The North American Imaging in Multiple Sclerosis Cooperative steering committee developed a uniform high-resolution 3T MR imaging protocol relevant to the quantification of cerebral lesions and atrophy and implemented it at 7 sites across the United States. They assessed intersite variability in scan data, by imaging a volunteer with relapsing-remitting MS with a scan-rescan at each site. In multicenter studies with consistent scanner field strength and manufacturer after protocol harmonization, systematic differences can lead to severe biases in volumetric analyses.
Abstract
BACKGROUND AND PURPOSE:
MR imaging can be used to measure structural changes in the brains of individuals with multiple sclerosis and is essential for diagnosis, longitudinal monitoring, and therapy evaluation. The North American Imaging in Multiple Sclerosis Cooperative steering committee developed a uniform high-resolution 3T MR imaging protocol relevant to the quantification of cerebral lesions and atrophy and implemented it at 7 sites across the United States. To assess intersite variability in scan data, we imaged a volunteer with relapsing-remitting MS with a scan-rescan at each site.
MATERIALS AND METHODS:
All imaging was acquired on Siemens scanners (4 Skyra, 2 Tim Trio, and 1 Verio). Expert segmentations were manually obtained for T1-hypointense and T2 (FLAIR) hyperintense lesions. Several automated lesion-detection and whole-brain, cortical, and deep gray matter volumetric pipelines were applied. Statistical analyses were conducted to assess variability across sites, as well as systematic biases in the volumetric measurements that were site-related.
RESULTS:
Systematic biases due to site differences in expert-traced lesion measurements were significant (P < .01 for both T1 and T2 lesion volumes), with site explaining >90% of the variation (range, 13.0–16.4 mL in T1 and 15.9–20.1 mL in T2) in lesion volumes. Site also explained >80% of the variation in most automated volumetric measurements. Output measures clustered according to scanner models, with similar results from the Skyra versus the other 2 units.
CONCLUSIONS:
Even in multicenter studies with consistent scanner field strength and manufacturer after protocol harmonization, systematic differences can lead to severe biases in volumetric analyses.
Conventional MR imaging is an established tool for measuring CNS lesions and tissue compartment volumes in vivo in individuals with multiple sclerosis. In the brain and spinal cord, inflammatory demyelinating lesions appear hyperintense on T2-weighted images. Total cerebral T2 lesion volume (T2LV) is a key metric for the longitudinal monitoring of disease severity, as well as a standard outcome in clinical trials of MS therapeutics.1–3 Many T2 lesions exhibit pulse-sequence-dependent hypointensity on T1-weighted images, which has been shown to be associated with more severe (destructive) histopathology and worse clinical outcomes.4–8 MR imaging is also used to measure cerebral atrophy, a commonly used supportive outcome measure of the neurodegenerative aspects of the disease in both relapsing-remitting and progressive forms of MS.9–18 Together, lesion and atrophy measures provide complementary quantitative information about disease progression that are considered central to patient assessment.19
Unfortunately, differences in acquisition methods have the potential to bias MR imaging metrics. Factors such as equipment manufacturer, magnetic field strength, and acquisition protocol can affect image contrast and resultant volumetric data. Indeed, several groups have investigated the reliability of volumetric measurements across scanners,20–27 but little is understood about the variability in volumetric measurements of lesions and atrophy in individuals with MS. Furthermore, many automated segmentation algorithms depend on statistical atlases or models that are built with healthy volunteers or that depend on registration, which can be compromised by the presence of MS pathology.28
The North American Imaging in Multiple Sclerosis Cooperative (NAIMS) was established to accelerate the pace of imaging research. As a consortium, our first aim was to facilitate multicenter imaging studies by creating harmonized MR imaging protocols across sites. In this article, we describe initial results from our pilot study, which tested the feasibility of multisite standardization of MR imaging acquisitions for the quantification of lesion and tissue volumes. We compare inter- to intrasite scan-rescan variability in various MR imaging output metrics with consistently acquired 3T acquisitions.
Materials and Methods
Participant
A 45-year-old man with clinically stable relapsing-remitting MS and mild-to-moderate physical disability was imaged at 7 NAIMS sites across the United States (Table). He developed the first symptoms of the disease 13 years before study enrollment and had been relapse-free in the previous year after starting dimethyl fumarate. His last intravenous corticosteroid administration was 5 years previously. His timed 25-foot walk at study entry was 5.3 seconds. His Expanded Disability Status Scale score was 3.5, both at study entry and exit, without any intervening relapses on-study. The participant signed informed consent for this study, which was approved by the institutional review board of each site.
