Abstract
Drug resistance is a prevalent phenomenon that decreases the efficacy of cancer treatments and contributes to cancer progression and metastasis. Weakening drug resistant cancer cells prior to chemotherapy is a potential strategy to combat chemoresistance. One approach to damage resistant cancer cells is modulation of nutritional intake. The combination of nutrient restriction with targeted compound treatment results in pronounced molecular changes. This study provides valuable information about augmenting existing chemotherapeutic regimes with simultaneous glucose restriction and autophagy inhibition in colorectal cancer cells. In this study, we explore the chemical pathways that drive the cellular response to nutrient restriction, autophagy inhibition and the chemotherapy irinotecan using global quantitative proteomics and imaging mass spectrometry. We determined that significant pathways were altered including autophagy and metabolism via glycolysis, gluconeogenesis, and sucrose degradation. We also found that period circadian clock 2 (PER2), a tumor suppressor protein, was significantly up regulated only when glucose was restricted with autophagy inhibition and chemotherapy. The upstream regulators of these differentially regulated pathways were determined to have implications in cancer, showing an increase of tumor suppressor proteins and a decrease in nuclear protein 1 (NUPR1) an important protein in chemoresistance. We also evaluated the phenotypic response of these cells and discovered autophagy inhibition and chemotherapy treatment increased apoptosis and decreased cell clonogenicity and viability. When glucose restriction was combined with autophagy inhibition and chemotherapy, all of the phenotypic results were intensified. In sum, our results indicate that glucose metabolism is of great importance in the ability of cancer cells to survive chemotherapy. By weakening cancer cells with glucose restriction and autophagy inhibition prior to chemotherapy, cancer cells become more sensitive to therapy.
Keywords: Nutrient restriction, autophagy, chloroquine, colorectal cancer, proteomics, iTRAQ, MALDI Imaging Mass Spectrometry, three-dimensional cell culture
Graphical Abstract
Introduction
One of the biggest hurdles in cancer treatment is that not all cancer cells succumb to chemotherapy.1 There are several reasons for this fact, including cancer cell chemoresistance and the inability of drugs to reach all cancer cells. When even a small portion of cancer cells survive treatment they continue to promote cancer progression and metastasis.2–4 Weakening cancer cells prior to chemotherapy has the potential to sensitize them to treatment. One way to incapacitate cancer cells is by subjecting them to cellular stress. Nutrient restriction is a simple, easily implemented, form of cellular stress.5–7
Nutrient restriction (NR), defined as a decrease of total caloric intake while sustaining life, is known to activate cellular processes including autophagy.8–10 Autophagy is the process in which cells maintain cellular metabolism by recycling their cellular components.11,12 Autophagy is induced, in particular, by glucose restriction. The autophagy pathway functions to support cell survival by recycling essential nutrients through lysosomal degradation.13–15 Autophagy is therefore vital for pathways that cancer cells utilize to support cell survival under conditions of glucose restriction.14 Thus, the inhibition of autophagy machineries that are required for these processes holds therapeutic potential.
Chloroquine (CQ) is a widely used anti-malarial drug that inhibits autophagy by blocking fusion of the autophagosomes with lysosomes.16,17 Sustained exposure to CQ halts late stage autophagy and drives cells to undergo apoptosis.17,18 Due to CQ’s role as an autophagy inhibitor, it is being considered as a potential cancer therapeutic. Clinical trials are currently ongoing to evaluate the effects of CQ as a chemotherapeutic. The PINC trial is presently recruiting patients to evaluate the ability of CQ to reduce the survival and spread of ductal carcinoma in situ (Clinical trial #NCT01023477). In addition, there is evidence that CQ can enhance the potency of 5-fluorouracil in colon cancer.18,19 While CQ shows promise as a standalone treatment option, or in combination with other drugs, there is intriguing evidence that CQ combined with nutrient restriction is also a potent therapeutic option.20 Cancer cells exposed to glucose restriction will further induce autophagy activation and the nutrient recovery normally afforded by autophagy is then blocked with the CQ treatment.19,21 We hypothesized that the combined treatment of cancer cells with glucose restriction and CQ would have a higher death rate when they undergo chemotherapy treatment.20 The proteomic changes that occur in conjunction with these treatments are currently unknown.
This study evaluates the synergistic effects of the chemotherapy drug irinotecan (IR) used in tandem with glucose restriction and CQ treatment in colorectal cancer three-dimensional cell cultures, known as spheroids. First, we examined the penetration of IR and CQ into spheroids under normal and glucose restriction conditions by imaging mass spectrometry (IMS). To analyze the chemical pathways that underwrite the cellular response to these different perturbations, we utilized global quantitative proteomics and identified up-regulation of tumor suppressor proteins due to treatment. This intriguing data indicates that exploiting the dependence of cancer cells on glucose is a possible therapeutic strategy for treating colorectal cancer.
Experimental Section
Cell Culture Spheroid Growth and Treatment
The human colon carcinoma cell line HCT 116 was obtained from American Type Culture Collection (ATCC) and grown in RPMI 1640 cell culture medium (Life Technologies) supplemented with 10% fetal bovine serum (FBS) (Thermo Scientific). The provider assured authentication of the cell line by cytogenetic analysis. In addition, the cell line was validated by short tandem repeat (STR) analysis within the last 18 months. Spheroid cell culture was carried out using agarose-coated 96-well plates, seeding 7000 cells into each well as previously described.22 Culture medium was changed every 48 h after 5 days in culture. After 10 days in culture, spheroids were treated with 25 μM chloroquine diphosphate (Sigma Aldrich). For spheroids treated with nutrient restriction, on day 10 of culture, media was changed to RPMI 1640 containing no glucose (Life Technologies) in addition to 25 μM CQ. On day 12 in culture, spheroids were treated with 47.8 μM irinotecan (Sigma Aldrich). All spheroids were harvested for analysis on day 14. Control untreated spheroids had full nutrient media changed every 48 h until harvested also on day 14. Flow cytometry analysis of spheroids was used to construct an IC50 dose response curve for CQ. The IC50 concentration of CQ was determined using the CYTO-ID® Autophagy Detection Kit (ENZO). with flow cytometry, as previously described23 (Supplemental Figure 1A). Similarly, flow cytometry was used to generate a dose response curve for IR. The IC50 concentration of IR was determined using a LIVE/DEAD® Fixable Dead Cell Stain (Thermo Scientific) with flow as previously described23 to determine an IC50 concentration of IR treatment of 48 hours (Supplemental Figure 2B).
MALDI-IMS Sample Preparation and Analysis
Gelatin Embedding and Sectioning
Spheroids were washed with phosphate buffered saline (PBS) at 37 °C following dosing. Washed spheroids were placed on a layer of solidified gelatin in a 24 well plate. A warm layer of gelatin was placed on top of the spheroids. The gelatin embedded spheroids were flash frozen at −80 °C. The embedded spheroids were sliced with a cryostat into 16 μm thick slices at −30 °C. These slices were thaw mounted onto indium tin oxide coated slides and stored at −80 °C prior to matrix application.
