google Scholar]
- 34.Mortuza R, Feng B, Chakrabarti S. miR-195 regulates SIRT1-mediated changes in diabetic retinopathy. Diabetologia. 2014;57(5):1037–1046. doi: 10.1007/s00125-014-3197-9. [DOI] [PubMed] [Google Scholar]
- 35.Lagosquintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA (New York, N.Y.) 2003;9(2):175. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen JK, Yang X, Xie B, Chen Y, Swaim M, Hackett SF, et al. MicroRNAs regulate ocular neovascularization. Mol Ther J Am Soc Gene Ther. 2008;16(7):1208. doi: 10.1038/mt.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryan DG, Oliveirafernandes M, Lavker RM. MicroRNAs of the mammalian eye display distinct and overlapping tissue specificity. Mol Vision. 2006;12(5):1175–1184. [PubMed] [Google Scholar]
- 38.Kwak N, Okamoto NJ, Campochiaro P. VEGF is major stimulator in model of choroidal neovascularization. Investig Ophthalmol Visual Sci. 2000;41(10):3158–3164. [PubMed] [Google Scholar]
- 39.Spoerri PE, Caballero S, Wilson SH, Shaw LC, Grant MB. Expression of IGFBP-3 by human retinal endothelial cell cultures: IGFBP-3 involvement in growth inhibition and apoptosis. Investig Ophthalmol Visual Sci. 2003;44(1):365–369. doi: 10.1167/iovs.02-0309. [DOI] [PubMed] [Google Scholar]
- 40.Hirota K, Keino H, Inoue M, Ishida H, Hirakata A. Comparisons of microRNA expression profiles in vitreous humor between eyes with macular hole and eyes with proliferative diabetic retinopathy. Graefe’s Arch Clin Exp Ophthalmol. 2015;253(3):335–342. doi: 10.1007/s00417-014-2692-5. [DOI] [PubMed] [Google Scholar]
- 41.de la Torre NG, Fernández-Durango R, Gómez R, Fuentes M, Roldán-Pallarés M, Donate J, et al. Expression of angiogenic microRNAs in endothelial progenitor cells from type 1 diabetic patients with and without diabetic retinopathy. Investig Ophthalmol Visual Sci. 2015;56(6):4090. doi: 10.1167/iovs.15-16498. [DOI] [PubMed] [Google Scholar]
- 42.Qing S, Yuan S, Yun C, Hui H, Mao P, Wen F, et al. Serum miRNA biomarkers serve as a fingerprint for proliferative diabetic retinopathy. Cell Physiol Biochem. 2014;34(5):1733–1740. doi: 10.1159/000366374. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi Y, Chen Q, Rajala RV, Ma JX. MicroRNA-184 modulates canonical wnt signaling through the regulation of frizzled-7 expression in the retina with ischemia-induced neovascularization. FEBS Lett. 2015;589(10):1143–1149. doi: 10.1016/j.febslet.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu J, Wang R, Ye Z, Sun X, Chen Z, Xia F, et al. Protective effects of methane-rich saline on diabetic retinopathy via anti-inflammation in a streptozotocin-induced diabetic rat model. Biochem Biophys Res Commun. 2015;466(2):155–161. doi: 10.1016/j.bbrc.2015.08.121. [DOI] [PubMed] [Google Scholar]
- 45.Yan B, Yao J, Liu JY, Li XM, Wang XQ, Li YJ, et al. LncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA novelty and significance. Circ Res. 2015;116(7):1143. doi: 10.1161/CIRCRESAHA.116.305510. [DOI] [PubMed] [Google Scholar]
- 46.Ye P, Jian L, He F, Wen X, Ke Y. Hypoxia-induced deregulation of miR-126 and its regulative effect on VEGF and MMP-9 expression. Int J Med Sci. 2014;11(1):17–23. doi: 10.7150/ijms.7329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li J, Riau AK, Setiawan M, Mehta JS, Ti SE, Tong L, et al. S100a expression in normal corneal-limbal epithelial cells and ocular surface squamous cell carcinoma tissue. Mol Vision. 2011;17(245–46):2263–2271. [PMC free article] [PubMed] [Google Scholar]
