Abstract
Biomedical ontologies often reuse content (i.e., classes and properties) from other ontologies. Content reuse enables a consistent representation of a domain and reusing content can save an ontology author significant time and effort. Prior studies have investigated the existence of reused terms among the ontologies in the NCBO BioPortal, but as of yet there has not been a study investigating how the ontologies in BioPortal utilize reused content in the modeling of their own content. In this study we investigate how 355 ontologies hosted in the NCBO BioPortal reuse content from other ontologies for the purposes of creating new ontology content. We identified 197 ontologies that reuse content. Among these ontologies, 108 utilize reused classes in the modeling of their own classes and 116 utilize reused properties in class restrictions. Current utilization of reuse and quality issues related to reuse are discussed.
Keywords: ontology reuse, content reuse, ontology modeling, BioPortal, ontology structural analysis
Graphical Abstract
1. Introduction
A commonly used ontology design principle is the reuse of content from other ontologies [1, 2]. Reusing content (e.g., classes and properties) of reliable quality can save an ontology author significant time and effort [3]. The author can focus on creating and managing content specific to his or her ontology, rather than modeling content that has already been created, and is maintained, in other sources. Reuse of content allows for a consistent representation of a domain among all ontologies that reuse the same content. Support for reusing ontology content is included as part of the Web Ontology Language (OWL) specification [4] (i.e., using owl:imports axioms) and is an important design principle in many ontologies, such as the members of the OBO Foundry [5].
The National Center for Biomedical Ontology (NCBO) [6] BioPortal [7] is an online repository of over 500 biomedical ontologies. Ontologies hosted in BioPortal (“BioPortal ontologies”) reuse content from other ontologies to enable the modeling of new classes, to cover a subject domain, and to support applications. Previous studies have investigated the existence of reused classes [2] and the inclusion of upper ontologies [1] in BioPortal. These prior studies were limited in that they did not investigate how reused content is utilized in ontologies; they only established the existence and extent of reuse in the BioPortal ontologies.
It important to understand the role of reused content in an ontology, especially when it is used to support the development of new classes and properties. In this study we describe a methodology for analyzing how reused content is utilized in a collection of ontologies. We apply our methodology on 355 BioPortal ontologies.
The goals of this study were to (1) identify which ontologies include classes and properties from other ontologies, (2) identify which ontologies have content reused by BioPortal’s ontologies, and (3) investigate how, and to what extent, BioPortal’s ontologies utilize reused classes and properties to model new classes and properties.
The majority of the ontologies in BioPortal are released in OWL or OBO format (472/506 = 93% as of January 2017) and in this study we only analyzed ontologies in those formats. OBO format ontologies can be directly converted into OWL [8]. Thus, throughout this paper, we describe our analysis using OWL ontology terminology (e.g., class, object property, equivalence).
2. Background
2.1 NCBO BioPortal
BioPortal [7], developed and maintained by the National Center for Biomedical Ontology (NCBO) [6], is a large online repository of over 500 biomedical ontologies. In addition to ontologies like SNOMED CT [9] and NCIt [10], BioPortal hosts the majority of the OBO Foundry [5] ontologies. As the largest online repository of biomedical ontologies, different aspects of the BioPortal ontologies have been analyzed in prior studies. Mortensen et al. [1] investigated the use of ontology design patterns among the BioPortal ontologies, including the use of upper-level ontologies such as the BFO. Ghazvinian et al. [11] looked at the orthogonality of the OBO Library [5] ontologies and their extent of reuse. Kamdar et al. [2] reviewed term (i.e., class) reuse among the BioPortal ontologies. Kamdar et. al’s approach used a combination of IRI matching, cross-reference (xref) annotations, and the UMLS [12] to identify matching terms in different ontologies. Kamdar et al. found that, while many ontologies include a term that exists in a different ontology, the overall number of reused terms is relatively small.
The research described in this paper expands on the type of empirical analysis described in these previous studies. In particular, we focus on the role reused classes play in the modeling of new classes. We also analyze how reused properties (i.e., object properties and data properties) are utilized among BioPortal’s ontologies. In this paper we will refer to most ontologies using their abbreviated name, as listed on BioPortal. Appendix A provides a glossary of abbreviations for the ontologies mentioned in this paper.
2.2 Content Reuse in Biomedical Ontologies
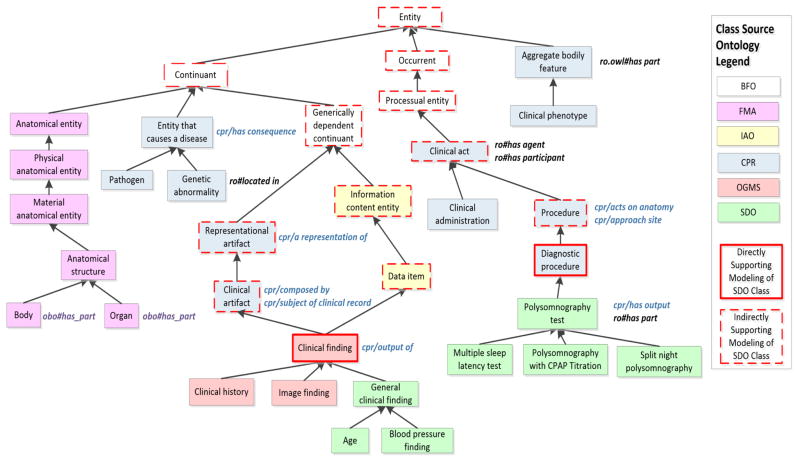
We will now briefly review the decisions ontology authors make when choosing to reuse content and we will briefly review how ontologies can include reused content. We define the ontology from which reused content originates as the source ontology. Figure 1 illustrates how reused content, from various source ontologies, is structured in the Sleep Domain Ontology (SDO) [13]. The SDO was chosen as a representative example due to our previous experience analyzing its content [14] and co-author (SA)’s role in developing the ontology. The SDO reuses content from several source ontologies covering various subject domains (e.g., anatomy, drugs, and clinical studies).
Figure 1.
An excerpt of 34 classes, included from various source ontologies, in the Sleep Domain Ontology (SDO). Classes are shown as boxes. Each class is color-coded according to its source ontology (see legend). Classes with a solid red outline are directly supporting the modeling of an SDO class and classes with a dashed outline are indirectly supporting the modeling of an SDO class. Upward directed arrows represent taxonomic (i.e., hierarchical) relationships. Properties that are used in a restriction on a class are shown to the right of the respective class and are color coded according to their source (e.g., properties from CPR are colored blue). We note that the SDO reuses the has part object property from multiple versions of the Relations Ontology (RO). We denote the different versions according to their IRI and color coding (obo#has_part, included via FMA, is purple and ro.owl/has part is black).
2.2.1 Including Upper Ontologies and Upper Domain Ontologies
Upper (or top-level) ontologies [15] often serve as a starting point for ontology development. One of the most widely used upper ontologies is the Basic Formal Ontology (BFO) [16], which aims to model the basic structures of reality and provides classes that categorize, at a high level, real-world entities. In practice, the BFO’s high level classes (e.g., continuant and occurrent) support categorization of classes in other ontologies. Use of the BFO (and the Relation Ontology (RO) [17], which provide high-level relations, e.g., part of) is a principle of the ontologies in the OBO Foundry [5].
An ontology author may include one or more upper domain (or top-domain) ontologies as a starting point for their ontology. Upper domain ontologies include general classes related to a subject domain. For example, the Ontology for General Medical Science (OGMS) [18] includes general classes that can be used in ontologies related to the human medical disciplines (e.g., classes like disease, disorder, and patient). The OGMS was built on top of the BFO and the Information Artifact Ontology (IAO) [19]. BioTop, another upper domain ontology that is also based on the BFO, includes high-level classes related to the life sciences (e.g., organism process, tissue process).
Figure 1 illustrates the integration of the BFO, OGMS, BioTop, IAO, and RO into the SDO.
2.2.2 Including Domain-specific Ontologies
An ontology may need to include classes that cover multiple subject domains. The coverage required may be more specific than what is provided in an upper domain ontology. A given subject domain may have already been modeled properly and sufficiently by a pre-existing ontology and can, thus, be reused.
Reusing domain ontologies to model content
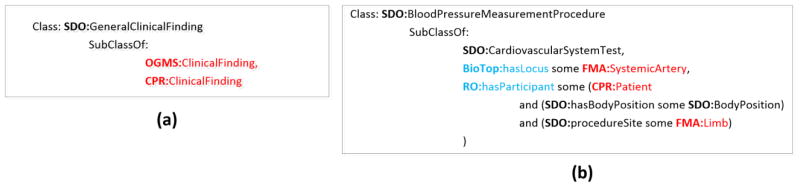
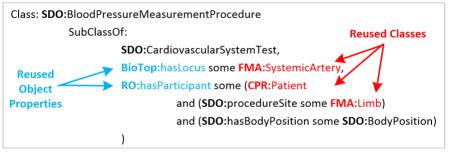
Domain-specific content may be included for different reasons, depending on the purpose of the ontology. As described in detail in Section 3.2, classes and properties may be reused to support modeling of new classes and properties. For example, the SDO class Blood pressure measurement procedure required a restriction procedure site (an object property defined in the SDO) with a range of Limb (see Figure 2). Hence, the modeling of some of the SDO’s classes required anatomy classes. Instead of designing a new anatomy subhierarchy specifically for the SDO, classes from a selected portion of the FMA were reused (Figure 1).