Matrix Application
The MALDI matrix 2,5-dihydroxybenzoic acid (DHB) (Sigma-Aldrich, St. Louis, MO, USA) was dissolved in 60:40 ACN/H20 with 0.2 % TFA (EMD, Billerica, MA, USA) to a final concentration of 30 mg/ml. The DHB solution was filtered through a 0.22 μm filter. The filtered matrix solution was applied to samples using a TM-Sprayer (HTX Technologies, Carrboro, NC, USA) with heated nitrogen sheath flow gas. The solvent pump flow rate was set to 0.1 ml/min and the temperature was set to 70 °C. Matrix was applied 4 times in a crisscross pattern. The samples were then dried in a desiccator for at least 1 hour prior to MALDI-MSI.
Mass Spectrometry Imaging
An ultrafleXtreme MALDI-TOF-TOF mass spectrometer (Bruker Daltonics, Bremen, Germany) was used for MALDI-MSI analysis. Mass spectra were acquired in reflectron positive ion mode with 600 laser shots per spot. Mass spectra were acquired in a mass range of 300–1000 m/z. The experiment was performed using a laser spot size of 35 μm. A sampling frequency of 2 kHz was used and the laser intensity was set at 65%. Images were processed with flexImaging 4.1software (Bruker Daltonics, Bremen, Germany) to generate ion maps with a semiquantitative color scale bar.
Flow Cytometry Sample Preparation and Analysis
For the apoptotic assay, the FITC Annexin V Apoptosis Detection Kit (BioLegend) was used. Twelve spheroids grown for 14 days were collected and prepared using the methods previously described.23 Samples were analyzed on a BD LSRFortessa X-20 flow cytometer in the green (FITC) channel. Forward scatter (FS) and side scatter (SS) were used to identify single cells. The blank (unstained) sample, and labeled control (untreated) samples were analyzed for adjustment of the parameters in each channel.
Clonogenic Assay Sample Preparation and Analysis
For the clonogenic assay, 60 spheroids per condition were grown and treated according to the treatment plan (Figure 1). On day 14 in culture, spheroids were homogenized using a mechanical pipet and filtered through a 40 μm cell strainer (Corning Incorporated) to obtain a single cell suspension. Live cells were counted using a hemocytometer and Trypan Blue (Thermo Scientific) according to manufacturer’s protocol. 2000 live cells were seeded in triplicate to an adherent 15 mm dish. The 15 mm dish was incubated at 37 °C in 5% CO2 for the equivalent of six cell divisions (5.25 days for HCT 116 cells). Media was removed from the dish and the cells were washed with 5 mL PBS. Cells were then fixed with 5 mL neutral buffered formalin solution (10 %) for 15 minutes and stained with 5 mL of 0.01% crystal violet in dH2O (Sigma Aldrich) for 120 minutes. Excess crystal violet was washed with dH2O and allowed to dry. Digital images were obtained and colonies were counted by hand and verified with ImageJ (Fiji Version 1.44a).
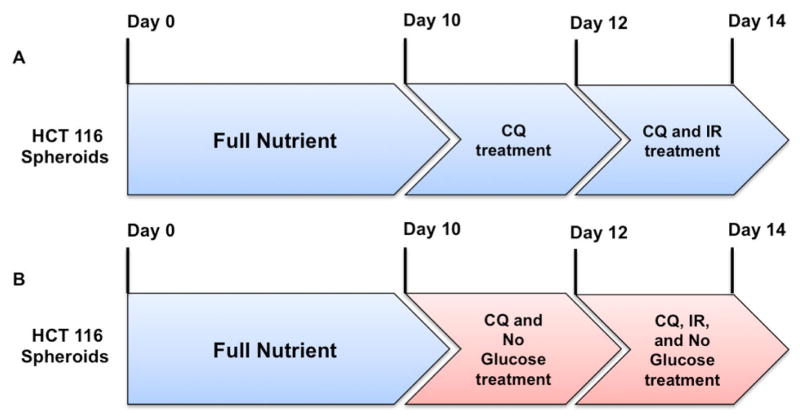
Figure 1.
Treatment plan for HCT 116 spheroids. A. HCT 116 spheroids are cultured for 10 days in full nutrient RPMI 1640 media. On day 10, media is changed to full nutrient RPMI 1640 with 25 μM CQ. On day 12, media is changed to full nutrient RPMI 1640 with 25 μM CQ and 47.8 μM IR. Spheroids are harvested for analysis on day 14. B. HCT 116 spheroids are cultured for 10 days in full nutrient RPMI 1640 media. On day 10, media is changed to RPMI 1640 that does not contain glucose with 25 μM CQ. On day 12, media is changed to RPMI 1640 that does not contain glucose with 25 μM CQ and 47.8 μM IR. Spheroids are harvested for analysis on day 14.
CellTiter-Blue® Cell Viability Assay Sample Preparation and Analysis
Treated spheroids were transferred to a 96 well plate containing 100 μL of PBS without agarose on day 14. Once spheroids were transferred, 20 μL of the CellTiter-Blue® (Promega) reagent was added to the 96 well plate and incubated for 1 h at 37 °C. Spheroids were analyzed using a Molecular Devices Spectramax M5 plate reader according to manufacturer’s protocol.
Sample Preparation for Proteomic Analysis
Whole spheroids (25 per biological replicate, 3 biological replicates analyzed) were homogenized and lysed using a procedure previously described.23 The Pierce BCA Protein Assay Kit (Thermo) was used to quantify total protein amount per sample. For all eight samples, 100 μg of protein was denatured for 1 h at 37 °C, reduced with 5 mM dithiothreitol (DTT) for 1 h at 37 °C, and alkylated with 14 mM iodoacetamide for 30 min at RT in the dark. The reaction was then quenched with 5 mM DTT for 25 min at RT. The urea in samples was diluted with 25 mM Tris-HCl (pH 8.2) to 1.5 M and samples were digested with trypsin in 1 mM CaCl2 overnight at 37 °C. Peptides were desalted with 50 mg C18 Sep-Paks (Oasis) and labeled with isobaric tags for relative and absolute quantification (iTRAQ) 8-plex reagents according to the manufacturer’s protocols (AB Sciex). Once labeled, samples were combined, vacuum dried, and desalted with 100 mg C18 Sep-Paks. The sample was then resuspended for strong cation exchange (SCX) liquid chromatography fractionation in 120 μL Buffer A (10 mM KH2PO4 in 20% ACN, pH 2.85).
SCX Fractionation
The iTRAQ combined sample was fractionated using SCX liquid chromatography and a Waters Alliance HPLC System (Waters). The sample (100 μL) was loaded onto an SCX guard column (2.1 mm i.d. X 50 mm length, 5 μM particles, Agilent Technologies). The mobile phase gradient was generated using Buffer A and Buffer B (1 M KCl in Buffer A, pH 2.85) at a flow rate 0.25 mL/min. Total run time was 60 minutes. The first 10 min of the method consisted of washing with 100% Buffer A to remove excess iTRAQ reagent. Following the wash, peptides were fractionated by a linear gradient from 100% Buffer A to 100% Buffer B collecting fractions every minute for a total of 31 fractions. The column was then washed with 100% Buffer B and equilibrated with 100% Buffer A. Fractions from 10–15 min were combined as one fraction, due to low concentration of sample eluting from the column in the first five minutes of the gradient.