3T brain MRI anatomic acquisition protocolsa
| 3D T2 FLAIR |
3D T1 MPRAGE |
|||||
|---|---|---|---|---|---|---|
| Siemens Skyra | Siemens Verio | Siemens Tim Trio | Siemens Skyra | Siemens Verio | Siemens Tim Trio | |
| Operation system version | syngo MR D13 | syngo MR B17 | syngo MR B17 | syngo MR D13 | syngo MR B17 | syngo MR B17 |
| Coil | 32 or 64 Channelb | 32 Channel | 32 Channel | 32 or 64 Channelb | 32 Channel | 32 Channel |
| Acceleration factor for parallel imaging | 2 | 2 | 2 | 2 | 2 | 2 |
| Orientation | Sagittal | Sagittal | Sagittal | Sagittal | Sagittal | Sagittal |
| FOV (cm) | 25.6 × 25.6 | 25.6 × 25.6 | 25.6 × 25.6 | 25.6 × 25.6 | 25.0 × 25.0 | 25.0 × 25.0 |
| Matrix size | 512 × 512 | 512 × 512 | 512 × 512 | 256 × 256 | 256 × 256 | 256 × 256 |
| No. of sections | 176 | 176 | 176 | 176 | 176 | 176 |
| TR (ms) | 4800 | 4800 | 4800 | 1900 | 1900 | 1900 |
| TE (ms) | 353 | 354 | 355 | 2.52 | 2.52 | 2.52 |
| Flip angle | 120° | 120° | 120° | 9° | 9° | 9° |
| Voxel size (mm) | 0.5 × 0.5 × 1.0 | 0.5 × 0.5 × 1.0 | 0.5 × 0.5 × 1.0 | 1.0 × 1.0 × 1.0 | 0.977 × 0.977 × 1.0 | 0.977 × 0.977 × 1.0 |
| Scan time (min:s) | 6:53 | 7:00 | 7:00 | 4:15 | 4:16 | 4:16 |
| No. of signal averages | 1 | 1 | 1 | 1 | 1 | 1 |
Each of the 7 sites used 1 of 3 different Siemens scanner models (Skyra, Verio, and Tim Trio), necessitating 3 model-specific protocols for the 2 pulse sequences.
University of California, San Francisco = 64-channel (the other Skyra sites = 32 channel).
Scan Acquisition
Through consensus agreement in the Cooperative, NAIMS developed a standardized high-resolution 3T MR imaging brain scan protocol. All imaging was acquired with Siemens scanners, which, at the time of the study, were used by most NAIMS sites. Scan-rescan pairs were acquired on these scanners; the most relevant acquisition sequences are shown in the Table. At each site, the scan-rescan experiment was performed on the same day, with the participant removed and repositioned between scans. None of the participant's scans were coregistered to each other, to replicate a “real world” clinical trial setting. The volunteer was also imaged at the National Institutes of Health NAIMS site at the beginning and end of the study (5 months later) to assess disease stability. Raw MR imaging scans were distributed to 4 NAIMS sites for postprocessing.
Expert Lesion Tracing
De-identified images underwent manual quantification to assess total cerebral T1-hypointense lesion volume (T1LV) and T2LV from the native 3D FLAIR and T1 images by the consensus of trained observers (G.K., F.Y.) under the supervision of an experienced observer (S.T.). For T2LV, this process involved manually identifying all lesions on the FLAIR images. For T1LV, lesions were required to show hypointensity on T1-weighted images and at least partial hyperintensity on FLAIR images. The lesions were then segmented by 1 observer (G.K.) with a semiautomated edge-finding tool in Jim (Version 7.0; http://www.xinapse.com/home.php) to determine lesion volumes. Images were presented to the same reading panel for all of the above steps in random order in 1 batch and mixed into a stack of 50 other MS images to reduce scan-to-scan memory effects and preserve blinding.
Automated Analysis
Several fully automated pipelines were also used to estimate T2LV and the volumes of total brain, normal-appearing white matter, and both cortical and deep gray matter structures. To prevent overfitting, we used all pipelines with their default settings, according to published recommendations for each method separately, in which appropriate images were inhomogeneity corrected, rigidly aligned across sequences from each scan session, processed for removal of extracerebral voxels for all processing pipelines, and intensity normalized. For lesion measurements, several algorithms were applied by the laboratories that developed or codeveloped the various methods: Lesion-TOADS (TOpology-preserving Anatomical Segmentation; https://www.nitrc.org/projects/toads-cruise/),29 a fuzzy C-means-based segmentation technique with topologic constraints; Automated Statistical Inference for Segmentation (OASIS),30 a logistic-regression-based segmentation method leveraging statistical intensity normalization; Subject Specific Sparse Dictionary Learning (S3DL; https://www.nitrc.org/projects/s3dl/),31 a patch-based dictionary learning multiclass method; and White Matter Lesion Segmentation (WMLS; https://www.nitrc.org/projects/wmls/),32 a local support vector machine-based segmentation algorithm developed for vascular lesions that also uses corrective learning. To estimate the volume of gray matter structures, we used Lesion-TOADS; FMRIB Integrated Registration and Segmentation Tool (FSL-FIRST; http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FIRST)33 (a Bayesian appearance method); Multi-atlas Segmentation with Brain Surface Estimation (MaCRUISE)34 (a combined multiatlas segmentation and cortical reconstruction algorithm); and MUlti-atlas region Segmentation utilizing Ensembles of registration algorithms (MUSE)35 (an ensemble multiatlas label-fusion method). The FSL-FIRST33 analysis was applied directly to the raw T1 images according to common practice, and OASIS30 was applied to the T1, FLAIR, and a 3D T2 high-resolution sequence after preprocessing; all other pipelines were applied to appropriately preprocessed T1 and FLAIR images. Not all algorithms measured volumes of the same set of structures. Lesion-filling was not performed. Lesion-TOADS, MaCRUISE, and MUSE also yielded estimates for total brain volume.