- 48.Murray AR, Takahashi Y, Ma J. Micro-RNA (miR)-200b, a regulator of oxidation resistance 1 (Oxr1), and miR-1224, a regulator of blue cone opsin (Opn1sw), are differentially expressed in diabetic retinopathy. Investig Ophthalmol Visual Sci. 2011;52:5941. doi: 10.1167/iovs.10-6894. [DOI] [PubMed] [Google Scholar]
- 49.Cao Y, Feng B, Chen S. Mechanisms of endothelial to mesen. Investig Ophthalmol Visual Sci. 2014;55(11):7321–7331. doi: 10.1167/iovs.14-15167. [DOI] [PubMed] [Google Scholar]
- 50.Wang Y, Li W, Zang X, Chen N, Liu T, Tsonis PA, et al. MicroRNA-204-5p regulates epithelial-to-mesenchymal transition during human posterior capsule opacification by targeting smad4. Investig Ophthalmol Visual Sci. 2013;54(1):323. doi: 10.1167/iovs.12-10904. [DOI] [PubMed] [Google Scholar]
- 51.Mcarthur K, Feng B, Wu Y, Chen S, Chakrabarti S. MicroRNA-200b regulates vascular endothelial growth factor-mediated alterations in diabetic retinopathy. Diabetes. 2011;60(4):1314–1323. doi: 10.2337/db10-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen JK, Yang X, Xie B, Chen Y, Swaim M, Hackett SF, et al. MicroRNAs regulate ocular neovascularization. Mol Ther. 2008;16(7):1208. doi: 10.1038/mt.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 54.Ebert MS, Sharp PA. MicroRNA sponges: progress and possibilities. RNA. 2010;16(16):2043–2050. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 56.Summerton J. Morpholino antisense oligomers: the case for an RNase H-independent structural type. Biochem Biophys Acta. 1999;1489(1489):141–158. doi: 10.1016/s0167-4781(99)00150-5. [DOI] [PubMed] [Google Scholar]
- 57.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2012;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hentze MW, Preiss T. Circular RNAs: splicing’s enigma variations. EMBO J. 2013;32(7):923–925. doi: 10.1038/emboj.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang G, Zhu H, Shi Y, Wu W, Cai H, Chen X. Cir-ITCH, plays an inhibitory role in colorectal cancer by regulating the Wnt/β-catenin pathway. PLoS ONE. 2015;10(6):e0131225. doi: 10.1371/journal.pone.0131225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Farr RJ, Januszewski AS, Joglekar MV, Liang H, Mcaulley AK, Hewitt AW, et al. A comparative analysis of high-throughput platforms for validation of a circulating microRNA signature in diabetic retinopathy. Sci Rep. 2015;5(4):10375. doi: 10.1038/srep10375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Devi TS, Lee I, Hüttemann M, Kumar A, Nantwi KD, Singh LP. TXNIP links innate host defense mechanisms to oxidative stress and inflammation in retinal muller glia under chronic hyperglycemia: implications for diabetic retinopathy. Exp Diabetes Res. 2012;1:438238. doi: 10.1155/2012/438238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perrone L, Devi TS, Hosoya KI, Terasaki T, Singh LP. Inhibition of TXNIP expression in vivo blocks early pathologies of diabetic retinopathy. Cell Death Dis. 2010;1(8):e65. doi: 10.1038/cddis.2010.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kadiyala CS, Zheng L, Du Y, Yohannes E, Kao HY, Miyagi M, et al. Acetylation of retinal histones in diabetes increases inflammatory proteins: effects of minocycline and manipulation of histone acetyltransferase (HAT) and histone deacetylase (HDAC) J Biol Chem. 2012;287(31):25869–25880. doi: 10.1074/jbc.M112.375204. [DOI] [PMC free article] [PubMed] [Google Scholar]