Peptide Desalting
The fractions resulting from SCX fractionation were dried down and resuspended in 60 μL of 0.1% Formic Acid (FA) in H2O. 20 μL of the resuspended sample was desalted with a 5 μg C18 ZipTip (ZTC18S096, Millipore), and dried down. Finally, peptides were resuspended in 5 μL of 0.1% FA in H2O and analyzed by ultra-performance liquid chromatography (UPLC)-ESI-MS/MS.
UPLC-ESI-MS/MS analysis
Peptide separation was performed by a nanoACQUITY UltraPerformance LC® (UPLC®) system (Waters) with mobile phases Buffer A (0.1% FA in H2O) and Buffer B (0.1% FA in ACN). For each sample, 2 μL of peptides were automatically loaded onto a commercial C18 reversed phase column (Waters, 100 μm × 100 mm, 1.7 μm particle, part No. 186003546, column temperature 40°C). The LC method was 75 minutes at a flow rate of 1 μL/min, with the following gradient: 0–8 min at 2% buffer B, followed by a linear gradient of 2–6% buffer B in 2 min, 6–25% in 42 min, 25–50% buffer B in 6 min, 50–85% buffer B in 5 min, and maintained at 85% B for 5 min. The column was equilibrated with 2% buffer B for 7 min. The peptides were then analyzed via electrospray ionization (ESI) by a Q-Exactive mass spectrometer (Thermo Fisher Scientific). The following parameters were used for the analysis: electrospray voltage was set to 2.1 kV and the ion transfer tube temperature was 280°C. The data acquisition was programmed in data dependent acquisition (DDA) mode with the S-Lens RF level of 50.00. A top 12 method was used and Full MS scans were acquired in the Orbitrap mass analyzer 350–1800 m/z range with resolution of 70,000 and the number of microscans set to 1. The target value was 1.00E + 06, and maximum injection time was 250 ms. The isolation window was set as 1.0 m/z. The normalized collision energy was 31.0%, and tandem mass spectra were acquired in the Orbitrap mass analyzer with resolution 17,500 with the fixed first mass m/z 100.0. The target value was 1.00E + 06 and maximum injection time was 120 ms. The number of microscans was set to 1 and the ion selection threshold was 5.0E + 04 counts. Peptide match and exclude isotopes were turned on. Dynamic exclusion was set as 40 s. All injections were run in triplicate.
Data Analysis
Raw data files were searched with MaxQuant version 1.5.6.0 with MASCOT version 2.2.4 and UniProt human and decoy databases. Trypsin with a maximum of two cleavage misses was selected. The precursor mass tolerance was set to 10–20 ppm, and fragment mass tolerance was set to 0.05 Da. Dynamic modification were set to be deamidation, oxidation, acetylation and iTRAQ 8-plex labels on protein N-terminal and tyrosine. A static modification was set to be carbamidomethylation. Peptides were searched against a reverse sequence data to determine false discovery rate (FDR) with the FDR set to 0.01 for peptide and protein identifications. iTRAQ 8-plex as the reporter ion quantitation type was set as well as instrument Orbitrap MS and MS/MS. Functional classification was performed using Ingenuity Pathway Analysis (IPA).
Statistical Analyses
All experiments were performed in biological triplicate. Data were analyzed using unpaired Students t test for comparison of means to detect differences compared with control as well as Pearson correlation coefficient to measure linear correlation between two variables. Differences were considered to be statistically significant at *P < 0.01.
Results and Discussion
Treatment and distribution of CQ and IR via MALDI-IMS
For this study, HCT 116 colon carcinoma cells were grown into three-dimensional cell cultures, or spheroids. Spheroids are an excellent model system for cancer research. Spheroids are relatively inexpensive, high throughput, and grow on a much shorter time scale than animal models. 24,25 Our laboratory has previously shown that HCT 116 spheroids display radially symmetric chemical gradient of nutrients and oxygen, leading to three distinct cellular populations, similar to a poorly vascularized tumor.26 The Sutherland lab also characterized lactate and pH gradients within these colon spheroids.27–29 Spheroids consist of an outer proliferative region, a middle quiescent region and an inner apoptotic/necrotic core. We previously established spheroids as an effective model system to study the global proteomic effects of nutrient restriction.23
HCT 116 spheroids were divided into two treatment groups: full nutrients and glucose restriction (Figure 1). Both groups were grown for 10 days in full nutrient media. On day 10, the glucose restriction group was subjected to media depleted of glucose and full nutrient spheroids were maintained in full nutrient media. Both treatment groups received CQ treatment on day 10 and IR treatment on day 12. Spheroids were harvested on day 14. Dose response curves and autophagic assays were used to calculate the half maximal inhibitory concentration (IC50) of IR and CQ (Supplemental Figure 1). Both treatment groups were dosed with the IC50 values for IR and CQ.
IR is a first and second line therapy for metastatic colorectal cancer.30 Our laboratory previously analyzed the penetration and metabolism of irinotecan in spheroids with the use of mass spectrometry.26,31–33 We found that penetration of irinotecan into spheroids in both treatment groups is observed following 48 hours of treatment (Figure 2B, D, Supplemental Figure 2 B, D). This result is consistent with previous findings from our research group that irinotecan can fully penetrate HCT 116 spheroids.
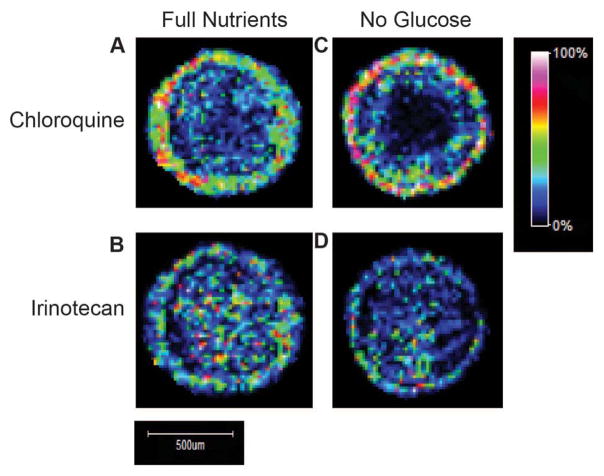
Figure 2.
MALDI-IMS analysis of treated HCT 116 spheroids (n=3). A. Imaging of CQ (m/z 320) in a spheroid treated with full nutrient media in addition to CQ and IR. B. Imaging of IR (m/z 587) in a spheroid treated with full nutrient media in addition to CQ and IR. C. Imaging of CQ in a spheroid treated with no glucose media in addition to CQ and IR. CQ is localized to the outer region of the spheroids in both conditions. D. Imaging of IR in a spheroid treated with no glucose media in addition to CQ and IR. Images are normalized to total ion current and intensity scale on right shows localization of ions. IR is distributed throughout the spheroid in both conditions.