Figure 2.
Examples of classes in the SDO, in Manchester OWL syntax [30]. Reused classes are highlighted in red and reused properties are highlighted in blue. (a) The modeling of SDO:General clinical finding. Two reused classes, OGMS:Clinical finding and CPR:Clinical finding, are direct superclasses. Thus, they are directly support the modeling of General clinical finding. (b) The SDO class Blood pressure measurement procedure. The reused properties BioTop:hasLocus and RO:hasParticipant are used in class restrictions. The reused classes FMA:SystemicArtery, FMA:Limb, and CPR:Patient, are in the ranges of these restrictions. Thus, these classes are directly support the modeling of Blood pressure measurement procedure in the SDO.
Reusing domain ontologies to support applications
Alternatively, reused classes may be included to cover a domain for an ontology-backed application. Reused properties, which may have been included to represent the relationships between a set of reused classes, may also be integrated into applications (e.g., to support querying in a user interface).
2.2.3 Methods of Reusing Ontology Content
An ontology author may include all, or just a small subset, of a source ontology’s contents. There are various tools and mechanism for including a source ontology’s content in an ontology. The majority of BioPortal’s ontologies are in OWL format, which defines the owl:imports functionality. Utilizing owl:imports enables the import of content “on the fly” when an ontology is opened. Most ontology tools (e.g., Protege [20]) open the full contents of the source ontology in the owl:imports statement when the importing ontology is opened. Alternatively, one can use Protégé to merge the contents of two ontologies into a single ontology.
One simple way of reusing a subset of content is to copy/paste the desired entities and their associated axioms (e.g., the unique identifier) from a source ontology. However, this is only practical if a specific entity, or a small number of specific entities, are being reused. Typically, automatic and semi-automatic software tools are used to include subsets of content (e.g., custom-built ontology modules [3] consisting of a desired subset of classes and properties). Various studies have reviewed the relationship between ontology modularization and content reuse (e.g., [21, 22]).
The Minimum Information to Reference an External Ontology Term (MIREOT) [23] technique was developed to support the reuse of individual classes from a source ontology and is used by many OBO Library ontologies. When a class is reused with MIREOT, an ontology module that contains the class’s unique identifier, superclasses, and annotations is built automatically. This module can then be imported into an ontology. MIREOT can also be used to include the ancestor classes of a selected class. OntoFox [24] is a web-based tool for reusing subsets of ontologies with MIREOT and other reuse methodologies. The BioPortal import plugin for Protégé [25] serves a similar purpose. Using the plugin, a Protégé user can search BioPortal for relevant terms and include individual classes (or subhierarchies of classes) along with a selected subset of their information (e.g., hierarchical relationships, annotations).
Beyond the methodologies utilized to reuse content, there are many design decisions made by ontology authors when reusing content. For example, ontology authors have to decide how much of an individual class or property to reuse (e.g., is the complete definition of a class preserved? Are only subclass relationships included?). An ontology author may only reuse a given entity’s unique identifier and label. Alternatively, the editor may need to include a logically complete subset of a source ontology based on of a given class. This choice would result in reusing all of the selected class’s ancestors (and/or descendants) and all of the axioms needed to model the subset of classes (restrictions, equivalences, properties, etc.).
3. Methods
Our analysis was conducted in two phases. In the first phase, using the technique described in Section 3.1, we identified which BioPortal ontologies are reusing content and which source ontologies are being reused. After we collected this information, we performed a structural analysis, described in Section 3.2, to determine how reused classes and properties are being used to model new classes and properties. In this study we did not consider reuse by transitivity (i.e., reusing content from an ontology that itself reuses content) differently than direct reuse. For example, the SDO reuses the BFO by transitivity by including the OGMS. Thus, our analysis captures reuse from the perspective of an end user, who browses or downloads an ontology from BioPortal (or some other source). Additionally, reused content that is included by transitivity can be used in the modeling of classes (e.g., the SDO uses the BFO to model several of its own classes) and thus it is important that it is included in our analysis. All Results in this manuscript, including Tables and Figures, include reuse by transitivity.
3.1 Detecting Content Reuse
Our approach to detecting the reuse of content is based on each entity’s Internationalized Resource Identifier (IRI). An entity’s IRI is its unique identifier in an ontology. For details on the properties of IRIs in OWL ontologies see [4]. IRIs can vary in style from ontology-to-ontology. Some ontologies, such as those in the OBO Foundry, follow a consistent IRI pattern [26]. We define the base IRI of an ontology as the portion of an IRI that uniquely identifies all of the entities that are defined in an ontology. OWL format ontologies typically include a unique identifier as part of the serialization of the ontology. This identifier typically corresponds to the base IRI of the ontology.
Given a set of ontologies, we identify, in a semi-automatic way, the base IRI of each ontology. For example, the base IRI of the SDO is http://mimi.case.edu/ontologies/2009/1/SDO.owl; the IRIs of all the entities defined in the SDO start with this identifier. Similarly, the base IRI for the Gene Ontology is http://purl.obolibrary.org/obo/GO_. If an entity’s IRI starts with an ontology’s base IRI then, in our analysis, that entity is identified as being defined in the ontology with the given base IRI (e.g., the class Polysomnography test, with the IRI http://mimi.case.edu/ontologies/2009/1/SDO.owl#Polysomnography, is defined in the SDO).
We note that the same ontology may have several base IRIs, often originating from different versions or release formats (e.g., http://www.ifomis.org/bfo/1.1/ and http://purl.obolibrary.org/obo/bfo, among others, for the BFO). To identify these cases, we manually reviewed each base IRI and mapped it to a single ontology (e.g., BFO for the above example). Each base IRI was assigned a number to uniquely identify which versions of an ontology were reused. Table 1 identifies various FMA base IRIs found in the BioPortal data set.
Table 1.
Unique IRI IDs assigned to the various FMA base IRIs found among the BioPortal ontologies. A* indicates the current IRI used by FMA classes in the currently version of the FMA.
| URI ID | FMA Base IRI | Ontologies that Include a Class with this Base IRI |
|---|---|---|
| 1 | http://sig.biostr.washington.edu/fma3.0 | MCBCC, OCRe |
| 2 | http://purl.org/obo/owl/FMA | COGPO, EP, HUPSON, Neomark4, SDO, VSO |
| 3* | http://purl.org/sig/ont/fma/ | FMA |
| 4 | http://sig.uw.edu/fma | BDO, CLO, HUPSON |
| 5 | http://purl.obolibrary.org/obo/fma_ | OBI BCGO, VO |
| 6 | http://purl.org/sig/fma/ | ONSTR |
Two ontologies can potentially have the same base IRI. For example, various versions of GO (e.g., GO Simple and GO Plus) [27] have a base IRI of http://purl.obolibrary.org/obo/GO_. In this case we treat the various versions of GO as the same ontology. Alternatively, there may be two completely different ontologies that have the same base IRI. For example, the Document Acts Ontology (D-ACTS) ontology (not hosted in BioPortal but in the OBO Library) defines some classes with the base IRI http://purl.obolibrary.org/obo/IAO_, the same base IRI as the Information Artifact Ontology (IAO) (which D-ACTs includes). In such a case, one can potentially identify the source of a class based on which ontology file it originated from (e.g., the class Grapheme, with IRI http://purl.obolibrary.org/obo/IAO_0020001, is defined in D-ACTs and is not found in the IAO ontology).
In this study we considered content reused if an entity had a different base IRI than a given ontology’s base IRI. For example, the object property http://www.obofoundry.org/ro/ro.owl#has_part has a base IRI of http://www.obofoundry.org/ro/ro.owl, indicating that it is from (a version of) RO. This base IRI is different from SDO’s base IRI http://mimi.case.edu/ontologies/2009/1/SDO.owl. Thus, for the SDO, this object property is considered reused. As noted before, we define the ontology from which reused content originates as the source ontology (e.g., RO is the source ontology for the above mentioned has part object property in the SDO).
For each ontology we extracted the IRI of every class and property. The base IRI (and thus, source ontology) of the entity was automatically identified by matching the IRI to the set of base IRIs we previously identified. In the event that the base IRI of an entity could not be correctly identified, or it was ambiguous (e.g., http://protegeuserexample and http://www.semanticweb.org/ontologies/2011/7/ontology1314368515010.owl), the source ontology was marked as “unknown” and the entity was excluded from the study.
3.2 Structural Analysis of Reused Content
The method described above identifies the existence of reused content and its source ontology. It does not indicate how the reused content is utilized. He et al. [28] introduced a methodology to identify redundant (i.e., unused) content from source ontologies. We identified unused content in the Drug Discovery Investigations (DDI) ontology [29], which includes content from several ontologies, to determine which classes are redundantly included.