Statistical Analysis
All statistical analyses were conducted in the R software environment (http://www.r-project.org/).36 To compare estimated volumes within and across sites, we computed mean volumes and SDs. T tests were also used for differences in within-site averages between scanner platforms. Correlations between these averages across segmentation algorithms were also explored. The proportion of variation explained by site was computed, and the association with site was assessed with permutation testing. The coefficients of variation were also estimated across sites. To assess associations between session-average measured total brain and lesional volumes and time of day (morning versus afternoon), we used Wald testing within a linear model framework, both marginally and adjusting for scanner platform.
Results
The participant was found to be stable regarding cerebral lesion load during the study. When we compared images acquired at the National Institutes of Health at study entry and exit, the manually measured T2LV in the participant was similar (17.9 mL in September 2015 versus 17.8 mL in February 2016). The T1LV was also stable (15.5 versus 15.1 mL). This imaging stability paralleled his clinical stability (see “Materials and Methods”).
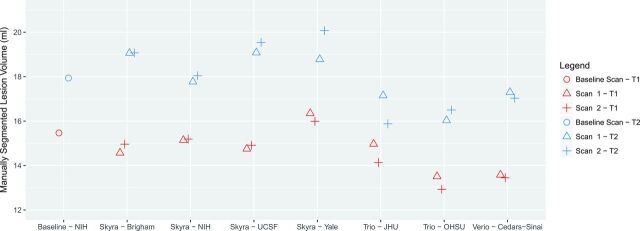
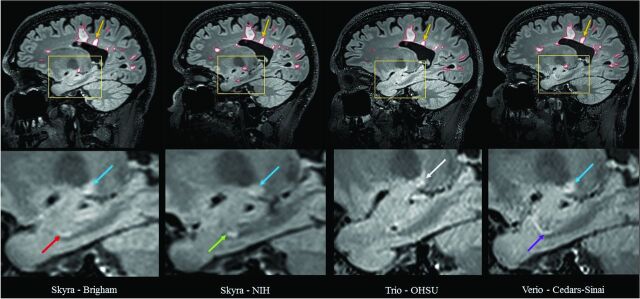
The manually estimated T1LV and T2LV for each scan is shown in Fig 1. Site explained 95% of the variation observed in the estimated T2LV and 92% of the variation in the estimated T1LV, indicating marked scanner-to-scanner differences despite protocol harmonization, which clearly exceeded scan-rescan variability within sites. The range of T2LVs was 15.9 to 20.1 mL, indicating that differences of up to 25% of the lesion volume were observed across sites. The range of T1LVs was similarly wide, ranging from 13.0 to 16.4 mL. Further inspection of these volumes across platforms indicated that Skyra (Magnetom Skyra; Siemens, Erlangen, Germany) scanners showed larger lesion volumes compared with other Siemens platforms both on T1LV (Skyra: mean T1, 15.2 mL compared with non-Skyra: mean T1, 13.8 mL; P < .05) and T2LV (Skyra: mean T2, 18.9 mL compared with non-Skyra: mean T2, 16.6 mL; P < .01). An example of the segmented lesions across scanners is provided in Fig 2.
Fig 1.
Manually measured T1 (red) and T2 (blue) lesion volumes for scan-rescan pairs at each of 7 NAIMS sites. Results from the baseline scan, acquired on the same Skyra scanner and subsequent imaging acquired at the National Institutes of Health, are shown with circles. Points have been slightly offset relative to one another for ease of visualization. UCSF indicates University of California, San Francisco; JHU, Johns Hopkins University; OHSU, Oregon Health & Science University.
Fig 2.
Comparison of manual segmentation of cerebral T2 hyperintense lesions at 4 NAIMS sites. 3T MR imaging scans on Siemens scanners from a single subject with multiple sclerosis showing T2 hyperintense lesions from sagittal fluid-attenuated inversion recovery sequences from 4 different North American Imaging in Multiple Sclerosis Cooperative sites and scanner models: Brigham and Women's Hospital, Skyra; National Institutes of Health, Skyra; Oregon Health & Science University (OHSU), Tim Trio; Cedars-Sinai, Verio. The upper panel shows the native images. The lower panel shows zoomed and cropped images to illustrate the key findings. The green arrow (lower panel) shows a possible lesion detected and traced on the National Institutes of Health scan; the red arrow shows the same lesion not detected by the expert procedure on the Brigham and Women's Hospital scan. The purple arrow shows a similar tubular area interpreted as a blood vessel on the Cedars-Sinai scan, which was not selected as a lesion by the expert tracing; no lesion was detected on the Oregon Health & Science University scan in this area on this section or any of the adjacent sections (not shown). The blue arrow shows a different lesion detected and traced on the Brigham and Women's Hospital, National Institutes of Health, and Cedars-Sinai scans but not detected by the expert review on the Oregon Health & Science University scan, appearing hazy/subtle (white arrow). The yellow arrow (upper panel) shows a lesion on all scans; however, when we added the tracing of all sections showing the lesion, the 3D volume of the lesion differed among sites: Brigham and Women's Hospital = 0.059 mL, National Institutes of Health = 0.053 mL, Oregon Health & Science University = 0.033 mL, Cedars-Sinai = 0.053 mL.