CQ is an established anti-malarial drug currently being evaluated in clinical trials as a possible therapeutic for cancer. Following 96 hours of treatment, CQ is concentrated in the periphery of the spheroids and shows an increased penetration in full nutrient spheroids (Figure 2A, C, Supplemental Figure 2 A, C). CQ treatment for autophagy inhibition began at the same time as autophagy induction by glucose restriction. Since CQ was administered at a higher dosage and longer time scale than irinotecan, the data does not support that CQs localization is concentration or time dependent. Weakly basic chemotherapeutics have been shown to have reduced penetration at the typical acidic pH of a tumor.34–36 CQ is weakly basic and has shown decreased cellular uptake in acidic environments.37 As previously described, HCT 116 spheroids show gradients of oxygen and pH giving rise to a hypoxic and acidic core.27 The acidic environment of the spheroid core explains the lack of CQ penetration into the core region of the spheroid. Our data indicates that increased glucose levels in the full nutrient spheroids lead to a more viable and less acidic core attributing to increased penetration of CQ.
Changes in apoptosis and proliferation in HCT 116 spheroids due to combinations of CQ, IR and NR treatment
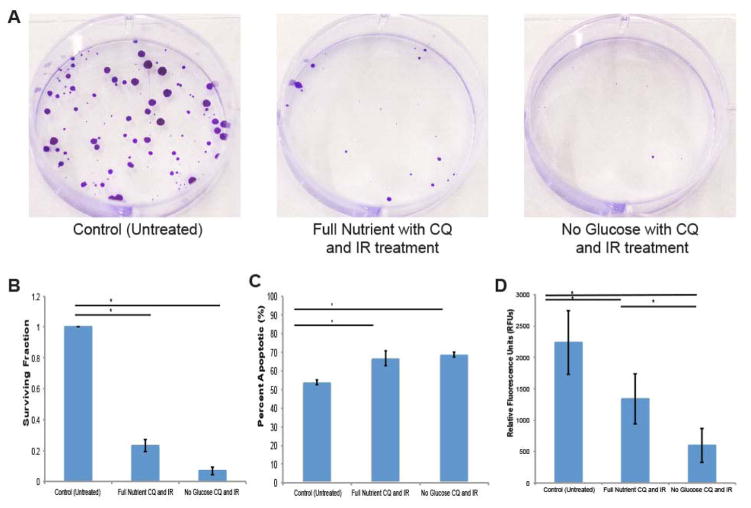
Treatment of CQ and IR significantly decreases the ability of HCT 116 cells to proliferate. Colony formation decreases to 23% when spheroids were treated with CQ and IR compared to the untreated control spheroids. When spheroids were subjected to glucose restriction in addition to CQ and IR, colony formation decreases to 7% compared to the untreated control spheroids (Figure 3A, B). The percentage of cell death increases with treatment as assessed by changes of early stage apoptotic cells stained by Annexin V (Figure 3C). There is a significant increase in the percentage of early apoptotic cells due to the combined treatment of glucose restriction, CQ, and IR. Spheroids treated with only CQ and IR show a 13% increase in apoptotic cells compared to the control, and spheroids treated with no glucose in addition to CQ and IR show a 15% increase of apoptotic cells. Assessing cell viability shows that CQ and IR treatment cause a decrease in cell viability (Figure 3D). Compared to the untreated control, spheroids treated with CQ and IR experience a 40% decrease in viability, while spheroids treated with no glucose in addition to CQ and IR experience a 73% decrease in viability. By treating cancer cells with autophagy inhibition in addition to chemotherapy, the cancer cells are unable to use autophagy to support survival, resulting in decreased cell survival. The addition of glucose restriction to the treatment of spheroid with CQ and IR intensifies the phenotypic results, as seen in Figure 3. These data support the idea that glucose metabolism can be exploited in the treatment of cancer.
Figure 3.
A. Images of clonogenic assay to assess cellular proliferation after treatment. HCT 116 spheroids were dissociated, counted, and plated (2000 cells/well, n=5). After six doublings (5.25 days) cells were fixed and stained with crystal violet and colonies were counted. B. Decrease in the surviving fraction of cells after treatment compared to untreated control for both full nutrient CQ and IR treated spheroids and no glucose CQ and IR treated spheroids (* = p<0.05). Data represented as average ± SD. C. Spheroids were trypsinized and stained with Annexin V to determine percentage of apoptotic cells after treatment. Both full nutrient CQ and IR treated spheroids and no glucose CQ and IR treated spheroids show an increase in apoptotic cells compared to untreated control (n=5, * = p<0.05). Data represented as an average ± SD. D. Cell viability assay of treated spheroids. Both full nutrient CQ and IR treated spheroids and no glucose CQ and IR treated spheroids show a decrease in viability compared to untreated control (n=10, * = p<0.05). Data represented as an average ± SD.
iTRAQ proteomic evaluation reveals similar protein regulation in both full nutrient and glucose restricted HCT 116 spheroids treated with CQ and IR
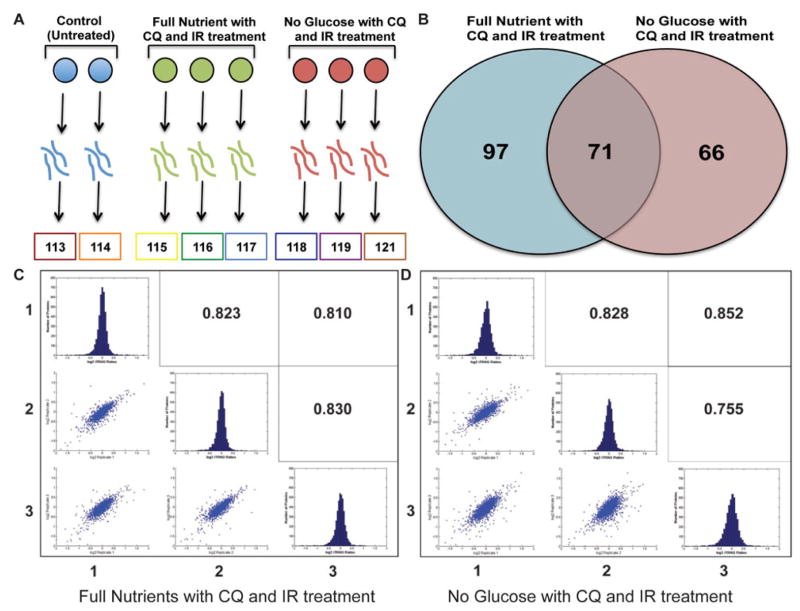
To determine the proteins that were differentially regulated by the treatment of glucose restriction, CQ and IR, we employed iTRAQ-based quantitative proteomics. The mass spectrometer is able to identify the peptide as well as the intensities of the iTRAQ reporter ions in the tandem spectra. The ability to do both measurements results in protein identification as well as fold change quantitation in treatment conditions compared to the untreated control. Proteins were harvested from spheroids from the untreated control in biological duplicate and the treated conditions in biological triplicate. The proteins were digested with trypsin and the subsequent peptides were labeled with eight-plex iTRAQ tags (Figure 4A). The combined sample mixture was then separated using strong cation exchange (SCX) fractionation into 30 fractions, then further fractionated by reversed-phase ultraperformance liquid chromatography. The sample was analyzed by a Q-Exactive mass spectrometer (UPLC-MS/MS).