In summary, the methodology described by He et al. identifies which classes are included from a specific source ontology and then searches the ontology to see if the reused classes appear in the definition of the ontology’s own classes. If a class did not appear in a definition then it was classified redundant. This methodology, and its associated algorithms, was modified to enable the study described in this paper.
Specifically, in this study, we modified the approach by (1) analyzing the inverse of He et al. (i.e., identifying the classes that do appear in a class definition), (2) considering all source ontologies that are included, not a specific one, (3) scaling the methodology so it can be applied to hundreds of ontologies, (4) analyzing reuse of properties in the modeling of classes, and (5) categorizing how each reused class and reused properties is utilized in the modeling of classes.
In general, our analysis focuses on reuse of classes and properties as it relates to the structure of an ontology (i.e., its hierarchy, restrictions, equivalences, etc.). Thus, in this study, we do not consider reuse of elements that do not affect the structure of the ontology (e.g., reuse of annotations), though our approach can be modified to do so.
A reused class directly supports modeling if it appears in the definition of a new class. This could be as a (stated or inferred) superclass, as part of the range of a class restriction, in an equivalence, or in any other kind of defining axiom. A reused class indirectly supports modeling if it does not directly support modeling for any class but it is an ancestor of a class that directly supports modeling. Figure 2 illustrates several examples of reused classes and properties being used to directly support the modeling of SDO classes. The ancestors of these classes, some of which are shown in Figure 1, indirectly support modeling
In this study these categorizations were treated as disjoint; if a reused class directly supports the modeling of any class it is not categorized as indirectly supporting modeling in the ontology. The distinction of a class directly or indirectly supporting modeling is a structural categorization. In both cases a reused class (or property) is part of the model of a new class. However, this distinction allows us to identify which reused classes (or properties) were explicitly utilized by an editor versus classes that were simply ancestors of a class that was utilized for modeling.
Reused properties can also be used to support the modeling of new ontology content (e.g., as part of class restrictions). OWL also allows hierarchies of properties and a reused property can be utilized as a super property of new properties.
To evaluate the correctness of the output of our methodology we manually reviewed the results for several ontologies and compared the raw metrics to other sources that identify reused classes (e.g., Ontobee).
4. Results
In April 2015, we collected the most recent release of 355 ontologies hosted in BioPortal. The 355 ontologies could be automatically downloaded using BioPortal’s public API, opened in Protege [20], opened with the most recent release of the OWL API [31], and processed with the HermiT reasoner [32] (to obtain the ontology’s inferred axioms) when appropriate. We developed a software tool using components from our previously developed Ontology Abstraction Framework (OAF) [33] to apply our analysis algorithms, described below, to detect how reused content is utilized among the 355 BioPortal ontologies.
Among the 355 BioPortal ontologies analyzed in this study we identified 435 base IRIs that cover 1,288,247 classes and properties. We could not identify the source ontology of only 1,877 classes and properties. We identified 28 ontologies that had multiple base IRIs in the sample (e.g., FMA in Table 1). Table 2 summarizes our findings. In Section 4.1 we review the existence of reused content in BioPortal. In Section 4.2 we review how reused content is utilized in the modeling of new content. Results are further separated according to (1) the ontologies that are reusing content (Section 4.1.1 and Section 4.2.1) and (2) source ontologies that are being reused (Section 4.1.2 and Section 4.2.2).
Table 2.
Summary of ontologies that reuse content and source ontologies reused.
| Reuse Metrics for BioPortal Ontologies (%, n = 355) | |
|---|---|
| # Including reused content | 197 (55.5%) |
| # Including reused classes | 149 (42.0%) |
| # Including properties | 160 (45.1%) |
| # Including the BFO | 77 (21.7%) |
| # Using reused classes in modeling of new classes | 108 (30.4%) |
| # Using reused properties in class restrictions | 116 (32.7%) |
| Source Ontologies (%, n = 147) | |
| # With reused classes | 144 (98.0%) |
| # With reused properties | 58 (39.5%) |
| # With classes supporting modeling | 91 (63.2%) |
| # With classes directly supporting modeling | 91 (63.2%) |
| # With classes indirectly supporting modeling | 54 (37.5%) |
| # With properties used in restrictions on new classes | 23 (15.6%) |
| # With properties used as a super property | 15 (10.4%) |
4.1 Existence of Reused Content
A total of 197 BioPortal ontologies included reused content and a total of 147 source ontologies were identified. The source ontologies were not necessarily from the 355 BioPortal ontologies analyzed in this study (e.g., classes from Friend of a Friend (FOAF) [34] are reused by several ontologies, but FOAF is not hosted in BioPortal).
4.1.1 Ontologies that Reuse Content
Class reuse
A total of 149 BioPortal ontologies were found to include at least one class from at least one source ontology. Ontologies typically include classes from several source ontologies (on average 5.44, standard deviation of 5.75). On a per-ontology basis, the number of source ontologies varies significantly. For example, HUPSON includes classes from 32 different source ontologies, CCONT includes classes from 28 source ontologies, and EFO includes classes from 26 source ontologies. On the other hand, OCRe, BSPO, and VTO reuse classes from only one source ontology each. In between, the BDO includes classes from six source ontologies (BFO, FMA, HP, NCIt, OGMS, and PATO) and NPO, that also includes classes from six source ontologies (BFO, ChEBI, FIX, GO, PATO, and UO). We note that there exist several ontologies that are composed entirely of reused classes.
Reuse of the BFO is fairly common among the ontologies that reused content. We found 77 ontologies that reuse one or more classes from the BFO. A total of 40 ontologies include a BFO class with the http://www.ifomis.org/bfo/1.1/ base IRI and 45 include a BFO class with the http://purl.obolibrary.org/obo/bfo base IRI. A total of 38 (/77 = 49.4%) ontologies include the entire BFO and the majority (41/77 = 53.2%) include over 30 BFO classes (77.0% of the classes in the BFO). A total of 31 (/77 = 40.3%) ontologies included fewer than ten BFO classes. In our analysis of how ontologies reuse the BFO, we found that 20 ontologies (e.g., ACGT, CanCo, and OGMS) use the owl:imports mechanism. The remaining ontologies included BFO classes using some other means (e.g., including it manually or through transitivity). A total of 19 (/197 = 9.64%) ontologies (e.g., BDO, SDO, OBI, and VO) include one or more classes from OGMS and only five ontologies (/197 = 2.54%) include one or more classes from BioTop (DCO, HUPSON, NATPRO, NTDO, and SDO).
Reuse of properties
We found 160 ontologies that include at least one property from a source ontology. An ontology that reuses a property, on average, reuses properties from 2.24 source ontologies (standard deviation of 1.69). A total of 73(/160 = 45.6%) ontologies include a property from only one source ontology. At the extreme, ENM includes properties from 13 different source ontologies (e.g., NPO, Uberon, and RO). Most reused properties are included from a very small number of source ontologies, primarily RO.
4.1.2 Source Ontologies
Reused classes
We identified a total of 144 source ontologies for reused classes. Most source ontologies have content reused by relatively few other ontologies (on average, at least one class is reused by 5.35 ontologies, standard deviation of 10.43). Almost all cases of reusing classes from non-BFO source ontologies are instances of partial reuse. As can be seen from Table 3, relatively few ontologies reuse a non-BFO source ontology in full.
Table 3.
Ten (non-BFO) source ontologies that have a class included in other ontologies. Note that all of these ontologies are found in the OBO Foundry.
| Source Ontology | # Classes in Source Ontology | # Ontologies that Include Any Class | # Ontologies that Include All Classes | Average # Classes Included (% of source ontology classes) [standard deviation] | Example Ontologies that Include a Class (# Classes reused) | Source Ontology Has BFO Classes | Source Ontology Subject Domain |
|---|---|---|---|---|---|---|---|
| IAO | 125 | 50 | 4 | 25.36 (20.3%) [37.32] | AERO (125), DDI (109), ERO (18) | Yes | Information entities |
| OBI | 2,309 | 44 | 0 | 66.6 (2.88%) [150.4] | ENM (632), ERO (524), CCONT (59) | Yes | Biomedical investigations |
| PATO | 1,570 | 43 | 6 | 279.9 (17.8%) [754.7] | NIFCell (1570), FYPO (128), CLO (10) | Yes | Phenotypes and traits |
| GO | 40,471 | 37 | 0 | 186.3(0.46%) [450.73] | FYPO (2346), OMIT (263), CLO (13) | No | Genes |
| ChEBI | 56,231 | 33 | 0 | 319.2 (0.57%) [732.0] | DINTO (3922), EFO (891), PORO (32) | No | Chemicals |
| NCBITaxon | 906,907 | 26 | 0 | 219.4 (0.00%) [415.0] | CCONT (1447), VO (405), IDOMAL (19) | No | Organismal taxonomy |
| UO | 331 | 25 | 3 | 96.0 (29.0%) [127.7] | MS (331), BAO (49), DDI (3) | Yes | Units and measurements |
| SO | 2,313 | 23 | 0 | 35.6 (1.54%) [79.5] | MIRNAO (348), OBI BCGO (35), EFO (3) | No | Biological sequencing |
| CL | 2,183 | 20 | 0 | 93.1 (4.26%) [195.4] | EFO (653), CLO (133), OBI (28) | Yes | Cell types |
| OGMS | 124 | 19 | 2 | 11.0 (8.87%) [20.6] | SDO (71), AERO (69), IDO (5), VSO (2) | Yes | Clinical encounters |
The use of the owl:imports mechanism is fairly uncommon outside of including the BFO. However, six ontologies were found to import PATO using owl:imports axioms (e.g., EP and NBO). One common use of owl:imports we observed was to include a custom subset of classes from a source ontology. For example, the Porifera Ontology (PORO) includes custom-built subsets of ChEBI, CL, GO, PATO, and Uberon using the owl:imports mechanism. Similar applications of owl:imports were observed in the BioAssay Ontology (BAO), the Human Interaction Network Ontology (HINO), VIVO, and others.