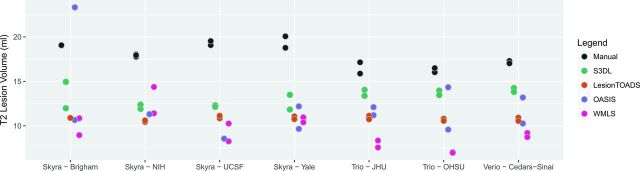
Results from the automated techniques for delineating and measuring T2LV are shown in Fig 3. The automated lesion segmentations showed marked disagreement in the average lesional volume measurements compared with the manually assessed volumes, and all methods showed large site-to-site differences (in some cases up to 7.5 mL, or almost 50% of the manually measured lesion volume), except for Lesion-TOADS (range, 10.5–11.0 mL), which was more stable. For all methods, site explained >50% of the observed variation; 53% of the variation was explained by site (permutation P = .36) for S3DL, 54% for Lesion-TOADS (P = .41), 44% for OASIS (P = .57), and 83% for WMLS (P = .002), which clearly was most prone to site-related variation.
Fig 3.
Comparison of manual and automated methods for measuring lesional volume. Scan-rescan imaging is shown by using multiple dots for each site and algorithm. UCSF indicates University of California, San Francisco; JHU, Johns Hopkins University; OHSU, Oregon Health & Science University.
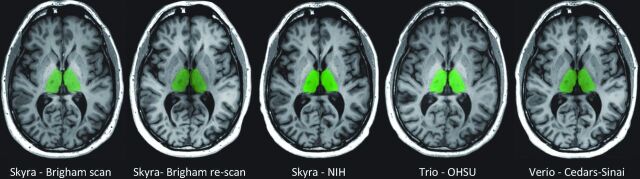
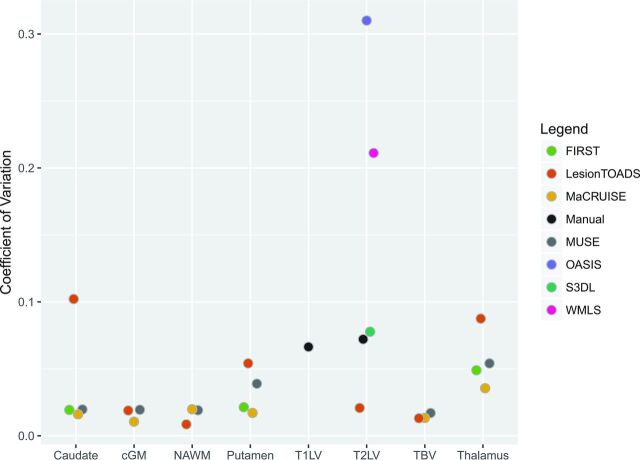
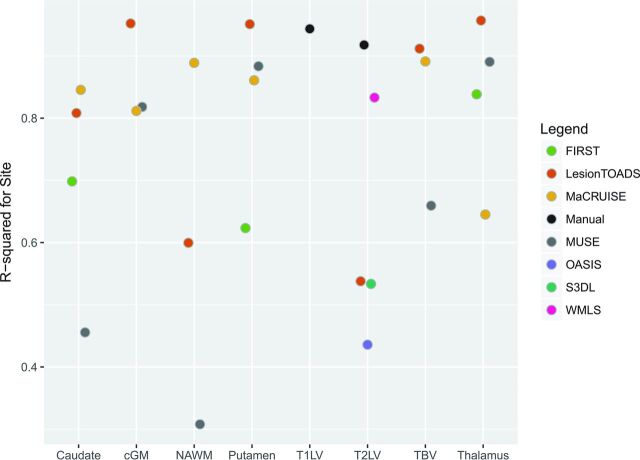
To measure brain structure volumes, we used several automated methods. As an example, results for the thalamus are shown in Figs 4 and 5. While Lesion-TOADS estimated smaller volumes, MUSE, FSL-FIRST, and MaCRUISE yielded similar average measurements. Nonetheless, site was strongly associated with measured thalamic volume, explaining 96% of the Lesion-TOADS volume variation (P < .01), 89% of MUSE (P < .01), 84% of FSL-FIRST (P = .04), and 65% of MaCRUISE (P = .17). Similar results for the putamen, caudate, cortical gray matter, normal-appearing white matter, and total brain volume were found, as provided in On-line Figs 1–5. Summaries of the coefficient of variation give an intuitive measure of the scale of the combined scan-rescan and across-site variation as shown in Fig 6. Finally, the proportion of variation explained by site is shown in Fig 7. Note that in almost all cases, site explained >50% of the variation, with most measurement techniques showing >80% variation due to site for all structures assessed.