Figure 4.
A. iTRAQ 8-plex experimental set-up. Spheroids (n=30) were homogenized, digested, and labeled with a specific reporter ion. Samples were mixed together, fractionated and analyzed by tandem mass spectrometry. B. Reporter ion intensities were compared to control and proteins outside the 1.5 log2 ratio were considered to be differentially up or down regulated. Full nutrient with CQ and IR treatment had 168 proteins that were differentially regulated and no glucose with CQ and IR treatment had 137 proteins differentially regulated, 71 of the proteins overlapped. C. and D. Log2 iTRAQ fold change ratios histograms were plotted of biological triplicates of treatment conditions compared to the untreated control. The biological triplicate conditions were compared with a Pearson’s correlation and scatter plot were used to measure linear correlation between replicates.
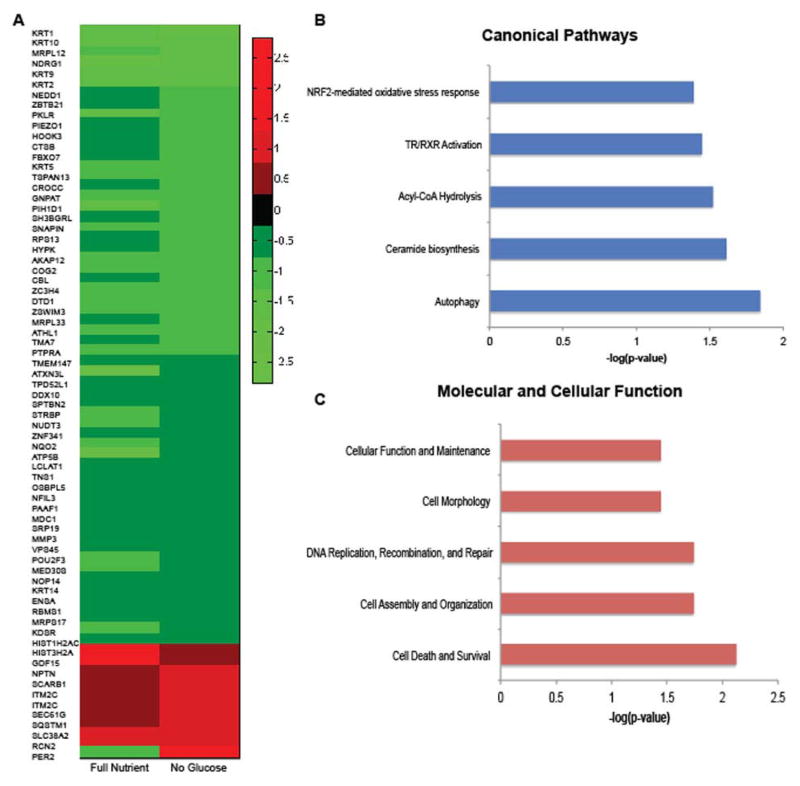
We identified and quantified 4,019 proteins at a 1% FDR. The average fold change distribution between treated and untreated spheroids was plotted on a log2 scale and found to have a tight distribution centered around zero, indicating that most proteins were not differentially regulated (Figure 4C, D). In addition, there is a positive linear relationship between the treated and untreated samples for all biological replicates as indicated by the Pearson correlation of approximately 0.8 (Figure 4C, D). Spheroids treated with CQ and IR had 168 proteins that were differentially regulated compared to the untreated control, while spheroids treated with no glucose in addition to CQ and IR had 137 proteins that were differentially regulated. A total of 71 proteins were found to be commonly differentially regulated by both treatments (Figure 4B). Only one of these proteins, period circadian clock 2 (PER2), is oppositely regulated. PER2 is down regulated in full nutrient, CQ, and IR treated spheroids and up regulated in glucose restricted, CQ, and IR treated spheroids (Figure 5A).
Figure 5.
A. A heatmap of the 71 common differentially regulated proteins between treatment conditions was plotted using MatLab. Only one protein, PER2, common to both conditions was oppositely regulated. B. The canonical pathways of the 71 common proteins were analyzed using IPA and the top 5 enriched pathways are displayed. C. The 71 common proteins were analyzed using IPA and the top 5 enriched molecular and cellular functions are displayed.
PER2 is a circadian rhythm protein. The circadian clock has been shown to control many cellular processes including metabolism, cell proliferation, cell death, and DNA damage response and repair.38,39 Most intriguing is that the overexpression of period circadian clock genes (PER1 and PER2) are associated with inhibiting cancer cell clonogenicity, and it has been suggested that PER2 should be a therapeutic target for treatment of malignant tumors.40 We found that when glucose is removed from HCT 116 spheroids that were treated with CQ and IR, PER2 protein expression is 2.5 fold increased (Figure 5A) and clonogenicity decreased (Figure 3A, B). This result suggests that glucose restriction increases expression of an important tumor suppressor protein. This increase could be a possible molecular mechanism of the phenotypic results.
To further characterize the proteins that were differentially regulated by both conditions, we performed functional classification using IPA software. The functional analysis categorizes proteins differentially regulated by both treatments into canonical pathways (Figure 5B) and molecular and cellular function (Figure 5C). In each dataset, we show the top five enriched pathways according to p value. This result indicates that HCT 116 colorectal cancer cells, with or without glucose restriction, have autophagy inhibited due to CQ treatment as predicted, and the differentially regulated proteins show enrichment of the autophagy pathway. Other pathways enriched included ceramide biosynthesis, acyl-CoA hydrolysis, Tr/RXR activation, and NRF2-mediated oxidative stress response. In addition, IPA identifies potential upstream regulators of the common differentially abundant proteins to be lysine-specific demethylase 3A (KDM3A), E3 ubiquitin-protein ligase RING2 (RNF2) and SP3 transcription factor. Many of these pathways are implicated in cancer progression.41
Treatment of CQ and IR on full nutrient spheroids enriches autophagy related pathways and stress response upstream regulators
The 168 proteins that were differentially abundant in full nutrient spheroids treated with CQ and IR were also analyzed using IPA. Functional analysis reveals that the canonical pathways that were enriched included autophagy and phagosome maturation (Supplemental Figure 2A). Phagosome maturation is the end of the phagocytic process seen both during autophagy and apoptosis.42 The enrichments of these pathways are explained by the treatment of CQ, which causes late stage autophagy inhibition. We see an increase in apoptotic cells due to treatment, which is logical as prolonged exposure to CQ causes cells to undergo apoptosis (Figure 3C). In addition, phosphatidylglycerol biosynthesis was enriched, which is involved in lipid metabolism. The molecular mechanisms that are enriched included cell death and survival, cellular development, and cell growth and proliferation, as expected since the treatment involved the chemotherapy drug IR (Supplemental Figure 2B).