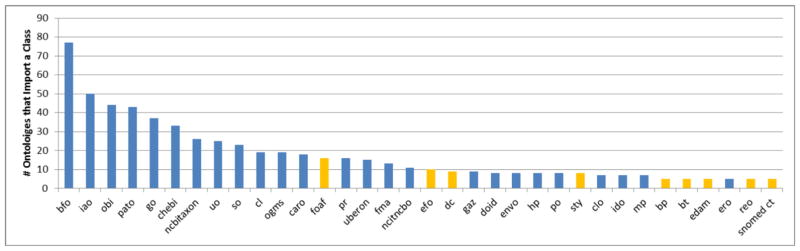
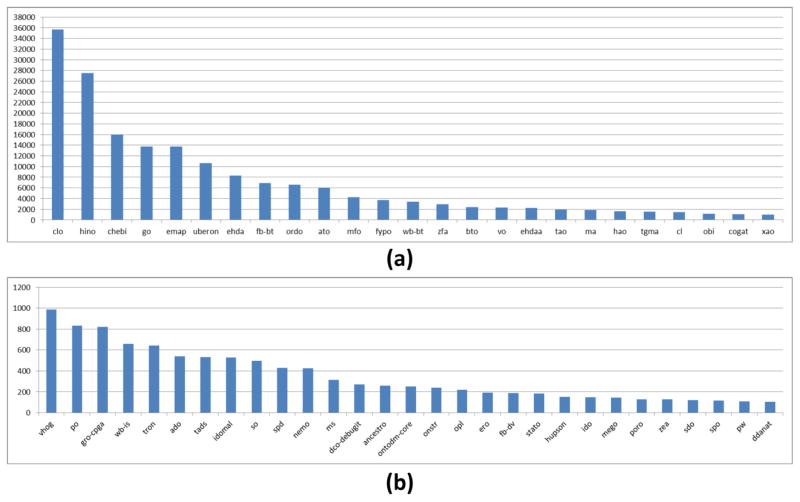
Figure 3 shows the number of ontologies that reuse at least one class from the most reused source ontologies. Table 3 provides expanded details for the ten most reused ontologies, excluding the BFO.
Figure 3.
Source ontologies that have a class reused by five or more ontologies. Ontologies in blue are found in the OBO Foundry.
In Table 3, source ontologies are ordered according to how many ontologies reuse at least one of their classes. However, alternative definitions for “most popular” source ontologies were investigated. One can look at, for example, how classes are reused from the source ontology. For example, all 125 IAO classes are reused by at least one ontology. On the other hand, only 608 (26.3%) SO classes are reused by at least one ontology. Alternatively, one can define how popular a source ontology is based on the proportion of its classes that are reused by ontologies. For example, on average, ChEBI classes make up 5.00% of ontologies that reuse a ChEBI class. Similarly, IAO, OBI, PATO, and GO classes make up, on average, 7.31%, 6.69%, 10.7%, and 8.30% of ontologies that reuse their classes, respectively. Similar metrics can be derived using maximum, minimum, median, or other statistics.
Reused properties
A total of 58 source ontologies have at least one property reused. Most properties are reused from a relatively small number of source ontologies (only five source ontologies have a property reused by five or more BioPortal ontologies). Table 4 lists the source ontologies that have the most reused properties. We note that, based on the IRIs in the ontology, the current version of the RO ontology includes RO, BFO, GO, and several other ontologies’ properties. However, for this study, we are treating the properties as coming from their respective ontologies. We note that RO originated from OBO REL, an ontology that, like RO, provided a set of commonly used properties. Since OBO REL originated from, and included, many of the same properties as RO (e.g., has part and part of), we combined the results for OBO REL and RO in this study. However, by looking at the base IRI of RO and OBO REL properties it is possible to determine which ontologies still include OBO REL.
Table 4.
The ontologies whose properties are included in the most ontologies.
| Source Ontology | # Ontologies that Include a Property | Example Properties (# Ontologies that include) | Example Ontologies that Include a Property |
|---|---|---|---|
| RO/OBO REL | 122 | Part of (67), Located in (39), Has participant (39), Derives from (39) | DDI, EFO, IDO, SDO |
| BFO | 65 | Has part (49), Part of (42), Occurs in (24), Realizes (23), Contains process (17) | CLO, GO, IAO, OGMS |
| OBI | 35 | Has specified output (19), Has specified input (17), Is specified output of (16) | BCO, CAO, ERO, IAO |
| IAO | 30 | Is about (27), Denotes (15), Has measurement unit label (13) | AERO, DDI, OBI, TMO |
| PATO | 10 | different in magnitude relative to (9), increased in magnitude relative to (9), has divisor quality (8) | CCONT, CMPO, ENM, EP |
| DC | 10 | Publisher (3), Creator (3), Contributor (3), Date (3), Provenance (2) | BCO, BIM, BRIDG, NPO |
| FOAF | 8 | Skype ID (4), MSN Chat ID (4), Yahoo chat ID (4), sha1sum of a personal mailbox URI name (4) | BIM, BRIDG, VIVO |
| PO | 5 | Participates in (3), Located in (2), Derives in manipulation from (2) | PAE, PSDS, VO |
| GO | 4 | Results in (2), Regulates (1), Positively regulates (1) | FYPO, MF, MIRNAO |
| SO | 3 | Variant of (2), Member of (2), Guided by (3) | OGSF, OGI, MIRNAO |
The large majority of reused RO properties have either the http://purl.obolibrary.org/obo/ro_ base IRI (456 properties reused by, on average, 2.37 ontologies) or the http://www.obofoundry.org/ro/ro.owl base IRI (48 properties reused by, on average, 14.9 ontologies). We note that, more recently, some object properties were transferred from RO to the current version of the BFO [35] (e.g., has part and part of), so we display them separately to identify which ontologies include these properties via the most recent version of BFO.
4.2 How are Reused Entities Utilized when Modeling New Classes?
4.2.1 Ontologies that Include Reused Content
Modeling with reused classes
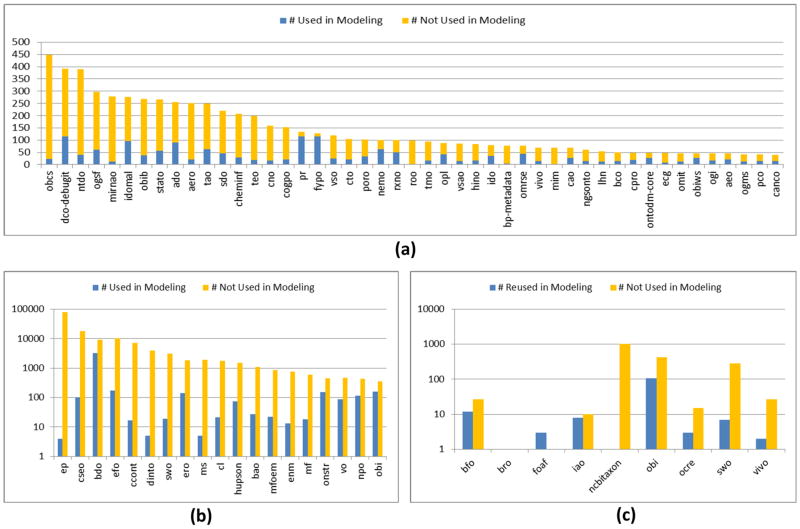
A total of 108 ontologies utilize reused classes in the modeling of their own classes. The most common uses of reused classes are as superclasses (101 ontologies) and in the range of someValuesFrom restrictions (64 ontologies). Other uses, such as in objectAllValuesFrom restrictions and in objectIntersectionOf axioms are less frequent (15 ontologies and 13 ontologies, respectively). On average, 34.8% of reused classes directly support modeling and 16.0% indirectly support modeling. The proportion of reused classes used to (directly or indirectly) support modeling of new classes is shown in Figure 4(a) and (b). Figure 4(c) shows the proportion of reused classes used in the modeling of ERO classes.
Figure 4.
(a) The ratio of reused classes that are utilized (directly or indirectly) or not utilized to support the modeling of new content for ontologies with fewer than 500 reused classes. (b) The same information, displayed in log scale, for ontologies with more than 500 reused classes. (c) The proportion of reused classes, organized by source ontology, used to model ERO classes.