Fig 4.
FSL-FIRST automated segmentation results: thalamus. Representative anatomic section showing segmentation of the thalamus (green) in the single subject. The segmentation maps are overlaid to the original raw 3D T1-weighted images after re-orientation to the axial plane. Segmentation was performed by the fully automated FSL-FIRST pipeline. The scan site and 3T Siemens model are shown for each image. The first 2 scans are from the scan/re-scan at Brigham and Women's Hospital. OHSU indicates Oregon Health & Science University.
Fig 5.
Comparison of automated methods for measuring thalamic volume. Scan-rescan imaging is shown by using multiple dots for each site and algorithm. UCSF indicates University of California, San Francisco; JHU, Johns Hopkins University; OHSU, Oregon Health & Science University.
Fig 6.
Estimated across-site coefficient of variation for each structure with various methods for volumetric measurement. cGM indicates cortical gray matter; NAWM, normal-appearing white matter; TBV, total brain volume.
Fig 7.
Estimated proportion of variation explained by site for using various segmentation methods for different structures in the brain. cGM indicates cortical gray matter; NAWM, normal-appearing white matter; TBV, total brain volume.
While all images were acquired on 3T Siemens scanners, the model type appeared to influence the results; there was evidence of systematic differences in many measurements between Skyra and non-Skyra scanners. Figure 8 shows the negative log P values for the comparison of volumes averaged across scan-rescan measurements, with larger values indicating more systematic differences between platforms. The largest platform-associated differences were observed in MaCRUISE measurements of normal-appearing white matter, cortical gray matter, and, consequently, total brain volume. Lesion-TOADS also showed large differences in total brain volume attributable to cortical gray matter, as did S3DL for T2LV measurements. MUSE showed major differences in thalamic volume across scanner models, and FSL-FIRST showed similar discrepancies in the thalamus and caudate. The correlation between site-averaged measurements varied dramatically, especially for lesional and total brain volume measurements (On-line Fig 6); this variation indicates that site differences resulted in contrasting effects on output from the different algorithms. While the other measurements showed less scanner model–related variation, most still showed prominent differences between Skrya and non-Skrya scanners.
Fig 8.
Negative logarithm (base 10) P value from t tests describing the difference in average volume between Skyra-versus-non-Skyra platforms explained by site with various segmentation methods for different structures in the brain. cGM indicates cortical gray matter; NAWM, normal-appearing white matter; TBV, total brain volume.
The time of day of scan acquisition was not associated with manually segmented T1 lesion volumes (t = 0.45) or T2 lesion volumes (t = 0.38) or total brain volume, as measured by any of the automated algorithms (On-line Figs 7 and 8).
Discussion
Clinical MS therapeutic trials have traditionally used 1.5T MR imaging platforms to provide metrics on cerebral lesions and atrophy as supportive outcome measures. However, there is growing interest in the use of high-resolution 3T imaging to assess disease activity and disease severity in MS. Such 3T imaging has the potential for increased sensitivity to lesions37,38 and atrophy,39 higher reliability,39,40 and closer relationships to clinical status,38,39 compared with scanning at 1.5T. The purpose of this study was to evaluate the consistency of metrics obtained from a single MS participant with a high-resolution 3T brain MR imaging protocol distributed to 7 sites. The results of our study indicate that even in multicenter acquisitions from the same scanner vendor after careful protocol harmonization, systematic differences in images led to severe biases in volumetric analyses. These biases were present in manually and automatically measured volumes of white matter lesions, as well as in automatically measured volumes of whole-brain and gray and white matter structures. These biases were also highly dependent on scanning equipment, which resulted from a higher sensitivity to lesions in newer scanners from the same manufacturer compared with earlier models, even at the same field strengths.
In comparison with past estimates of reliability of volumetric measurements of brain structures, our findings point to higher between-site variation than previously documented. In particular, Cannon et al27 reported that between 3% and 26% of the observed variation in global and subcortical volumes were attributable to site; this was a study of 8 healthy participants imaged on 2 successive days across 8 sites with 3T Siemens and GE Healthcare scanners. However, the proportion of explained variation has a different interpretation from that reported here. The total variation in Cannon et al consisted of 4 contributors to variance: first, across-site differences; second, across-scan differences; third, across-day differences; and fourth, across-subject differences. In our single-participant study, we isolated only the first 2 variance components, allowing us to compare variation because it is relevant for precision medicine (subject-specific) applications.
