Overview
Advances in tumor genome sequencing have enabled discovery of actionable alterations leading to novel therapies. Currently, there are approved targeted therapies across various tumors that can be matched to genomic alterations, such as point mutations, gene amplification, and translocations. Tools to detect these genomic alterations have emerged as a result of decreasing costs and improved throughput enabled by next-generation sequencing (NGS) technologies. NGS has been successfully utilized for developing biomarkers to assess susceptibility, diagnosis, prognosis, and treatment of cancers. However, clinical application presents some potential challenges in terms of tumor specimen acquisition, analysis, privacy, interpretation, and drug development in rare cancer subsets. Although whole-genome sequencing offers the most complete strategy for tumor analysis, its present utility in clinical care is limited. Consequently, targeted gene capture panels are more commonly employed by academic institutions and commercial vendors for clinical grade cancer genomic testing to assess molecular eligibility for matching therapies, whereas whole-exome and transcriptome (RNASeq) sequencing are being utilized for discovery research. This review discusses the strategies, clinical challenges, and opportunities associated with the application of cancer genomic testing for precision cancer medicine.
Genomic sequencing technologies have enabled identification of actionable targets (e.g., BRAF in melanoma, EGFR in lung cancer) thus facilitating treatment selection beyond what is offered by conventional histopathologic methods (Fig. 1). Although NGS has helped identify genomic alterations and uncover novel targets for therapies, there are several barriers for translating this into clinical practice, such as informed consent, choosing a scalable cost-effective testing strategy, turnaround time, and clinical interpretation of results. Several pilot studies have addressed some of these hurdles and demonstrated the feasibility of offering genomic testing for patients with advanced cancer within a clinically relevant time frame and interpreting the results to facilitate new treatment options for patients.1–6 Today, cancer genomic testing has become more widely available in academic cancer centers and commercial testing labs.
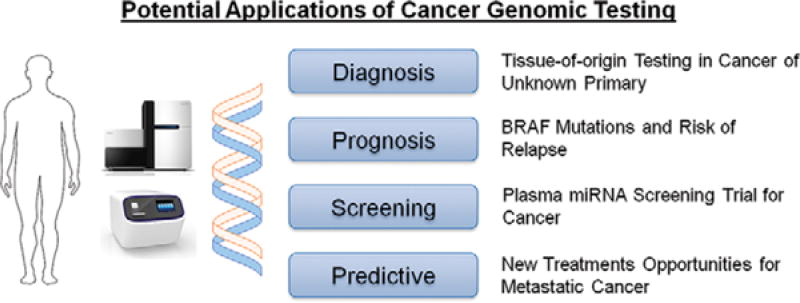
FIGURE 1. Potential Applications of Cancer Genomic Testing.
Cancer genomic sequencing assays can aid clinical decision making with potential implications for diagnosis, prognosis, and treatment. Several assays are available to aid in identifying tissue-of-origin in cancer of unknown primary, which may lead to identification of potential favorable subsets and their appropriate treatment options. For patients with metastatic or refractory cancer, multiple testing strategies are available to identify genomic alterations that may provide molecular eligibility for novel targeted therapies in clinical trials.
Although whole-genome, whole-exome, and whole-transcriptome sequencing offer an unbiased approach and opportunities for discovery, their immediate effect on clinical decision making is limited, as only a fraction of cancer genes are well characterized in terms of biology and therapeutic relevance. Further, these unbiased sequencing approaches remain expensive and time consuming and are burdensome for computational analysis. All of these limitations make these approaches less amenable to meet standards required for clinical testing, such as Clinical Laboratory Improvement Amendments (CLIA) and College of American Pathology certification. Instead, many academic cancer centers and commercial testing labs have developed focused cancer gene panels ranging from 25 to 400 genes. These cancer gene panels are more cost-effective, have faster turnaround times, and are more scalable for clinical grade testing (Sidebar 1). As an example, Foundation Medicine offers a targeted approach for the entire coding sequence of 315 cancer-related genes plus selected introns from 28 genes often rearranged in solid tumors.7 Caris provides an assay that combines immunohistochemistry, fluorescence in situ hybridization (FISH), and NGS for selected hotspot mutations involving select exons. However, ARUP Labs offers a targeted DNA sequencing assay for solid tumors for hotspot mutations in 48 genes.
SIDEBAR 1. Pros and Cons for In-House versus Outsourced Cancer Genomic Testing.