The upstream regulator analysis tool in IPA allows for the prediction of activated and inhibited transcriptional regulators using an activation z-score. The treatment of full nutrient spheroids with CQ and IR causes a putative activation of tumor protein p53 (TP53) and cyclin dependent kinase inhibitor 1A (CDKN1A) based on the activation z-score (Table 1A). TP53 is the key regulator of apoptosis in cancer, and over 50% of all cancers have p53 mutations.43 CDKN1A expression is tightly controlled by p53 and can explain the putative activation of CDKN1A in response to CQ and IR treatment. The activation z-scores also indicate a putative inhibition of transforming growth factor beta 1 (TGFB1), tumor protein p63 (TP63), mitogen-activated protein kinase 9 (MAPK9), and nuclear protein 1 (NUPR1) (Table 1A), all of which are implicated in cancer. We find inactivation of NUPR1 to be very interesting. NUPR1 is found to be over expressed in many cancer types and is involved in chemoresistance and metastases.44–46 There have been studies that show that overexpression of NUPR1 enhances progression of cancer cells.47 Interestingly, NUPR1 is found to be unregulated in many cancers and prevents stress-induced death.45
Table 1. IPA analysis of upstream regulators due to treatment.
IPA upstream regulators tool was used to determine proteins putatively up or down regulated based on the differential abundance of the proteins from the iTRAQ dataset. Activation z-score is used to represent the likelihood of a protein to be activated or inactivated based on A. full nutrients with CQ and IR treatment and B. no glucose with CQ and IR treatment.
| A Full Nutrient with CQ and IR treatment | |
|---|---|
|
| |
| Upstream Regulator | Activation z-score |
| TP53 | 1.138 |
| CDKN1A | 1.000 |
| TGFB1 | −0.817 |
| TP63 | −0.927 |
| MAPK9 | −1.387 |
| NUPR1 | −1.387 |
|
| |
| B No Glucose with CQ and IR treatment | |
|
| |
| Upstream Regulator | Activation z-score |
|
| |
| TP53 | 1.315 |
| NUPR1 | −0.632 |
Glucose restriction of spheroids treated with CQ and IR causes enrichment of metabolism pathways and increased p53 activity
Removing glucose, the main energy for cancer cells,7,48 in addition to CQ and IR treatment, alters the top canonical pathways that are enriched. In addition to the autophagy pathway, IPA analysis of the 137 differentially regulated proteins show that the metabolism pathways of glycolysis, gluconeogenesis, and sucrose degradation are also enriched (Supplemental Figure 2C). This result is likely due to the Warburg Effect, the notion that cancer cells utilize glucose at a higher rate than normal cells and generate ATP primarily anaerobically through glycolysis.49,50 In a solid tumor, where oxygen is low, this increase in glucose consumption is evident.51 Exploiting the dependence of cancer cells on glucose by nutrient restriction or fasting is a standalone treatment option.8,52,53 Our data indicates that adding glucose restriction to autophagy inhibition and chemotherapy is a powerful way to impair cancer cells and inhibit cancer cell proliferation. Similar to the full nutrient treatment, the molecular and cellular functions shown to be enriched by IPA when spheroids are also treated with no glucose are cellular development, cell death and survival, and cellular growth and proliferation (Supplemental Figure 2D).
The upstream regulator tool in IPA revealed that TP53 is likely activated by no glucose, CQ, and IR treatment with a z-score of 1.315, and NUPR1 is putatively inhibited with a z-score of −0.632 (Table 1B). Our data supports the notion that glucose metabolism is linked to the expression of tumor suppressor genes as seen with the putative activation of p53 and the 2.5 fold up regulation of PER2 (Figure 5A). The combined treatment of CQ and IR with or without glucose restriction inactivates this potent cancer promoter.
Conclusions
This study compared autophagy inhibition and chemotherapy treatment with or without glucose restriction. We found that when glucose is restricted in addition to CQ and IR treatment in HCT 116 spheroids there is a decrease in cell viability and clonogenicity and an increase in cell death that may be tied to the up regulation of tumor suppressor proteins. The only protein, the tumor suppressor protein PER2, common to both conditions was found to be differentially regulated. In addition, pathway analysis using the proteomic data from these studies leads to the identification of upstream regulators. Many of these upstream regulators are tumor suppressor proteins, such as p53, that are found to be putatively activated, or cancer promoters such as NURP1 found to be putatively inactivated. PER2 activation has been shown to lead to a decrease in cancer cell clonogenicty.39,54 NURP1 is also implicated in affecting cancer cell clonogenicity.44,46
To our knowledge there are no proteomic studies that address the use of nutrient restriction and autophagy inhibition to potentiate the effects of chemotherapy. In 2012, Harhaji et al. showed that CQ treatment and calorie restriction in vivo resulted in a decrease of melanoma tumor weight and volume. This study concluded that CQ has a strong anticancer effect on calorically restricted tumors.20 More recently, Lashinger et al. investigated the metabolic effects of in vivo caloric restriction and autophagy inhibition using Atg5−/− (an autophagy related protein required for autophagy) in Ras-driven tumors.55 In this ground-breaking paper, the authors concluded that combining caloric restriction with autophagy inhibition reduced tumor growth more so than either treatment alone. This finding further promotes the use of CQ and nutrient restriction in combination as an anticancer therapy. While our results support these studies, we included the treatment of the cancer cells with a common chemotherapeutic to evaluate the anticancer activity of glucose deprivation and autophagy inhibition.
Previously, the mechanistic role of glucose metabolism and autophagy in colorectal cancer has been poorly understood. Our results indicate that adding glucose restriction and autophagy inhibition to treatment regimes for colorectal cancer can enhance the efficacy of the treatment. The downstream effect of this action is an increase in abundance of tumor suppressor proteins. An increase in the abundance of tumor suppressor proteins can lead to a decrease of clonogenicity and viability and an increase of cellular death through apoptosis.
The cellular processes of autophagy and glucose metabolism through glycolysis allow for cancer cells in solid tumors to survive chemotherapy. Targeting these cellular processes prior to chemotherapy treatment will weaken cancer cells making them more susceptible to treatment.
Supplementary Material
Supplemental Figure 1: Dose response curve for IR treatment, and autophagy assay for CQ treatment used to determine concentrations of IR and CQ treatments.
Supplemental Figure 2: MALDI-IMS analysis of treated HCT 116 spheroids replicates.
Supplemental Figure 3: Pathway analysis of Full nutrient with CQ and IR treatment spheroids and No glucose with CQ and IR treatment spheroids using IPA.
XLXS: MaxQuant Data Excel Sheet containing the raw data from MaxQuant for quantification of proteins using iTRAQ as well as the data analysis for determining up and down regulated proteins in treatment compared to the untreated control.
Acknowledgments
MS was supported by National Institutes of Health Training Grant –Chemistry Biochemistry Biology Interface Program (T32GM075762). ABH was supported by the National Institutes of Health (R01GM110406), and the National Science Foundation (CAREER Award, CHE-1351595). The UltrafleXtreme instrument (MALDI-TOF-TOF) was acquired through National Science Foundation award #1625944. We gratefully acknowledge the assistance of Dr. Susan Skube for her edits and the Notre Dame Mass Spectrometry and Proteomics Facility (MSPF).