Previous work indicated that the observed variation attributable to scanning occasion was small25,27; indeed, Cannon et al27 found this to constitute <1% of the variation. Thus, we did not scan our participant on subsequent days but rather simply repositioned the participant between scans during the same imaging session. A notable difference between our study and that of Cannon et al is that we did not use data from a standardized phantom concurrently acquired for correction of between-scanner variations in gradient nonlinearity and scaling. Cannon et al found that this correction improved between-site intraclass correlations and greatly reduced differences between scanner manufacturers. Similarly, Gunter et al41 reported the usefulness of a phantom for scanner harmonization and quality control in the Alzheimer's Disease Neuroimaging Initiative (http://www.adni-info.org/). In future studies, we will focus on applying phantom calibrations across NAIMS sites to extend our current observations. Despite the growing literature on the importance of diurnal variation and hydration status for volumetric analyses,42–45 we found no significant associations between time of day and measured volumes. This may indicate that in single-participant analyses, time of day and day-to-day variation may be of less concern than the much larger source of variation of scanner platform. Most interesting, Cannon et al also found that measurements acquired with scanners from the same manufacturer and similar receive coils had higher reliability. In our study, we found that even scanner models (ie, Skyra versus non-Skyra) from the same manufacturer varied markedly in their estimates of lesion volume; this variation highlights the importance of between-scanner differences for assessing MS-related structural changes.
To assess differences across processing pipelines, we used a variety of techniques for automated segmentation of lesion and white and gray matter volumes. Different segmentation algorithms showed a range of variability in their estimates, as well as their sensitivity to differences between scanners. For example, Lesion-TOADS showed much less variable lesion measurements than any other technique and was not as sensitive to differences in scanner platform. Lesion-TOADS was the only unsupervised lesion-segmentation technique used. Contrast differences between the participant data and the training data of the other supervised methods could be associated with greater sensitivity to scanner differences, and this might be mitigated by specific (albeit potentially laborious) tuning to individual platforms. However, while sensitivity to biologic change is generally higher for methods yielding less noisy estimates, because only a single individual was studied here, our data cannot be taken to indicate that Lesion-TOADS is superior to other methods of estimating thalamic volume, for example. Additionally, both purely intensity-based segmentation algorithms, OASIS and WMLS, appeared to be more sensitive to site differences, which may indicate that methods that rely more on topology, shape, or spatial context may be more stable across scanners. This finding indicates that across-scanner differences may be driven by contrast differences rather than geometric distortions. Future investigation to extend these findings could involve quantitative contrast-to-noise and signal-to-noise comparisons across scanners. Allowing segmentation parameters to vary across sites could also help stability.
A limitation of this study is its single-subject and single time point design, which makes the generalizability of the findings dependent on further investigation. In particular, the degree to which across-site differences might vary by lesion burden and degree of atrophy, as well as demographic variables, requires additional study. Future larger studies of multiple participants across disease stages, including longitudinal measurements, are necessary for understanding the implications of the biases described in this pilot study. Indeed, such studies would also allow the assessment of the trade-off between stability in measures across sites, with sensitivity to biologic differences. Differences between scanning equipment and scanner software versions have also been noted in past studies of reliability,23,25,27,46,47 but their implications for the assessment of pathology remain unclear. In particular, repeat acquisitions on scanners with different receive coils could provide additional insight concerning reliability. In addition, our study was from a single time point across scanners, whereas clinical trials rely on the quantification of intrasubject longitudinal change.48 Each participant is typically scanned on the same platform, which may limit the variability in on-study change between participants. Further studies are necessary to assess whether scan platform introduces the same level of acquisition-related variability when assessing longitudinal changes.
Given the intersite differences observed in lesional measurements, across-site-inference statistical adjustment for site is clearly necessary when analyzing volumetrics from multisite studies, even when images are acquired with a harmonized protocol on 3T scanners produced by the same manufacturer. From a single participant, it is unclear what the role of differential sensitivity to lesions might be across individuals with heterogeneity in lesion location. For example, while lesion detection in the supratentorial white matter might be more straightforward and comparable across individuals, detection of lesions in the brain stem, cerebellum, and spinal cord may be more sensitive to differences in equipment. New statistical methods for measuring and correcting systematic biases are warranted, especially for studies in which patient populations may differ across sites. Indeed, intensity normalization and scan-effect removal techniques49–55 (akin to batch-effect removal methods in genomic studies56) are an active area of methodologic research and promise to improve comparability of volumetric estimates from automated segmentation methods. After volumes are measured, statistical techniques for modeling estimated volumes from multicenter studies are also rapidly evolving.18,57 These techniques bring the potential to mitigate site-to-site biases in group-level analyses, with better external validity at the cost of increased sample size.
Conclusions
By imaging the same subject with stable relapsing-remitting MS during 5 months, we assessed scanner-related biases in volumetric measurements at 7 NAIMS centers. Despite careful protocol harmonization and the acquisition of all imaging at 3T on Siemens scanners, we found significant differences in lesion and structural volumes. These differences were especially pronounced when comparing Skyra scanners with other Siemens 3T platforms. The results from this study highlight the potential for interscanner and intersite differences that, unless properly accounted for, might confound MR imaging volumetric data from multicenter studies of brain disorders.