Customizable gene panel content
Access to quality metrics and variant frequencies
Cost-effective in the long run
Integration of reports into electronic health records
Unified platform for clinical use and discovery
Although genomic tumor testing has become available for patients and oncologists, there are several limitations to consider in practice including specimen quality, distinguishing driver and passenger mutations, tumor heterogeneity, and incidental germ-line mutations. Genomic testing and interpretation can be limited by tumor content and the quality of small, formalin-fixed tumor samples. Although formalin-fixed paraffin embedded (FFPE) samples may be subject to degradation complicating sequencing, strategies that accommodate FFPE samples have been successfully developed.8,9 At times, there may be a need for repeat or fresh frozen tumor biopsies as these may provide better quality for sequencing and can support specialized research assays. Although cancer gene panels focus on identification of potentially targetable driver mutations that provide a selective growth advantage for tumors, it is often hard to discern the role of passenger or bystander mutations. Also, pretreatment of tumors with cytotoxic therapies can often lead to increased genomic instability and alterations, making interpretation of tumor evolution and heterogeneity an arduous task.10 Gerlinger et al highlighted the challenges associated with tumor heterogeneity and the potential limitations of a single biopsy, as different sites of metastases may have different tumor subclones and mutations.11 However, it is expected that the dominant mutations exist as “trunk” mutations and would be the priority target for therapy. Finally, to aid in distinguishing driver and passenger mutations, it may also be helpful to have tumor and normal genomic testing by assessing germ-line DNA obtained from blood or buccal smears to allow distinction of somatic variants from germ line. However, germ-line sequencing also presents issues including the need for informed consent, counseling, expertise to interpret germ-line findings, and disclosure.12 This requires substantial resources and time and limits the broader use of germ-line tissue in oncology practices. Consequently, most commercial vendors offer tumor only testing.
STRATEGIES FOR MOLECULAR PROFILING
Whole-genome sequencing offers the most comprehensive strategy for tumor genomic analysis; however, it is currently limited in its clinical applicability because of cost and turnaround time for sequencing and analysis. Therefore, strategies that incorporate targeted gene sequencing are preferred for clinical applications, reducing cost, and offering a faster turnaround time. Nonetheless, since these panels test for select genes, they may miss opportunities for discovery that are afforded by other intermediate approaches, such as whole exome and transcriptome sequencing, which focuses on the expressed components of the genome.
Targeted DNA Sequencing
Although comprehensive approaches are necessary to achieve a complete genomic characterization of a patient's tumor, many clinical laboratories have employed targeted DNA sequencing approaches as a practical alternative.7,13–15 Targeted sequencing of hundreds of genes selected according to their relevance to cancer enables cheaper and higher-throughput molecular profiling of patients' tumors and incurs more manageable computational requirements with regard to data storage and analysis. Further, the deeper sequence coverage afforded by targeted sequencing can result in increased detection sensitivity for mutations in heterogeneous or low purity tumors. Consequently, large numbers of patients can be screened for genomic alterations, predicting response to approved and investigational targeted therapies, with high confidence that all clinically significant mutations will be detected.
Several products are available for capturing and sequencing genomic regions of interest—all compatible with FFPE tumor tissue. Target capture methods fall into two main classes: enrichment by amplification and enrichment by hybridization. Enrichment by amplification, or amplicon capture, relies on a highly multiplexed polymerase chain reaction involving locus-specific primer pairs simultaneously amplifying target regions in the genome. Amplicon capture can produce deep sequence coverage with very little DNA, but it is typically suitable only for a limited number of genes. Further, amplicon capture methods may not be reliable for the detection of structural alterations such as copy number gains or losses and translocations. Panels based on amplicon capture often target hotspots of recurrent somatic mutations rather than the entire coding sequence of the genes and are typically coupled with benchtop DNA sequencers, such as the Illumina MiSeq and the Ion Torrent Personal Genome Machine. Enrichment by hybridization, or hybridization capture, utilizes synthetic DNA or RNA probes that bind to and enrich for complementary genomic DNA. Hybridization capture usually requires more input DNA than amplicon capture, but it can scale to a larger number of genes (up to the whole exome). Panels based on hybridization capture typically target all coding sequences of all genes and can be coupled with either benchtop or production sequencers, such as the Illumina HiSeq and the Ion Torrent Proton. Importantly, hybridization capture methods enable the accurate detection of copy number alterations and selected structural rearrangements.16 Both amplicon and hybridization capture methods benefit greatly from the use of sample barcodes, which permit many tumors to be profiled in a single NGS run.