A total of 63 ontologies included a reused class as a subclass. For example, the EFO has four BFO classes (Material entity, Material property, Process, and Site) as subclasses of the EFO class Experimental factor. This occurs much less frequently than utilizing reused classes as superclasses. On average, only 2.29 ontologies utilize a reused class as a subclass.
The number of source ontologies used to support modeling of new content varies significantly (e.g., OBI uses classes from 17(/20 = 85%) of its included source ontologies to model new classes but CCONT includes classes from 28 source ontologies but uses classes from only five in the modeling of its classes). A total of 48 ontologies utilize at least one class from every one of their source ontologies to model classes (average of 2.65 source ontologies included, standard deviation of 2.08) and 61 ontologies utilize classes from only a subset of the source ontologies they included. These ontologies include, on average, 8.75 source ontologies (standard deviation of 6.50) and, on average, utilize content from 59.1% of their source ontologies (standard deviation of 19.9%) in the modeling of new content. A total of 40 ontologies do not utilize any reused classes in the modeling of new classes (average number of source ontologies is 2.73, standard deviation of 4.48).
Reuse of object properties in restrictions
We analyzed how reused object properties are used in restrictions and equivalences. We focused on object properties because of their important role in expressing relations between classes and because their use is more common than data properties in the BioPortal ontologies [36]. A total of 116 ontologies utilized at least one reused object property in class restrictions and/or equivalences. These object properties originate from a relatively small number of source ontologies (mainly RO). On average, ontologies that utilized reused object properties in restrictions reuse object properties from 1.76 source ontologies and reuse, on average, 6.62 different object properties in class restrictions.
As with the other results in this study, there are extreme differences in the number of object properties used in restrictions. For example, Uberon utilizes a total of 89 different reused object properties in class restrictions (primarily from RO/BFO) while PATO and ChEBI only utilize one each (has part and has role from RO, respectively). As illustrated in Figure 5 and in the last column of Table 6, the number of classes modeled with a reused object property varies significantly. For example, CLO has 99,330 instances of a reused object property utilized in restrictions on 35,643 classes and ChEBI has 26,969 instances on 16,025 classes.
Figure 5.
The number of classes modeled with restrictions or equivalences that include a reused object property. (a) Ontologies where over 1000 classes have a restriction or equivalence with a reused property. (b) Ontologies with between 100 and 1000 classes that have a restriction or equivalence with a reused property.
Table 6.
The five most common source ontologies with properties used in class restrictions and equivalences.
| Reused Ontology | # Ontologies that Reuse Property for Modeling | # Classes that with Reused Property in Modeling | Example Properties (# Restrictions with the reused property) | Example Ontologies (# Classes with a restriction that include a reused property) |
|---|---|---|---|---|
| RO/OBO REL | 95 | 155,546 | Part of (50,900), Has part (46,321), Derives from (35,339), Has role (27,242) | CLO (34,544), HINO (23,595), ChEBI (16,025), EMAP (13,730) |
| BFO | 42 | 70,099 | Part of (75,949), Has quality at some time (26,605), Has part (3,042) | CLO (33,430), Uberon (9,007), GO (6,365), FB- BT (5,948) |
| OBI | 25 | 4,791 | Is specified input of (3,004), Is manufactured by (1,077) | CLO (2,917), VO (1,116), ERO (169), STATO (120) |
| IAO | 19 | 1,178 | Is about (1,081), Has measurement unit label (212) | OBI (602), ONSTR (180), NEMO (102) |
| BioTop | 3 | 327 | Bearer of (104), Has patient (95), Has Agent (66) | DCO-DEBUGIT (271), NTDO (39), SDO (17) |
A total of 43 ontologies were found to add restrictions on reused classes. For example, the Nano Particle Ontology (NPO) [37] adds restrictions (with NPO object properties) on 216 ChEBI classes and 39 GO classes, among others. The extreme case is PR, which adds restrictions onto 23,730 classes from nine source ontologies. A total of 30 of the ontologies only add restrictions onto one source ontology’s classes. For example, ACGT only adds restrictions onto eight BFO classes and DINTO adds restrictions onto 3,020 ChEBI classes.
Reused properties (both object properties and data properties) are rarely used as super properties of new properties. There are a total of 43 ontologies that defined a reused property as a super property of a new property (average number of such properties is 6.5, standard deviation of 10.1). Examples include ERO, which has 39 properties that are subproperties of a reused property (from OBI, SWO, and RO) and EFO, which has two properties that are subproperties of RO properties.
4.2.2 Source Ontologies Supporting Modeling
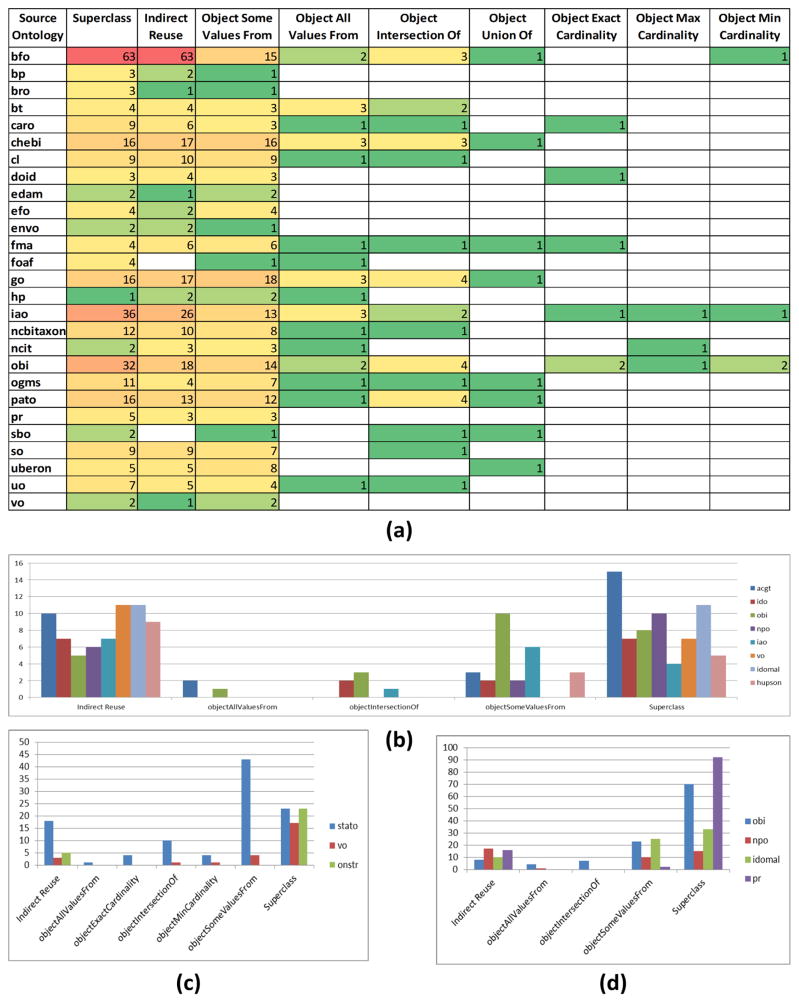
Classes from 92 source ontologies were directly or indirectly used in the modeling of classes in other ontologies. Classes from 73 different source ontologies are used as superclasses and classes from 64 source ontologies are used as ranges of objectSomeValuesFrom restrictions. Figure 6(a) is a heat map that details how many ontologies are reusing classes from a given source ontology, separated by the type of axiom(s) the reused classes appear in. From Figure 6(a) we observe that the majority of source ontologies serve as a source for superclasses. Figure 6(b) shows examples of the various ways classes from the BFO are used in the modeling of several example ontologies. Figures 6(c) and (d) show similar examples for OBI and GO. Table 5 lists metrics about the ten most subclassed source ontologies.
Figure 6.
(a) A heat map capturing how many ontologies in BioPortal are utilizing classes from source ontology in the modeling of their own classes. The 27 source ontologies reused by the most ontologies for modeling purposes are shown. (b) Use of BFO classes for modeling classes in eight ontologies (e.g., 15 BFO classes are superclasses of ACGT classes). (c) Use of OBI classes. (d) Use of GO classes.
Table 5.