Our findings raise a key issue of the interpretability of MR imaging measurements in the context of personalized medicine, even in carefully controlled studies with harmonized imaging protocols.
Supplementary Material
Acknowledgments
The following is a full list of individuals who contributed to this NAIMS study—Brigham and Women's Hospital, Harvard Medical School (Boston, Massachusetts): Rohit Bakshi, Renxin Chu, Gloria Kim, Shahamat Tauhid, Subhash Tummala, Fawad Yousuf; Cedars-Sinai Medical Center (Los Angeles, California): Nancy L. Sicotte; Henry M. Jackson Foundation for the Advancement of Military Medicine (Bethesda, Maryland): Dzung Pham, Snehashis Roy; National Institutes of Health (Bethesda, Maryland): Frances Andrada, Irene C.M. Cortese, Jenifer Dwyer, Rosalind Hayden, Haneefa Muhammad, Govind Nair, Joan Ohayon, Daniel S. Reich, Pascal Sati, Chevaz Thomas; Johns Hopkins University (Baltimore, Maryland): Peter A. Calabresi, Sandra Cassard, Jiwon Oh; Oregon Health & Science University (Portland, Oregon): William Rooney, Daniel Schwartz, Ian Tagge; University of California (San Francisco, California): Roland G. Henry, Nico Papinutto, William Stern, Alyssa Zhu; University of Pennsylvania (Philadelphia, Pennsylvania): Christos Davatzikos, Jimit Doshi, Guray Erus, Kristin Linn, Russell Shinohara; University of Toronto (Toronto, Ontario, Canada): Jiwon Oh; Yale University (New Haven, Connecticut): R. Todd Constable, Daniel Pelletier.
ABBREVIATIONS:
- NAIMS
North American Imaging in Multiple Sclerosis Cooperative
- T1LV
T1-hypointense lesion volume
- T2LV
T2 lesion volume
Footnotes
Disclosures: Russell T. Shinohara—RELATED: Grant: National Institutes of Health*; Support for Travel to Meetings for the Study or Other Purposes: Race to Erase MS, Comments: travel to consortium meetings; UNRELATED: Board Membership: Genentech, Comments: Scientific Advisory Board; Consultancy: Hoffmann-La Roche, Comments: expert legal consulting; Grants/Grants Pending: Gates Foundation*; Travel/Accommodations/Meeting Expenses Unrelated to Activities Listed: Government of Canada–Banff Research Institute–European Committee for Treatment and Research in Multiple Sclerosis, Comments: conference travel.* Jiwon Oh—UNRELATED: Consultancy: Consortium of Multiple Sclerosis Centers, EMD Serono, Novartis, Hoffmann-La Roche, Biogen Idec, Teva Pharmaceuticals; Grants/Grants Pending: MS Society of Canada, National MS Society, Biogen Idec, Genzyme*; Support for Travel to Meetings for the Study or Other Purposes: Consortium of Multiple Sclerosis Centers. Peter Calabresi—RELATED: Grant: Race to Erase MS, Comments: foundation grant*; Support for Travel to Meetings for the Study or Other Purposes: Race to Erase MS, Comments: The foundation pays for my travel to semiannual meetings; UNRELATED: Consultancy: Biogen Idec, Vertex Pharmaceuticals; Grants/Grants Pending: Biogen Idec, Teva Pharmaceuticals, Annexon Biosciences, Novartis, Medimmune*; Royalties: Cambridge Press, Comments: for editing a book on optical coherence tomography. Christos Davatzikos—RELATED: Grant: National Institutes of Health/National Institute on Aging computational neuroanatomy of aging and Alzheimer disease via pattern analysis, Comments: R01-AG014971.* Roland G. Henry—RELATED: Grant: Race to Erase MS, Comments: nominal/standard cost for MRI scans*; UNRELATED: Consultancy: Hoffmann-La Roche, AbbVie, Novartis, Genzyme, StemCells Inc*; Grants/Grants Pending: Hoffmann-La Roche*; Payment for Lectures Including Service on Speakers Bureaus: Genzyme.* Daniel Pelletier—UNRELATED: Consultancy: Genzyme, Novartis, EMD-Serono, Genentech; Grants/Grants Pending: Biogen Idec, Comments: investigator-initiated research grant.* Dzung L. Pham—RELATED: Grant: National MS Society, Comments: RG-1507–05243.* Daniel S. Reich—RELATED: Support for Travel to Meetings for the Study or Other Purposes: Race to Erase MS.*William Rooney—RELATED: Grant: Race to Erase MS, Comments: This organization provided pilot funds for the study*; UNRELATED: Employment: Oregon Health & Science University, Comments: employs me as professor/director; Patents (Planned, Pending, or Issued): Oregon Health & Science University, Brookhaven National Laboratory; Royalties: Oregon Health & Science University. Rohit Bakshi—RELATED: Grant: Race to Erase MS.* Nancy L. Sicotte—RELATED: Grant: Race to Erase MS.* *Money paid to the institution.