One of the most important decisions in the development of any cancer gene panel is what the content should be. Ideally the test will encompass all genes with “actionable” mutations that may affect a patient's treatment course. Additional genes may be considered if they have demonstrated biologic importance based on preclinical evidence. Custom panels afford the flexibility to target noncoding sequence, such as promoters and regulatory regions in the assay. By capturing introns of recurrently rearranged genes, it is possible to detect genomic breakpoints that produce gene fusions—many of which may be targeted by approved or experimental therapies. Ultimately, decisions about which and how many genes to sequence are best made by individual clinical laboratories according to their anticipated volume of cases, desired turnaround time and cost, and whether matched normal tissue is available for companion analysis.
RNA Sequencing in the Clinic
In addition to targeted DNA sequencing, RNA sequencing (RNAseq) can be supplemented by profiling the cancer transcriptome. RNAseq can provide data on gene expression, mutations, and gene fusions in cancer. Gene fusions have long been recognized as important in the oncogenesis of hematologic malignancies (e.g., BCR-ABL1 fusion in chronic myeloid leukemia); however, these were not well studied in solid tumors until the advent of NGS. RNASeq can be utilized for detection of novel fusions at a fraction of the cost of whole-genome sequencing. This has resulted in the characterization of novel oncogenic fusions with matching targeted therapies across various tumors (e.g., ALK, ROS1, RET fusions in lung cancers).17,18 Also, RNASeq can detect cryptic or novel translocations or gene fusions in leukemia that are not detectable by standard karyotyping or FISH.19 Nevertheless, RNAseq sequencing remains expensive and not easily implemented in clinical labs. Therefore, strategies that utilize targeted RNASeq may be preferable for rapid turnaround and reduced cost.20 For example, Zheng et al implemented an anchored multiplex RNAseq strategy to detect selected gene fusions in cancer. For sarcomas, Qadir et al demonstrated the feasibility of targeted RNAseq to detect prototypic fusions, which could replace costly FISH assays and facilitate detection of novel fusions.21 Several commercial and academic entities are developing additional clinical grade RNAseq strategies. Consequently, we anticipate a combination of DNA and RNA sequencing may have synergy as a clinical tumor sequencing strategy, incorporating data on DNA mutations and gene expression.
In addition to gene expression and fusions, RNASeq permits broader profiling of the cancer transcriptome, including detection of noncoding RNAs (ncRNA) such as microRNAs (miRNAs), small interfering RNAs, ribosomal RNAs, small nucleolar RNAs, and long noncoding RNAs. In fact, more than half of the cancer transcriptome consists of ncRNAs.22 These ncRNAs have been shown to be important in multiple cellular processes, such as gene silencing, DNA replication, and regulation of transcription and translation.23 Although the majority of these RNAs may not be applicable for clinical use, there is abundant potential for biomarker discovery and development. For example, miRNA profiling has been utilized to develop cancer-specific signatures that could be developed as diagnostic, early screening, prognostic, and treatment predictive biomarkers.24 A commercially available test has been developed to help classify tissue of origin in patients with cancer of unknown primary. These tissue-of-origin tests assess miRNA signatures that were derived from miRNA profiling 20 to 40 cancer subtypes and can help clinicians identify favorable subsets for cancer of unknown primary that may lead to treatment decisions.25,26 In another example, Sozzi et al developed a signature of miRNA from plasma of patients with lung cancer, and they are studying whether this test can improve accuracy of screening in combination with CT scans in a prospective screening study for lung cancer.27 Thus, there is great potential for ncRNA-specific signatures to aid diagnostic dilemmas and potentially identify new treatment options as we learn more about ncRNA biology.
IMPLEMENTATION OF A CLINICAL SEQUENCING WORKFLOW
Challenges and Considerations
When developing clinical sequencing workflows, academic and commercial laboratories must confront many challenges. In contrast to the research setting, where large, high-purity, fresh-frozen tumors can be prioritized for analysis, clinical laboratories must be equipped to analyze specimens of all sizes and qualities. These may include small biopsies, fine-needle aspirates, or FFPE samples that are heterogeneous or admixed with normal tissue. Sequencing protocols must be optimized for low-quality specimens and low-input quantities and still deliver deep coverage sequence data for the reliable detection of mutations with low allele frequency. Further, laboratories must be able to deliver results with a rapid turnaround time at a reasonable cost. The use of germ-line DNA from blood or healthy tissue as a normal control has major benefits for the analysis and interpretation of somatic mutations, but it creates logistic challenges involving tracking and transporting separate samples from the same patient. As multiple tumor samples from a given patient may occasionally be sequenced to monitor tumor progression and acquired resistance to therapy, flexible workflows are necessary to accommodate longitudinal analysis. Though the nature of these issues may vary for different laboratories depending on their throughput and sequencing platforms, they represent technical challenges common to all clinical laboratories.