The ten ontologies that are subclassed by the most ontologies.
| Source Ontology | # Ontologies that add a subclass | # Classes that are a subclass | Example Reused Classes (# Subclasses added in other ontologies) | Example Ontologies (# Subclasses added to reused ontology classes) |
|---|---|---|---|---|
| BFO | 63 | 1280 | Role (212), Quality (174), Process (156), Material entity (146) | OBI (109), IDOMAL (85), ADO (84), ACGT (81), IDO (80) |
| IAO | 36 | 1014 | Algorithm (173), Programming language (87), Data item (81) | SWO (249), NEMO (138), OBI (124), STATO (82), ERO (69) |
| OBI | 32 | 1042 | Data transformation (98), Planned process (79), Organism (41) | ERO (343), CHMO (190), STATO (104), ONSTR (60) |
| PATO | 16 | 3452 | Quality (1,046), Decreased occurrence (447), Lacking processual parts (307) | FYPO (3,268), PORO (41), HUPSON (39), NEMO (29) |
| ChEBI | 16 | 300 | Alkaloid (64), Biological role (30), Protein (25), Organic functional class (17), Molecular entity (15) | NATPRO (77), NPO (68), EFO (53) BAO (33), RNAO (23) |
| GO | 16 | 632 | Protein complex (152), Biological process (42), Behavior (20), Response to stimulus (12) | PR (328), IDOMAL (90), OBI (79), EFO (61), NPO (23) |
| OGMS | 11 | 116 | Clinical finding (58), Disease (10), Disorder (10), Laboratory finding (8), Bodily process (4) | AERO (54), SDO (18), NEOMARK (11), ONSTR (7) |
| NCBITaxon | 12 | 335 | Arabidopsis thaliana (159), Caenorhabditis elegans (54), Mus musculus (22) | EFO (269), OPL (34), IDOMAL (8), CCONT (6),VO (5), OBI (4) |
| CARO | 9 | 381 | Anatomical cluster (69), Cell (52), Multi-tissue structure (49) | TAO (297), AEO (52), VSAO (19), PORO (5), IDOMAL (3) |
| SO | 9 | 243 | RNA (124), Protein coding gene (50), DNA (34) | HINTO (158), PR (50), SNPO (19), TYPON (6) |
Object properties from 23 different source ontologies are used in class restrictions and equivalences. The overwhelming majority of reused properties come from RO/OBO REL, BFO, OBI, and IAO (Table 9). The remaining source ontologies have object properties that are used to model classes in only one or two ontologies. There were only 15 source ontologies with a property being used as a super property in another ontology. In total, 83 (/132=63.0%) of the reused properties utilized for this purpose are from various versions of RO.
4.3 Reuse and the OBO Foundry Ontologies
Established ontology design principles, such as those followed by the OBO Foundry ontologies, can play a significant role in the choice to reuse content, along with what content is reused. The OBO Foundry grew out of the contributors to the OBO Library. The curators of OBO Foundry ontologies have a commitment to ontological realism, as defined in the OBO Foundry principles [5]. These principles have had a strong influence on elements of the biology community and on how biological ontologies are developed within those segments. The OBO community has embraced the BFO (and RO) in part because the chief proponents of OBO created these ontologies [16]. There is a social expectation that ontologies that aim to be admitted into the OBO Foundry will reuse the BFO and/or RO. There also is a strong culture of reuse within the OBO community.
A large portion of the source ontologies used in the modeling of new classes are found in the OBO Foundry (e.g., 20/27=74.0% of the top source ontologies reused in modeling shown in Figure 6(a) and 4/5=80% of the source ontologies with properties reused for modeling). Some of these source ontologies make use of reused content in the modeling of their own classes, for example, out of the 27 source ontologies listed in Figure 6(a), 10 reuse classes from one or more other ontologies (commonly the BFO and/or IAO, e.g., by BioTop, OGMS, VO, and OBI).
Outside of the OBO Foundry ontologies, we found that reuse of content, including the reuse of BFO, is significantly less common. Less than half of the ontologies analyzed in this study (42.0%) reuse a class from another ontology and the majority of ontologies (278/355=78.5%) do not reuse any classes from the BFO (as noted above, reuse of properties from the RO is more common than class reuse from BFO). The most widely accessed ontologies in BioPortal (e.g., SNOMED CT, NCIt) do not reuse any content from other ontologies and they are also not widely reused as source ontologies. These self-contained ontologies generally predate the OBO community, and often have been developed by institutions rather than by groups of biomedical investigators.
Based on the results of Section 4.2 and 4.3, we can categorize ontologies based on which source ontologies they reuse. For example, we can characterize an ontology that includes BFO classes as a BFO-based ontology and we can characterize an ontology that does not directly include any BFO classes as a non BFO-based ontology. According to these characterizations, we observed that there are 59 BFO-based ontologies that reuse a BFO-based ontology (e.g., ERO and STATO). In contrast, there are only 10 non-BFO-based ontologies that reuse a BFO-based ontology (e.g., VSAO). Interestingly, a total of 67 BFO-based ontologies reuse non-BFO based ontologies (e.g., INO).
Based on class reuse, it appears that the OBO Foundry principles, including reuse, are well adhered to in the OBO Foundry community but have not been adopted by the wider biomedical ontology community. Thus, there is a divide in the BioPortal ontologies according to their design principles.
5. Discussion
The results of this study provide a high level picture of how BioPortal’s ontologies are utilizing reused classes and properties. In general, we observe high variability in how (and how much) content is reused among the BioPortal ontologies. While the majority of ontologies that reuse content utilize it in the modeling of new classes, the proportion of reused content utilized for this purpose differs significantly from ontology to ontology (as seen in Figure 6(a) and (b)). Even within individual ontologies, the percentage of reused content from different source ontologies utilized in the modeling of classes varies significantly (as illustrated by Figure 6(c)
This study identified which source ontologies have content being reused by other ontologies. It was not surprising that the BFO is used to support ontology development; it was designed specifically for that purpose. On the other hand, ontologies that were not necessarily designed to be extended or reused, such as GO and ChEBI, are also commonly used to model new classes (e.g., they have relatively many subclasses added in other ontologies, as shown in Table 5), apparently utilizing their specialized knowledge in genomics and chemistry, respectively.
In this study we did not investigate why ontologies do not utilize reused content in the modeling of their own content, or why only a small portion of reused content is utilized for that purpose. One likely possibility is that these ontologies are reusing content to cover a domain for an application. In some cases, content may have been erroneously included and could potentially be removed.
The picture of how ontology content is reused, as derived from the BioPortal ontologies, is somewhat limited. First, many of the ontologies in BioPortal have not been updated in several years (e.g., GO-EXT hasn’t been updated since 2011, even though new versions are released frequently). Additionally, we are aware of various ontologies that are hosted on other ontology repositories, such as the OBO Library and Ontobee, but are not found on BioPortal. Due to these limitations the picture of reuse derived from this study may be partially incomplete, especially with the prevalence of reuse in the OBO Foundry. Additionally, BioPortal only hosts ontology files. It does not include other reuse-related information, such as MIREOT [23] files, which would give more details about how ontologies are reusing other ontologies. These files can often be obtained from the web pages for individual ontologies, but gathering this information is labor-intensive.
In this study we did not investigate the social aspects that contribute to ontology reuse. We hypothesize that ontology authors are more likely to reuse content from ontologies that emerge from communities with which the authors are associated. When designing the SDO, co-author (SA) reused the CPR and OGMS ontologies in part because he was involved in the development of those ontologies.
5.1 Ontology Reuse Observations
Let us review some of the general findings of content reuse. First, we found that content reuse is relatively common in BioPortal (55.5% of ontologies in our study included at least one reused class or property). However, as discussed in Section 4.3, the majority of reuse is among ontologies in the OBO Foundry. Second, the reuse of properties was found to be slightly more common than reuse of classes (Table 2). However, properties are reused from relatively few source ontologies (mainly RO, Table 4). Third, we found that the BFO is the most reused ontology in BioPortal. In contrast, top-domain ontologies, and other ontologies that general domains, are reused significantly less (e.g., only 19 ontologies reuse the OGMS). Fourth, we found that, outside of the BFO, most ontologies do not include the full contents of a source ontology (see Table 3). Most ontologies include only a subset of a source ontology.
The number, and percentage, of classes that are reused from a source ontology also varied significantly among the ontologies we analyzed. For example, the eagle-i Research Resource Ontology (ERO) [38], which is composed of over 50% reused classes, reuses various numbers of classes from eleven separate source ontologies (e.g., 286 from Uberon, 524 from OBI, and 286 from SWO). The Vital Sign Ontology (VSO) [39], which is composed of 69% reused classes, also reuses content from eleven source ontologies (e.g., 25 from FMA and 12 from PATO).
The reuse of classes and properties to support to modeling of new classes and properties is a relatively common practice among the ontologies in BioPortal that reuse content. In the 355 ontologies analyzed in this study, 30.4% and 32.7% utilized reused classes and properties in the modeling of their own classes, respectively. As mentioned in Section 4.2.1, the reuse of properties in the creation of new properties is significantly less common (43 ontologies total); reused properties are primarily used in class restrictions and equivalences.
In general, reused classes are utilized as superclasses. From Figure 6(a), we see that the source ontologies that are used the most were designed for such a purpose (e.g., BFO, OGMS) or are ontologies that cover general domain topics that can be extended by ontologies in a variety of subdomains (e.g., IAO, OBI, UO). We also observe that the utilization of reused classes in the ranges of restrictions on new classes is relatively common for source ontologies that cover specific topic domains (e.g., GO, ChEBI, PATO, IAO, OBI).
In ontologies that utilized reused classes in the modeling of their own classes, most reused classes directly support modeling (on average, 34.8% vs 16.0% indirectly supporting modeling).