Abbreviations
- CQ
chloroquine
- IR
irinotecan
- DHB
2,5-dihydroxybenzoic acid
- FA
Formic Acid
- ESI
electrospray ionization
- IPA
Ingenuity Pathway Analysis
- iTRAQ
isobaric tags for relative and absolute quantitation
- SCX
strong cation exchange
- PER2
period circadian clock 2
- KDM3A
lysine specific demethylase 3A
- RNF2
E3 ubiquitin-protein ligase RING2
- TP53
tumor protein p53
- CDKN1A
cyclin dependent kinase inhibitor 1A
- TGFB1
transforming growth factor beta 1
- TP63
tumor protein p63
- MAPK9
mitogen-activated protein kinase 9
- NUPR1
nuclear protein 1
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Persidis A. Cancer multidrug resistance. Nat Biotechnol. 1999;17:94–95. doi: 10.1038/5289. [DOI] [PubMed] [Google Scholar]
- 3.Lee C, Raffaghello L, Longo VD. Starvation, detoxification, and multidrug resistance in cancer therapy. Drug Resist Updat. 2012;15:114–122. doi: 10.1016/j.drup.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sui X, et al. Autophagy and chemotherapy resistance: a promising therapeutic target for cancer treatment. Cell Death Differ. 2013;4:1–12. doi: 10.1038/cddis.2013.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Senichkin VV, Kopeina GS, Zamaraev AV, Lavrik IN, Zhivotovsky BD. Nutrient restriction in combinatory therapy of tumors. Mol Biol. 2016;50:362–378. doi: 10.7868/S0026898416030101. [DOI] [PubMed] [Google Scholar]
- 6.Norheim F, et al. Molecular nutrition research-The modern way of performing nutritional science. Nutrients. 2012;4:1898–1944. doi: 10.3390/nu4121898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klement RJ, Kämmerer U. Is there a role for carbohydrate restriction in the treatment and prevention of cancer? Nutr Metab (Lond) 2011;8:75. doi: 10.1186/1743-7075-8-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raffaghello L, Safdie F, Bianchi G, Dorff T, Longo VD. Fasting and differential chemotherapy protection in patients. Cell Cycle. 2010;9:4474–4476. doi: 10.4161/cc.9.22.13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Longo VD, Mattson MP. Review Fasting: Molecular Mechanisms and Clinical Applications. Cell Metab. 2014;19:181–192. doi: 10.1016/j.cmet.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lv M, Zhu X, Wang H, Wang F, Guan W. Roles of Caloric Restriction, Ketogenic Diet and Intermittent Fasting during Initiation, Progression and Metastasis of Cancer in Animal Models: A Systematic Review and Meta-Analysis. PLoS One. 2014;9:e115147. doi: 10.1371/journal.pone.0115147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.White E. The role for autophagy in cancer. J Clin Invest. 2015;125:42–46. doi: 10.1172/JCI73941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Espina V, et al. Malignant Precursor Cells Pre-Exist in Human Breast DCIS and Require Autophagy for Survival. PLoS One. 2010;5:1–15. doi: 10.1371/journal.pone.0010240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niekerk G, Van Hattingh SM, Engelbrecht A. Enhanced Therapeutic Efficacy in Cancer Patients by Short-term Fasting: The Autophagy Connection. Front Oncol. 2016;6:1–7. doi: 10.3389/fonc.2016.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang ZJ, Chee CE, Huang S, Sinicrope FA. The role of autophagy in cancer: therapeutic implications. Mol Cancer Ther. 2011;10:1533–41. doi: 10.1158/1535-7163.MCT-11-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amaravadi RK, Thompson CB. The Roles of Therapy-Induced Autophagy and Necrosis in Cancer Treatment. Clin Cancer Res. 2007;13:7271–7280. doi: 10.1158/1078-0432.CCR-07-1595. [DOI] [PubMed] [Google Scholar]
- 16.Projean D, et al. In vitro metabolism of chloroquine: Identification of CYP2C8, CYP3A4, and CYP2D6 as the main isoforms catalyzing N-desethylchloroquine formation. Drug Metab Dispos. 2003;31:748–754. doi: 10.1124/dmd.31.6.748. [DOI] [PubMed] [Google Scholar]
- 17.Kimura T, Takabatake Y, Takahashi A, Isaka Y. Chloroquine in Cancer Therapy: A Double-Edged Sword of Autophagy. Cancer Res. 2013;73:3–9. doi: 10.1158/0008-5472.CAN-12-2464. [DOI] [PubMed] [Google Scholar]
- 18.Liang X, Tang J, Liang Y, Jin R, Cai X. Suppression of autophagy by chloroquine sensitizes 5-fluorouracil-mediated cell death in gallbladder carcinoma cells. Cell Biosci. 2014;4:1–11. doi: 10.1186/2045-3701-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.YZ, et al. Chloroquine Inhibits Colon Cancer Cell Growth In Vitro and Tumor Growth In Vivo via Induction of Apoptosis. Cancer Invest. 2009;27:286–292. doi: 10.1080/07357900802427927. [DOI] [PubMed] [Google Scholar]
- 20.Harhaji-Trajkovic L, et al. Chloroquine-mediated lysosomal dysfunction enhances the anticancer effect of nutrient deprivation. Pharm Res. 2012;29:2249–2263. doi: 10.1007/s11095-012-0753-1. [DOI] [PubMed] [Google Scholar]
- 21.Fan C, Wang W, Zhao B, Zhang S. Chloroquine inhibits cell growth and induces cell death in A549 lung cancer cells. Bioorg Med Chem. 2006;14:3218–3222. doi: 10.1016/j.bmc.2005.12.035. [DOI] [PubMed] [Google Scholar]
- 22.Li H, Hummon AB. Imaging Mass Spectrometry of Three-Dimensional Cell Culture Systems. Anal Chem. 2011;83:8794–8801. doi: 10.1021/ac202356g. [DOI] [PubMed] [Google Scholar]
- 23.Schroll MM, Liu X, Herzog SK, Skube SB, Hummon AB. Nutrient Restriction of Glucose or Serum Results in Similar Proteomic Expression Changes in 3D Colon Cancer Cell Cultures. Nutr Res. 2016 doi: 10.1016/j.nutres.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pampaloni F, Reynaud EG, Stelzer EHK. The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mol Cell Biol. 2007;8:839–845. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- 25.Yamada KM, Cukierman E. Modeling Tissue Morphogenesis and Cancer in 3D. Cell. 2007;130:601–610. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 26.Liu X, Weaver EM, Hummon AB. Evaluation of therapeutics in three-dimensional cell culture systems by MALDI imaging mass spectrometry. Anal Chem. 2013;85:6295–6302. doi: 10.1021/ac400519c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sutherland R. Cell and Enivronment Interactions in Tumor Microregions: The Spheroid Model. Science (80- ) 1988;240:177–240. doi: 10.1126/science.2451290. [DOI] [PubMed] [Google Scholar]
- 28.Freyer JP, Sutherland RM. Selective Dissociation and Characterization of Cells from Different Regions of Multicell Tumor Spheroids Selective Dissociation and Characterization Regions of Multiceli Tumor Spheroids1. Cancer Res. 1980;40:3956–3965. [PubMed] [Google Scholar]