Bioinformatics and analysis requirements collectively represent another important challenge in establishing a clinical sequencing workflow. The bioinformatics algorithms and software required to call different classes of genomic alterations (sequence mutations, insertions, deletions, copy number gains and losses, and structural rearrangements) are constantly evolving, and there remains no consensus on the best approach or standardized pipeline. Laboratories performing NGS have the choice of utilizing third-party software for data analysis, which can be costly and limits flexibility, or developing custom pipelines in-house, which requires considerable time and expertise to build and maintain. Either way, a significant informatics infrastructure is needed to manage, store, and archive the data generated by the sequencing instruments and the results produced by bioinformatics pipelines. Access to high-performance computing resources for processing and analyzing sequence data is required. Laboratory information management systems and associated databases are typically also needed to track samples, experiments, and results. For hospitals and clinical laboratories that have not traditionally employed many computational biologists and software engineers, the recruitment and training of bioinformatics staff is challenging yet essential.
Regulatory requirements, including the up-front analytic and clinical validation of assays, must also be met in any clinical sequencing workflow. Clinical laboratories producing results that are returned to patients and used in treatment decisions are subject to legal obligations designed to ensure that tests are reproducible and adhere to high standards of sensitivity and specificity. Such labs, whether commercial or academic, must be compliant with the CLIA, under the oversight of the Centers for Medicare & Medicaid Services. Accordingly, extensive documentation and technical validation of diagnostic assays are a requirement for patient testing and subsequent billing to insurance companies and Medicare. The model that some institutions have adopted wherein large-scale sequencing is performed in research labs, followed by confirmation in CLIA-compliant labs, is unsustainable over the long-term if the costs of NGS cannot be recovered through reimbursement.
To achieve maximum clinical benefit from a diagnostic sequencing assay, results must be reported to clinicians in a clear and easily digestible way, yet with all supporting information necessary to interpret the significance of the collection of genomic alterations that were detected.28 The interpretation of results is frequently dependent on tumor type and other clinical factors and must be considered in that context. Also, although the goal is to identify actionable driver mutations, clinical sequencing assays typically turn up far more passenger mutations with unclear biologic and clinical significance. This is especially true in tumors with a high background mutation rate because of environmental exposures or abnormalities in DNA mismatch repair. It can be very difficult for a clinician to distinguish between key driver alterations that should affect treatment and passenger mutations with no apparent significance. Many academic cancer centers have created genomic or molecular “tumor boards” to collectively discuss and interpret challenging cases and recommend a course of action that the treating physician can take.4,29 As this process does not easily scale to accommodate the large number of tumors being sequenced today, groups have attempted to curate and codify biologic and clinical information about mutations into databases that can be queried or utilized to annotate molecular diagnostic reports.9,30 These “knowledge bases” must be granular enough to account for the fact that different sequence variants in the same gene may have opposite effects, and the same variant in different tumor types may have different clinical consequences. They should also enable the enumeration of clinical trials that might be beneficial to the patient, given their molecular profile. Nevertheless, no knowledge base is comprehensive or will ever include information on all possible alterations that a sequencing assay may reveal. Further, the co-occurrence of multiple driver mutations may have implications that cannot be inferred from the functions or clinical consequences of each individual mutation alone.
With the exception of targeted panels focused on mutational hotspots, germ-line DNA is typically used as a normal control to distinguish somatic mutations from inherited variants. In the absence of germ-line DNA, variants identified from tumor sequencing must be filtered according to databases of common single nucleotide polymorphisms. This can lead to false-positive mutation calls at sites of rare inherited single nucleotide polymorphisms, including cancer susceptibility alleles. The inclusion of germ-line DNA enables somatic mutations to be unambiguously called; yet it also enables the detection of pathogenic variants in the genes that are sequenced. This has considerable ethical and logistic implications. Incidental findings may emerge as a result of tumor sequencing that relate to a patient's inherited susceptibility to cancer or other diseases, with unanticipated yet significant consequences for family members who share these variants.31,32 At present, tests specifically designed to search for inherited genetic variants typically require that patients sign informed consent and are properly educated of the benefits and risks of the test by a genetic counselor. For laboratories setting up large-volume tumor sequencing initiatives, individual pretest genetic counseling of all patients may be untenable. Computational subtraction of germ-line variants during mutation calling may circumvent the requirement for pretest counseling, but it also has the consequence that inherited variants with potential clinical significance are willfully disregarded.33 Regardless of the circumstances, when an inherited predisposition to cancer (or another disease) is discovered, care must be taken to ensure the privacy and autonomy of patients and their families and to help manage the emotional and psychologic consequence of such a diagnosis.