The amount of reused classes from a source ontology that are utilized for modeling was also found to vary significantly among the 355 ontologies. For example, on average, 54.2% of BFO classes reused in an ontology are utilized model new classes (27.3% directly, on average). In contrast, on average 60.1% of reused OBI classes in an ontology are utilized in modeling (51.6% directly, on average) and on average 53.7% of GO classes reused in an ontology are utilized to support modeling (35.6% directly, on average).
5.2 Quality Issues due to Reuse of Content
In our current line of research we are investigating family-based quality assurance methodologies for ontologies [36, 40]. The goal of this research program is to develop quality assurance techniques that are applicable to families of structurally similar ontologies. As mentioned by Ochs et al. [36], the reuse of content is an important aspects of an ontology’s design that can be used to classify ontologies into families. Ontologies that reuse the same content often have similar structures and may suffer from similar quality assurance issues. The results of the current study have shed some light on reuse among the BioPortal ontologies and suggest an investigation of reuse-related quality assurance issues in ontologies.
Interestingly, it seems that until recently “import” or “reuse” was not identified as a cause of errors in ontologies. For example, the subject is not mentioned in a review of terminology QA studies by Zhu et al. [41], that appeared in a journal special issue that focused on auditing of terminologies [42]. Zhu et al. did not identify any quality assurance studies for the 197 ontologies that were found to reuse content in this study. Furthermore, they reviewed quality assurance techniques for only four source ontologies found in this study (FMA, GO, NCIt, and SNOMED CT). Additionally, reuse of SNOMED CT was not mentioned in a survey of SNOMED CT users by Elhanan et al. [43].
Ochs et al. [14] identified various quality issues in the Sleep Domain Ontology (SDO). Most of the issues, such as duplicated classes and properties, inconsistent utilization of reused properties, and redundant (i.e., unused) class hierarchies, were caused by content reuse. Based on the results of the current study, we observe the potential for similar quality issues in other BioPortal ontologies. We have found that, based on a preliminary review, there are a number of ontologies that include duplicated classes and properties from different versions of the same ontology (e.g., there are eleven ontologies that reuse multiple versions of the BFO). Other instances of duplicated classes and properties were found when several ontologies that cover a similar domain are reuse. For example, CLO reuses anatomy classes from FMA, EFO, and Uberon, a proportion of which appear to represent the same entities. Similarly, CSEO reuses parts of NCIt’s Disease subhierarchy while defining its own Finding subhierarchy, both of which contain similar classes.
Another potential quality issue is the lack of consistency concerning which version of an ontology is being reused. As shown in Table 1, there are six different versions of FMA classes found in the 355 BioPortal ontologies analyzed in this study. We found several ontologies that include classes from different versions of the same ontology. Similarly, we found 14 different version of RO relationships spread among the various BioPortal ontologies, many of which are not used consistently, even within the same ontology (e.g., the SDO includes multiple versions of the part of property from different versions of RO, see Figure 1).
We observed that many ontologies do not utilize all of the entities that they reuse to model classes (directly or indirectly). This raises several questions. For example, if an ontology only reuses a small number of BFO classes, does it need to import the whole BFO? In a studies of the CanCo and DDI ontologies we found that significant portions of the BFO did not play any role in the modeling of new content or in the applications supported by these ontologies [28, 40]. Since, on average, only 54.2% of imported BFO classes are used to model content, it may make sense for ontologies to only reuse a portion of the BFO.
One can find additional modeling irregularities caused by reusing content (e.g., identifying imported classes that have subclasses added). For example, the Electrophysiology ontology (EP) has three new classes (Cell, Organ, and Tissue) as subclasses of the FMA class Anatomical structure. However, in the current version of FMA, there are subclasses of Anatomical structure named Cell and Organ. Similarly, there is a subclass of Anatomical structure called Portion of tissue, which has a synonym Tissue. Thus, it is conceivable that these FMA classes can be reused in EP.
Non-reuse of content also raises questions about the design of an ontology. For example, the SDO has a custom designed Units Ontology that could have been replaced by the Measurements and Units Ontology (UO) once the UO was released. Among the BioPortal ontologies, in this study we identified various ontologies that define, for example, their own part of object properties. While this may be an intentional part of their design, it may have been possible to reuse RO’s part of instead.
Indeed, all of the above quality issues may be part of the intended design of the respective ontologies and the design decisions may have been made to support some specific applications. However, these preliminary findings suggest that there are reuse-related issues affecting a portion of the BioPortal ontologies.
5.3 Future Work
We note that this study produced significant amounts of data about how ontologies reuse content from other ontologies, much of which is not presented in this paper. However, rather than performing a broader analysis, our planned future studies will focus on a group of key ontologies that are reused the most, updated relatively frequently, and are accessed more frequently by BioPortal’s users. Based on the results of this study, we are planning to investigate two important aspects of ontology reuse within a subset of BioPortal’s ontologies: the structure of reused content from a source ontology and quality issues that result from reusing content. Additionally, we will expand on the category-based analysis described in Section 4.3 by using other criteria beyond including the BFO (e.g., including RO, including FMA, etc).
5.3.1 Structure of Reused Content
In this study we focused our analysis on how ontologies utilize reused classes and properties in the modeling of their own content. In a future study we will investigate how reused classes are integrated into the reusing ontology, with a focus on the completeness of reuse for each reused class. In this planned study we will investigate which subhierarchy (or subhierarchies) are reused from a source ontology and determine how the classes from the source ontology are modeled in the reusing ontology. For example, in a given ontology that includes a classes from a source ontology, we will determine if all of the axioms used to define the reused class (e.g., the superclasses, restrictions, and equivalences) also reused. Alternatively, only a class name and IRI may be included. The reusing ontology may also make changes to the modeling of a reused class (e.g., the SDO adds a CPR superclass to an OGMS class).
As a preliminary part of this investigation we have analyzed the structure of reused classes from the Gene Ontology source ontology in FYPO, IDOMAL, and OBI. For each reused class we compared the modeling as it is defined in the current version of GO with how the class is modeled in the current version of each reusing ontology. We observed that, for example, among the 2714 GO classes in the current FYPO release, all of them retained their original superclasses and restrictions from GO. In contrast, in OBI a total of 62 (/143=43.3%) classes had superclasses removed and in IDOMAL 24(/45=53.3%) of classes had superclasses removed. Similarly, in IDOMAL and OBI, two and 11 classes, respectively, had one or more class restrictions removed in relation to their modeling in the current version of GO.
These differences may be caused by using an older version of GO or by only including a selected subhierarchy of GO and ignoring the hierarchical relationships that link outside of the selected subhierarchy.
To assist in performing this analysis we will design methodologies, based on our previous work on abstraction networks [44], to summarize, in a visual way, how content from source ontologies is integrated into other ontologies. Subhierarchies of reused classes could be summarized and visualized in way that captures where (and how) they are integrated into other ontologies. Using techniques based on our previously developed diff abstraction networks [45] we could identify structural differences from the current version of a source ontology with how the classes are modeled in the reusing ontology.
5.3.2 Temporal Analysis of Reuse
In this study we utilized the most recent release available, as of April 2015, for each of the 355 ontologies analyzed. This study did not include temporal information, such as the date of release of the ontologies (e.g., the SDO was from 2010 while the most recent release of GO was from 2015). Temporal information will be analyzed in a future study to identify reuse trends across time (e.g., is it now more common for ontologies to reuse content?). We will also analyze multiple releases of the same ontology to determine how utilization of reuse changes within an ontology across time. In a study of the eagle-i research resource ontology (ERO), we analyzed [45] how its content changed due a merge with VIVO. The merge resulted in many changes to the structure of ERO, including its reused content.
5.3.3 Quality Assurance Guidelines for Reuse
In a planned future study we will investigate, in detail, quality issues caused by reusing content. This future study is being driven by our prior experiences with ontology errors caused by reuse and the findings of this study. Such a study will identify the types and causes of errors, categorize reuse-related errors, and describe their potential consequences. In collaboration with several ontology curators, who have developed ontologies that reuse content, we will also develop a set of guidelines that ontology authors can follow to avoid errors caused by reuse.
6. Conclusions
We investigated the reuse of classes and properties in a sample of 355 BioPortal ontologies hosted on the NCBO BioPortal. A total of 197 ontologies were found to include classes and/or properties from another ontology. A total of 108 ontologies utilized reused classes to model their own classes and 116 ontologies utilized reused properties in class restrictions. Reused classes were found to typically be utilized as superclasses and in the range of restrictions. The reuse of content was observed to be more prevalent among the ontologies in the OBO Foundry. Quality issues related to reuse, such as duplicated classes and properties, and inconsistencies related to the version(s) of an ontology that were reused, were discussed. Future studies to investigate completeness of reuse and to further explore quality issues caused by reuse will be conducted.
Supplementary Material
Highlights.
We analyze content reuse in 355 biomedical ontologies from the NCBO BioPortal.
A total of 197 ontologies are found to reuse classes and/or properties.
We identify how reused content is utilized in the modeling of new classes.
Trends in the utilization of reused ontology content are discussed.