- 29.Sutherland RM, Sordat B, Bamat J, Gabbert H, Bourrã B. Oxygennation and Differentiation in Multicellular Spheroids of Human Colon Carcinoma. Cancer Res. 1986;46:5320–5329. [PubMed] [Google Scholar]
- 30.Rougier P, et al. Randomised trial of irinotecan versus fluorouracil by continuous infusion after fluorouracil failure in patients with metastatic colorectal cancer. Lancet. 1998;352:1407–1412. doi: 10.1016/S0140-6736(98)03085-2. [DOI] [PubMed] [Google Scholar]
- 31.Liu X, Hummon AB. Quantitative determination of irinotecan and the metabolite SN-38 by nanoflow liquid chromatography-tandem mass spectrometry in different regions of multicellular tumor spheroids. J Am Soc Mass Spectrom. 2015;26:577–586. doi: 10.1007/s13361-014-1071-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.LaBonia GJ, Lockwood SY, Heller AA, Spence DM, Hummon AB. Drug penetration and metabolism in 3D cell cultures treated in a 3D printed fluidic device: assessment of irinotecan via MALDI imaging mass spectrometry. Proteomics. 2016;16:1814–1821. doi: 10.1002/pmic.201500524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X, Hummon AB. Mass Spectrometry Imaging of Therapeutics from Animal Models to Three-Dimensional Cell Cultures. Anal Chem. 2015;87:9508–9519. doi: 10.1021/acs.analchem.5b00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raghunand N, Mahoney BP, Gillies RJ. Tumor acidity, ion trapping and chemotherapeutics: II. pH-dependent partition coefficients predict importance of ion trapping on pharmacokinetics of weakly basic chemotherapeutic agents. Biochem Pharmacol. 2003;66:1219–1229. doi: 10.1016/s0006-2952(03)00468-4. [DOI] [PubMed] [Google Scholar]
- 35.Tannock IF, Rotin D. Acid pH in tumors and its potential for therpeutic exploitation. Cancer Res. 1989;49:4373–4384. [PubMed] [Google Scholar]
- 36.Pellegrini P, et al. Acidic extracellular pH neutralizes the autophagy-inhibiting activity of chloroquine: Implications for cancer therapies. Autophagy. 2014;10:562–571. doi: 10.4161/auto.27901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jensen PB, et al. Targeting the Cytotoxicity of Topoisomerase 11-directed Epipodophyllotoxins to Tumor Cells in Acidic Environments’. Cancer Res. 1994;54:2959–2963. [PubMed] [Google Scholar]
- 38.Wood PA, Du-Quiton J, You S, Hrushesky WJM. Circadian clock coordinates cancer cell cycle progression, thymidylate synthase, and 5-fluorouracil therapeutic index. Mol Cancer Ther. 2006;5:2023–2033. doi: 10.1158/1535-7163.MCT-06-0177. [DOI] [PubMed] [Google Scholar]
- 39.Wood Pa, Yang X, Hrushesky WJM. Clock genes and cancer. Integr Cancer Ther. 2009;8:303–8. doi: 10.1177/1534735409355292. [DOI] [PubMed] [Google Scholar]
- 40.Hua H, et al. Inhibition of tumorigenesis by intratumoral delivery of the circadian gene mPer2 in C57BL/6 mice. Cancer Gene Ther. 2007;14:815–8. doi: 10.1038/sj.cgt.7701061. [DOI] [PubMed] [Google Scholar]
- 41.Melchior F, Schergaut M, Pichler A. SUMO: Ligases, isopeptidases and nuclear pores. Trends Biochem Sci. 2003;28:612–618. doi: 10.1016/j.tibs.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 42.Kinchen JM, Ravichandran KS. Phagosome maturation: going through the acid test. Nature reviews Molecular cell biology. 2008;9 doi: 10.1038/nrm2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Munro AJ, Lain S, Lane DP. P53 abnormalities and outcomes in colorectal cancer: a systematic review. Br J Cancer. 2005;92:434–44. doi: 10.1038/sj.bjc.6602358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chowdhury UR, Samant RS, Fodstad O, Shevde LA. Emerging role of nuclear protein 1 (NUPR1) in cancer biology. Cancer Metastasis Rev. 2009;28:225–232. doi: 10.1007/s10555-009-9183-x. [DOI] [PubMed] [Google Scholar]
- 45.Hamidi T, et al. Nuclear protein 1 promotes pancreatic cancer development and protects cells from stress by inhibiting apoptosis. J Clin Invest. 2012;122:2092–2103. doi: 10.1172/JCI60144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cano CE, Iovanna JL. Stress proteins and pancreatic cancer metastasis. ScientificWorldJournal. 2010;10:1958–1966. doi: 10.1100/tsw.2010.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vasseur S, et al. Cloning and expression of the human p8, a nuclear protein with mitogenic activity. Eur J Biochem. 1999;259:670–675. doi: 10.1046/j.1432-1327.1999.00092.x. [DOI] [PubMed] [Google Scholar]
- 48.Panieri E, et al. Nutrient withdrawal rescues growth factor-deprived cells from mTOR-dependent damage. Aging (Albany NY) 2010;2:487–503. doi: 10.18632/aging.100183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Warburg O. Die Naturwissenschaften: Organ der Gesellschaft Deutscher Naturforscher und Ärzte. Über den Stoffwechsel der Carcinomzelle. 1924:1131–1137. [Google Scholar]
- 50.Heiden MG, Vander, et al. Understanding the Warburg Effect: Cell Proliferation. 2009;324:1029–1034. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beger R. A Review of Applications of Metabolomics in Cancer. Metabolites. 2013;3:552–574. doi: 10.3390/metabo3030552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Longo VD, Mattson MP. Fasting: Molecular Mechanisms and Clinical Applications. Cell Metab. 2014;19:181–192. doi: 10.1016/j.cmet.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mattson MP, Wan R. Beneficial effects of intermittent fasting and caloric restriction on the cardiovascular and cerebrovascular systems. J Nutr Biochem. 2005;16:129–137. doi: 10.1016/j.jnutbio.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 54.Gery S, Virk RK, Chumakov K, Yu A, Koeffler HP. The clock gene Per2 links the circadian system to the estrogen receptor. Oncogene. 2007;26:7916–7920. doi: 10.1038/sj.onc.1210585. [DOI] [PubMed] [Google Scholar]
- 55.Lashinger LM, et al. Starving cancer from the outside and inside: separate and combined effects of calorie restriction and autophagy inhibition on Ras-driven tumors. Cancer Metab. 2016;4:18. doi: 10.1186/s40170-016-0158-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Dose response curve for IR treatment, and autophagy assay for CQ treatment used to determine concentrations of IR and CQ treatments.
Supplemental Figure 2: MALDI-IMS analysis of treated HCT 116 spheroids replicates.
Supplemental Figure 3: Pathway analysis of Full nutrient with CQ and IR treatment spheroids and No glucose with CQ and IR treatment spheroids using IPA.
XLXS: MaxQuant Data Excel Sheet containing the raw data from MaxQuant for quantification of proteins using iTRAQ as well as the data analysis for determining up and down regulated proteins in treatment compared to the untreated control.