As discussed above, the sustainability of clinical NGS initiatives depends on reimbursement from insurance companies and Medicare. However, at present, molecular diagnostic testing is only reimbursed in a small number of tumor types where there are approved drugs whose administration depends on a positive or negative test result and where there are clear clinical guidelines mandating the use of molecular testing. Examples include lung adenocarcinoma, colorectal cancer, and melanoma. In other tumor types where comparable guidelines do not exist, more data are needed to determine the clinical utility of NGS-based molecular profiling. As a result, large academic cancer centers are investing considerable philanthropic and other institutional funds into clinical sequencing of nonbillable tumor types. Demonstrating the clinical utility of tumor sequencing across additional cancer types is essential to ensure greater reimbursement and promote broader access to testing outside of the largest centers. A related issue emerges when actionable mutations are detected in unexpected tumor types, and insurance companies are not always willing to reimburse the cost of the drug for an off-label indication. The emergence of molecularly guided clinical trials encompassing multiple tumor types, or “basket” clinical trials, may help some patients in this situation.
In-house versus Outsourcing
With all of these challenges inherent to the establishment of clinical sequencing infrastructure, it is tempting for academic cancer centers and community oncology practices to outsource the entire process to a commercial reference laboratory. For many centers, this may be the best decision given financial, volume, and staffing considerations. However, there are many advantages to setting up these processes in-house that can justify the effort and expense from an institutional perspective (Table 1).
TABLE 1.
Commercial Targeted DNA Tumor Sequencing
| Vendor | Name | No. of Genes | Results | Time |
|---|---|---|---|---|
| Foundation Medicine | Foundation One | 315 (plus introns from 28 genes) | SNVs. CNVs. fusions | 12–14 days |
| Caris Life Sciences | MI Profile | 46 | Hotspot mutations | 14 days |
| ParadigmDx | PCDx | 114 | SNVs. CNVs. fusions | 4–5 days |
| ARUP Labs | Solid Tumor Mutation Panel | 48 | Hotspot mutations | 14 days |
| PathGroup | SmartGenomics | 35 | Hotspot mutations | 7–10 days |
| Knight Diagnostic Labs | GeneTrails Cancer Gene Panel | 38 | Hotspot mutations | 10–14 days |
| Life Technologies | Pervenio Lung NGS Assay | 25 | SNVs. fusions | 7 days |
Abbreviations: SNV, single nucleotide variation or point mutation; CNV, copy number variation; NGS, next-generation sequencing.
First, developing and validating a tumor sequencing test in-house gives the laboratory ultimate control over the content of the assay. Panels can be designed to include targets of all clinical trials open within the institution as well as genes and noncoding regions suggested by preclinical research studies. Further, as new clinical knowledge emerges and new clinical trials are developed, additional genes and biomarkers can rapidly added. Second, the laboratory and collaborating investigators will have access to all raw data, including sequence quality metrics and mutation allele frequencies, which one would not expect to receive from an outside provider. This can enable the development of custom bioinformatics pipelines to explore additional features of the data such as tumor heterogeneity and clonal evolution, and the reanalysis of data as new tools and algorithms are created. Importantly, data access will also facilitate data mining initiatives through integration of clinical and phenotypic data for the patients whose tumors were sequenced. Third, though the establishment of clinical NGS tests requires a large up-front investment, it may ultimately lead to lower costs than commercial providers will offer, especially for large-volume laboratories. Additionally, as discussed above, institutions can use philanthropic and institutional funds to pay for nonbillable tests that will produce data to possibly justify reimbursement in the future. Fourth, analysis results and molecular reports can be integrated directly with the hospital information systems. This can facilitate rapid reporting, deposition into institutional databases, and the screening and selection of patients for clinical trials. As clinical trials in oncology increasingly require the presence of particular (often rare) genomic alterations, an institutional molecular database can help identify the patients that meet all eligibility criteria and are most likely to benefit from a new drug. Fifth, the same test that is validated and approved for clinical use can also be used to retrospectively analyze archived tumors, such as those obtained from “exceptional responders,” for research purposes.34,35 This allows data from a single platform to be merged and mined with greater power to discover biomarkers that correlate with clinical outcomes and/or response or resistance to therapy. It also provides additional flexibility to further develop and optimize the assay for other types of specimens, such as cell-free DNA from plasma or cerebrospinal fluid.