Acknowledgments
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number R01CA190779. The content is solely the responsibility of the authors and does not necessarily represent the views of the National Institutes of Health. BioPortal has been developed by the National Center for Biomedical Ontology, supported by the NIH Common Fund under grant U54 HG004028.
Footnotes
The authors have no conflicts of interest. All authors are aware of the submission of this manuscript
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mortensen JM, Horridge M, Musen MA, et al. Applications of Ontology Design Patterns in Biomedical Ontologies. Proc AMIA Annu Symp. 2012:643–52. [PMC free article] [PubMed] [Google Scholar]
- 2.Kamdar MR, Tudorache T, Musen MA. A Systematic Analysis of Term Reuse and Term Overlap across Biomedical Ontologies. Semantic Web journal. 2016 doi: 10.3233/sw-160238. (Accepted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pathak J, Johnson TM, Chute CG. Survey of modular ontology techniques and their applications in the biomedical domain. Integrated computer-aided engineering. 2009;16(3):225–42. doi: 10.3233/ICA-2009-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Motik B, Patel-Schneider PF, Parsia B. W3C -- World Wide Web Consortium. 2009. OWL 2 Web Ontology Language Structural Specification and Functional Style Syntax. [Google Scholar]
- 5.Smith B, Ashburner M, Rosse C, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nature biotechnology. 2007;25(11):1251–5. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Musen MA, Noy NF, Shah NH, et al. The national center for biomedical ontology. J Am Med Inform Assoc. 2012;19(2):190–5. doi: 10.1136/amiajnl-2011-000523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whetzel PL, Noy NF, Sham NH, et al. BioPortal: Enhanced Functionality via New Web services from the National Center for Biomedical Ontology to Access and Use Ontologies in Software Applications. Nucleic Acids Research (NAR) 2011;39(Web Server issue):W541–5. doi: 10.1093/nar/gkr469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tirmizi SH, Aitken S, Moreira DA, et al. Mapping between the OBO and OWL ontology languages. Journal of biomedical semantics. 2011;2(Suppl 1):S3. doi: 10.1186/2041-1480-2-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stearns MQ, Price C, Spackman KA, et al. SNOMED clinical terms: overview of the development process and project status. Proc AMIA Annu Symp. 2001:662–6. [PMC free article] [PubMed] [Google Scholar]
- 10.Fragoso G, de Coronado S, Haber M, et al. Overview and utilization of the NCI thesaurus. Comp Funct Genomics. 2004;5(8):648–54. doi: 10.1002/cfg.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghazvinian A, Noy NF, Musen MA. How orthogonal are the OBO Foundry ontologies? Journal of biomedical semantics. 2011;2(2):1. doi: 10.1186/2041-1480-2-S2-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic acids research. 2004;32(Database issue):D267–70. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arabandi S, Ogbuji C, Redline S, et al. Developing a Sleep Domain Ontology. AMIA Clinical Research Informatics Summit. 2010 [Google Scholar]
- 14.Ochs C, He Z, Perl Y, et al. Choosing the Granularity of Abstraction Networks for Orientation and Quality Assurance of the Sleep Domain Ontology. Proceedings of the 4th International Conference on Biomedical Ontology; 2013. pp. 84–9. [Google Scholar]
- 15.Mascardi V, Cordì V, Rosso P. A Comparison of Upper Ontologies. WOA. 2007:55–64. [Google Scholar]
- 16.Grenon P, Smith B, Goldberg L. Biodynamic Ontology: Applying BFO in the Biomedical Domain. In: Pisanelli DM, editor. Ontologies in Medicine. IOS Press; 2004. pp. 20–38. [PubMed] [Google Scholar]
- 17.Smith B, Ceusters W, Klagges B, et al. Relations in biomedical ontologies. Genome biology. 2005;6(5):R46. doi: 10.1186/gb-2005-6-5-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldfain A. Ontology for General Medical Science (OGMS) 2015 Feb 10; Available from: http://code.google.com/p/ogms/
- 19.Ceusters W. An information artifact ontology perspective on data collections and associated representational artifacts. MIE. 2012:68–72. [PubMed] [Google Scholar]
- 20.Musen MA. The protégé project: a look back and a look forward. AI Matters. 2015;1(4):4–12. doi: 10.1145/2757001.2757003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doran P, Tamma V, Iannone L. Ontology module extraction for ontology reuse: an ontology engineering perspective. Proceedings of the sixteenth ACM conference on Conference on information and knowledge management; 2007. pp. 61–70. [Google Scholar]
- 22.Grau BC, Horrocks I, Kazakov Y, et al. Modular reuse of ontologies: Theory and practice. Journal of Artificial Intelligence Research. 2008:273–318. [Google Scholar]
- 23.Courtot M, Gibson F, Lister AL, et al. MIREOT: The minimum information to reference an external ontology term. Applied Ontology. 2011;6(1):23–33. [Google Scholar]
- 24.Xiang Z, Courtot M, Brinkman RR, et al. OntoFox: web-based support for ontology reuse. BMC Research Notes. 2010;3(1):175–86. doi: 10.1186/1756-0500-3-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nair J, Tudorache T, Whetzel T, et al. The BioPortal Import Plugin for Protege. ICBO. 2011:298–9. [Google Scholar]
- 26. [7 February 2017];OBO Foundry Identifier Policy. 2017 Available from: http://www.obofoundry.org/id-policy.html.
- 27.Ashburner M, Ball CA, Blake JA, et al. Gene Ontology: tool for the unification of biology. Nature Genetics. 2000;25(1):25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He Z, Ochs C, Soldatova L, et al. Auditing Redundant Import in Reuse of a Top Level Ontology for the Drug Discovery Investigations Ontology. VDOS. 2013 [Google Scholar]
- 29.Da Q, King R, Hopkins A, et al. An ontology for description of drug discovery investigations. J Integrative Bioinformatics. 2010;7(3):126–39. doi: 10.2390/biecoll-jib-2010-126. [DOI] [PubMed] [Google Scholar]
- 30.Horridge M, Drummond N, Goodwin J, et al. The Manchester OWL Syntax. OWLed. 2006:216. [Google Scholar]
- 31.Horridge M, Bechhofer S. The OWL API: A Java API for Working with OWL 2 Ontologies. OWLED. 2009;529:11–21. [Google Scholar]
- 32.Shearer R, Motik B, Horrocks I. HermiT: a highly-efficient OWL reasoner. Proc 5th International Workshop on OWL: Experiences and Directions (OWLED); 2008. [Google Scholar]
- 33.Ochs C, Geller J, Perl Y, et al. A Unified Software Framework for Deriving, Visualizing, and Exploring Abstraction Networks for Ontologies. J Biomed Inform. 2016;62:90–105. doi: 10.1016/j.jbi.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Graves M, Constabaris A, Brickley D. Foaf: Connecting people on the semantic web. Cataloging & classification quarterly. 2007;43(3–4):191–202. [Google Scholar]
- 35.ROAndBFO. 2016 Jul 21; Available from: https://github.com/oborel/obo-relations/wiki/ROAndBFO.
- 36.Ochs C, He Z, Zheng L, et al. Utilizing a structural meta-ontology for family-based quality assurance of the BioPortal ontologies. J Biomed Inform. 2016;61:63–76. doi: 10.1016/j.jbi.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas DG, Pappu RV, Baker NA. NanoParticle Ontology for cancer nanotechnology research. J Biomed Inform. 2011;44(1):59–74. doi: 10.1016/j.jbi.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Torniai C, Essaid S, Lowe B, et al. Finding common ground: integrating the eagle–i and VIVO ontologies. ICBO 2013. 2013:46–9. [Google Scholar]
- 39.Goldfain A, Smith B, Arabandi S, et al. Vital sign ontology. Proceedings of the Workshop on Bio-Ontologies; 2011. pp. 71–4. [Google Scholar]
- 40.He Z, Ochs C, Agrawal A, et al. A Family-Based Framework for Supporting Quality Assurance of Biomedical Ontologies in BioPortal. Proc AMIA Annu Symp. 2013:581–90. [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu X, Fan JW, Baorto DM, et al. A review of auditing methods applied to the content of controlled biomedical terminologies. J Biomed Inform. 2009;42(3):413–25. doi: 10.1016/j.jbi.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geller J, Perl Y, Halper M, et al. Special issue on auditing of terminologies. J Biomed Inform. 2009;42(3):407–11. doi: 10.1016/j.jbi.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 43.Elhanan G, Perl Y, Geller J. A survey of SNOMED CT direct users, 2010: impressions and preferences regarding content and quality. J Am Med Inform Assoc. 2011;18(Suppl 1):i36–44. doi: 10.1136/amiajnl-2011-000341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Halper M, Gu H, Perl Y, et al. Abstraction Networks for Terminologies: Supporting Management of “Big Knowledge”. Artificial intelligence in medicine. 2015;64(1):1–16. doi: 10.1016/j.artmed.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ochs C, Perl Y, Geller J, et al. Summarizing and Visualizing Structural Changes during the Evolution of Biomedical Ontologies Using a Diff Abstraction Network. Journal Of Biomedical Informatics. 2015;56:127–44. doi: 10.1016/j.jbi.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.