IMPACT OF CANCER GENOMIC TESTING IN THE CLINIC
Although the availability of cancer genomic testing in the clinic has led to opportunities in oncology such as drug target discovery, it has also led to challenges including how to develop targeted therapies for small populations of patients with rare mutations. For example, in contrast to the frequency of ERBB2 amplification in breast cancer and BRAF mutations in melanoma (20% and 45%, respectively), the majority of actionable genomic alterations revealed by clinical tumor sequencing typically occur at frequencies of 2% to 5%. This has raised several challenges for delivering treatment to patients and developing novel therapies in clinical trials.
For example, cancer genomic sequencing enabled discovery of novel activating somatic point mutations the ERBB2 gene in patients with breast cancer who were negative for ERBB2 gene (HER2) amplification, but these mutations are only found in 1% to 2% of patients with breast cancer.36 In vitro studies demonstrated that these activating mutations conferred resistance to reversible inhibitor lapatinib but were sensitive to neratinib, an irreversible ERBB2 inhibitor.36 This has led to a phase II study of neratinib for patients with metastatic ERBB2-mutant breast cancer.2 Although this strategy appears rational for a disease- and mutation-specific trial, moving forward for other uncommon genomic alterations within a single tumor type, typical large randomized phase III trials will not be pragmatic for each disease and each mutation subset. Meanwhile predictive biomarker selection has led to exceptional tumor responses to matching therapies, and phase II trials may provide convincing evidence of clinical activity and benefit.37–39 The low prevalence of actionable oncogenic mutations has led to the evolution of “basket trials.” Unlike a conventional tumor histology–based clinical trial, patient selection is based on a specific genomic alteration and not on tumor type.40 This is different from BATTLE or I-SPY trials in which adaptive design is utilized to enrich patients into specific molecular cohorts based on initial efficacy results, while restricted to a single tumor type. Presently, basket trial approaches will unlikely lead to regulatory drug approval in a specific tumor type, but they help assess whether all or selected cancer types with specific genomic alterations (e.g., FGFR, BRAF) would indeed respond to a matching targeted therapy, and they help consider other endpoints, such as magnitude of response, duration of responses, and the study of novel predictive biomarkers for sensitivity and resistance.41 Finally, basket trials enable enrollment of multiple tumor types and facilitate patient accrual for both rare cancers and genomic alterations.
In addition to prospective trials, cancer genomic testing may help us understand clinical responses retrospectively based on patients who have exceptional responses to therapy. Iyer et al observed a complete, durable response of more than 2 years in a single patient with metastatic bladder cancer with everolimus treatment on a clinical trial that did not meet its primary endpoint. They performed whole-genome sequencing of the tumor, which revealed a mutation in TSC1, a gene involved in the mTOR pathway. They subsequently demonstrated a basis for clinical response to everolimus, an mTOR inhibitor.34 Similarly, whole-exome sequencing on a patient with metastatic anaplastic thyroid cancer, who had an exceptional response to everolimus, identified a mutation in TSC2, a negative regulator of the mTOR pathway.42 These individual patients highlight an application of genomic sequencing to understand how we can learn from exceptional responders to guide further development of targeted therapies. The National Cancer Institute and academic cancer centers are actively seeking to apply this approach in ongoing clinical trials across the country to guide drug development based on novel predictive biomarkers.
Finally, as we learn to identify the correct genomic alteration that can predict response to a therapy, we must also prospectively consider how cancers become resistant to therapy. Although targeted therapies may lead to remarkable initial responses for patients with selected biomarkers, metastatic cancers inevitably acquire resistance. NGS has been utilized to characterize mechanisms of secondary resistance to identify potential combinatorial therapies that can prevent or delay emergence of resistance. Wagle et al performed NGS in a patient with metastatic melanoma who developed resistance to vemurafenib after showing initial response. They identified an acquired mutation in MEK1, conferring resistance to RAF or MEK inhibition.43 Tumor sequencing has also helped identify resistance mechanisms in long-established therapies, such as estrogen blockade in breast cancer, where ligand-binding domain mutations in ESR1 (the estrogen receptor) mediate acquired resistance to antihormonal therapy.44,45 Similarly, whole-exome sequencing has been utilized to study acquired resistance to the recently approved BTK inhibitor ibrutinib in relapsed chronic lymphocytic leukemia and has identified a mutation in BTK that limits drug binding.46
CONCLUSION
Although the application of clinical tumor sequencing has enabled identification of actionable genomic alterations that could provide molecular eligibility to matching targeted therapies, clinical application and interpretation does have some challenges. Intratumor heterogeneity, discerning drivers from passenger mutations, lack of sustained response, and acquired resistance to targeted therapies are some of the issues that limit the potential of genomics-driven targeted therapies. Further, responses to targeting tend to vary across tumors and within a tumor depending on the treatment context. Biopsies at multiple time points, rational combination therapies, and basket trial designs can help address some of these issues. As oncology is migrating to a more molecularly matched therapy paradigm, a strong collaboration between basic scientists, molecular pathologists, bioinformaticians, and oncologists is paramount in an effort to identify novel cancer therapies that lead to improvement in patient survival.
Acknowledgments
Thanks to Cassie Hershberger for administrative support. Thanks to ASCO for organizing this education session.
References
- 1.Weiss GJ, Liang WS, Izatt T, et al. Paired tumor and normal whole genome sequencing of metastatic olfactory neuroblastoma. PLoS One. 2012;7:e37029. doi: 10.1371/journal.pone.0037029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. NCT01670877. Neratinib in Metastatic HER2 Non-amplified But HER2 Mutant Breast Cancer. [Accessed February 8, 2015]; https://clinicaltrials.gov/ct2/show/NCT01670877.
- 3.Welch JS, Westervelt P, Ding L, et al. Use of whole-genome sequencing to diagnose a cryptic fusion oncogene. JAMA. 2011;305:1577–1584. doi: 10.1001/jama.2011.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roychowdhury S, Iyer MK, Robinson DR, et al. Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci Transl Med. 2011;3:111ra121. doi: 10.1126/scitranslmed.3003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Link DC, Schuettpelz LG, Shen D, et al. Identification of a novel TP53 cancer susceptibility mutation through whole-genome sequencing of a patient with therapy-related AML. JAMA. 2011;305:1568–1576. doi: 10.1001/jama.2011.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones SJ, Laskin J, Li YY, et al. Evolution of an adenocarcinoma in response to selection by targeted kinase inhibitors. Genome Biol. 2010;11:R82. doi: 10.1186/gb-2010-11-8-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xuan J, Yu Y, Qing T, et al. Next-generation sequencing in the clinic: promises and challenges. Cancer Lett. 2013;340:284–295. doi: 10.1016/j.canlet.2012.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Allen EM, Wagle N, Stojanov P, et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat Med. 2014;20:682–688. doi: 10.1038/nm.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shyr D, Liu Q. Next generation sequencing in cancer research and clinical application. Biol Proced Online. 2013;15:4. doi: 10.1186/1480-9222-15-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Everett JN, Gustafson SL, Raymond VM. Traditional roles in a non-traditional setting: genetic counseling in precision oncology. J Genet Couns. 2014;23:655–660. doi: 10.1007/s10897-014-9698-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cottrell CE, Al-Kateb H, Bredemeyer AJ, et al. Validation of a next-generation sequencing assay for clinical molecular oncology. J Mol Diagn. 2014;16:89–105. doi: 10.1016/j.jmoldx.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pritchard CC, Salipante SJ, Koehler K, et al. Validation and implementation of targeted capture and sequencing for the detection of actionable mutation, copy number variation, and gene rearrangement in clinical cancer specimens. J Mol Diagn. 2014;16:56–67. doi: 10.1016/j.jmoldx.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wagle N, Berger MF, Davis MJ, et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;2:82–93. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Won HH, Scott SN, Brannon AR, et al. Detecting somatic genetic alterations in tumor specimens by exon capture and massively parallel sequencing. J Vis Exp. 2013:e50710. doi: 10.3791/50710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lipson D, Capelletti M, Yelensky R, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012;18:382–384. doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takeuchi K, Soda M, Togashi Y, et al. RET. ROS1 and ALK fusions in lung cancer. Nat Med. 2012;18:378–381. doi: 10.1038/nm.2658. [DOI] [PubMed] [Google Scholar]
- 19.Roberts KG, Li Y, Payne-Turner D, et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med. 2014;371:1005–1015. doi: 10.1056/NEJMoa1403088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng Z, Liebers M, Zhelyazkova B, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med. 2014;20:1479–1484. doi: 10.1038/nm.3729. [DOI] [PubMed] [Google Scholar]
- 21.Qadir MA, Zhan SH, Kwok B, et al. ChildSeq-RNA: a next-generation sequencing-based diagnostic assay to identify known fusion transcripts in childhood sarcomas. J Mol Diagn. 2014;16:361–370. doi: 10.1016/j.jmoldx.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 22.Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. doi: 10.1038/ng.3192. Epub 2015 Jan 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quinodoz S, Guttman M. Long noncoding RNAs: An emerging link between gene regulation and nuclear organization. Trends Cell Biol. 2014;24:651–663. doi: 10.1016/j.tcb.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenfeld N, Aharonov R, Meiri E, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26:462–469. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- 26.Pentheroudakis G, Pavlidis N, Fountzilas G, et al. Novel microRNA-based assay demonstrates 92% agreement with diagnosis based on clinicopathologic and management data in a cohort of patients with carcinoma of unknown primary. Mol Cancer. 2013;12:57. doi: 10.1186/1476-4598-12-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sozzi G, Boeri M, Rossi M, et al. Clinical utility of a plasma-based miRNA signature classifier within computed tomography lung cancer screening: a correlative MILD trial study. J Clin Oncol. 2014;32:768–773. doi: 10.1200/JCO.2013.50.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Allen EM, Wagle N, Levy MA. Clinical analysis and interpretation of cancer genome data. J Clin Oncol. 2013;31:1825–1833. doi: 10.1200/JCO.2013.48.7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tran B, Brown AM, Bedard PL, et al. Feasibility of real time next generation sequencing of cancer genes linked to drug response: results from a clinical trial. Int J Cancer. 2013;132:1547–1555. doi: 10.1002/ijc.27817. [DOI] [PubMed] [Google Scholar]
- 30.Levy MA, Lovly CM, Pao W. Translating genomic information into clinical medicine: lung cancer as a paradigm. Genome Res. 2012;22:2101–2108. doi: 10.1101/gr.131128.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Green RC, Berg JS, Grody WW, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Catenacci DV, Amico AL, Nielsen SM, et al. Tumor genome analysis includes germline genome: are we ready for surprises? Int J Cancer. 2015;136:1559–1567. doi: 10.1002/ijc.29128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bombard Y, Robson M, Offit K. Revealing the incidentalome when targeting the tumor genome. JAMA. 2013;310:795–796. doi: 10.1001/jama.2013.276573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iyer G, Hanrahan AJ, Milowsky MI, et al. Genome sequencing identifies a basis for everolimus sensitivity. Science. 2012;338:221. doi: 10.1126/science.1226344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wagle N, Grabiner BC, Van Allen EM, et al. Activating mTOR mutations in a patient with an extraordinary response on a phase I trial of everolimus and pazopanib. Cancer Discov. 2014;4:546–553. doi: 10.1158/2159-8290.CD-13-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bose R, Kavuri SM, Searleman AC, et al. Activating HER2 mutations in HER2 gene amplification negative breast cancer. Cancer Discov. 2013;3:224–237. doi: 10.1158/2159-8290.CD-12-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaw AT, Kim DW, Mehra R, et al. Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med. 2014;370:1189–1197. doi: 10.1056/NEJMoa1311107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ledermann J, Harter P, Gourley C, et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N Engl J Med. 2012;366:1382–1392. doi: 10.1056/NEJMoa1105535. [DOI] [PubMed] [Google Scholar]
- 40.Willyard C. ‘Basket studies’ will hold intricate data for cancer drug approvals. Nat Med. 2013;19:655. doi: 10.1038/nm0613-655. [DOI] [PubMed] [Google Scholar]
- 41.Sharma MR, Schilsky RL. Role of randomized phase III trials in an era of effective targeted therapies. Nat Rev Clin Oncol. 2012;9:208–214. doi: 10.1038/nrclinonc.2011.190. [DOI] [PubMed] [Google Scholar]
- 42.Wagle N, Grabiner BC, Van Allen EM, et al. Response and acquired resistance to everolimus in anaplastic thyroid cancer. N Engl J Med. 2014;371:1426–1433. doi: 10.1056/NEJMoa1403352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagle N, Emery C, Berger MF, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29:3085–3096. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toy W, Shen Y, Won H, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet. 2013;45:1439–1445. doi: 10.1038/ng.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robinson DR, Wu YM, Vats P, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet. 2013;45:1446–1451. doi: 10.1038/ng.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Woyach JA, Furman RR, Liu TM, et al. Resistance mechanisms for the Bruton's tyrosine kinase inhibitor ibrutinib. N Engl J Med. 2014;370:2286–2294. doi: 10.1056/NEJMoa1400029. [DOI] [PMC free article] [PubMed] [Google Scholar]