Major support for this study was provided by the Race to Erase MS. Additional support came from RO1NS085211, R21NS093349, R01EB017255, and S10OD016356 from the National Institutes of Health and RG-1507-05243 from the National Multiple Sclerosis Society. The study was also partially supported by the Intramural Research Program of the National Institute of Neurological Disorders and Stroke.
Paper previously presented in preliminary form at: Annual Meeting of the European Committee on Treatment and Research in Multiple Sclerosis, September 14–17, 2016; London, UK.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
REFERENCES
- 1. García-Lorenzo D, Francis S, Narayanan S, et al. Review of automatic segmentation methods of multiple sclerosis white matter lesions on conventional magnetic resonance imaging. Med Image Anal 2013;17:1–18 10.1016/j.media.2012.09.004 [DOI] [PubMed] [Google Scholar]
- 2. Lublin FD, Reingold SC, Cohen JA, et al. Defining the clinical course of multiple sclerosis: the 2013 revisions. Neurology 2014;83:278–86 10.1212/WNL.0000000000000560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Simon JH, Jacobs LD, Campion M, et al. Magnetic resonance studies of intramuscular interferon beta-1a for relapsing multiple sclerosis: the Multiple Sclerosis Collaborative Research Group. Ann Neurol 1998;43:79–87 10.1002/ana.410430114 [DOI] [PubMed] [Google Scholar]
- 4. Bagnato F, Jeffries N, Richert ND, et al. Evolution of T1 black holes in patients with multiple sclerosis imaged monthly for 4 years. Brain 2003;126(pt 8):1782–89 10.1093/brain/awg182 [DOI] [PubMed] [Google Scholar]
- 5. Sahraian MA, Radue EW, Haller S, et al. Black holes in multiple sclerosis: definition, evolution, and clinical correlations. Acta Neurol Scand 2010;122:1–8 10.1111/j.1600-0404.2009.01221.x [DOI] [PubMed] [Google Scholar]
- 6. Giorgio A, Stromillo ML, Bartolozzi ML, et al. Relevance of hypointense brain MRI lesions for long-term worsening of clinical disability in relapsing multiple sclerosis. Mult Scler 2014;20:214–19 10.1177/1352458513494490 [DOI] [PubMed] [Google Scholar]
- 7. van Walderveen MA, Kamphorst W, Scheltens P, et al. Histopathologic correlate of hypointense lesions on T1-weighted spin-echo MRI in multiple sclerosis. Neurology 1998;50:1282–88 [DOI] [PubMed] [Google Scholar]
- 8. Truyen L, van Waesberghe JH, van Walderveen MA, et al. Accumulation of hypointense lesions (“black holes”) on T1 spin-echo MRI correlates with disease progression in multiple sclerosis. Neurology 1996;47:1469–76 [DOI] [PubMed] [Google Scholar]
- 9. Evangelou N, Esiri MM, Smith S, et al. Quantitative pathological evidence for axonal loss in normal appearing white matter in multiple sclerosis. Ann Neurol 2000;47:391–95 [PubMed] [Google Scholar]
- 10. Evangelou N, Konz D, Esiri MM, et al. Regional axonal loss in the corpus callosum correlates with cerebral white matter lesion volume and distribution in multiple sclerosis. Brain 2000;123(pt 9):1845–49 [DOI] [PubMed] [Google Scholar]
- 11. Sastre-Garriga J, Ingle GT, Chard DT, et al. Grey and white matter volume changes in early primary progressive multiple sclerosis: a longitudinal study. Brain 2005;128(pt 6):1454–60 10.1093/brain/awh498 [DOI] [PubMed] [Google Scholar]
- 12. Sanfilipo MP, Benedict RH, Weinstock-Guttman B, et al. Gray and white matter brain atrophy and neuropsychological impairment in multiple sclerosis. Neurology 2006;66:685–92 10.1212/01.wnl.0000201238.93586.d9 [DOI] [PubMed] [Google Scholar]
- 13. Ge Y, Grossman RI, Udupa JK, et al. Brain atrophy in relapsing-remitting multiple sclerosis: fractional volumetric analysis of gray matter and white matter. Radiology 2001;220:606–10 10.1148/radiol.2203001776 [DOI] [PubMed] [Google Scholar]
- 14. Fisher E, Lee JC, Nakamura K, et al. Gray matter atrophy in multiple sclerosis: a longitudinal study. Ann Neurol 2008;64:255–65 10.1002/ana.21436 [DOI] [PubMed] [Google Scholar]
- 15. Fisniku LK, Chard DT, Jackson JS, et al. Gray matter atrophy is related to long-term disability in multiple sclerosis. Ann Neurol 2008;64:247–54 10.1002/ana.21423 [DOI] [PubMed] [Google Scholar]
- 16. De Stefano N, Matthews PM, Filippi M, et al. Evidence of early cortical atrophy in MS: relevance to white matter changes and disability. Neurology 2003;60:1157–62 [DOI] [PubMed] [Google Scholar]
- 17. Losseff NA, Wang L, Lai HM, et al. Progressive cerebral atrophy in multiple sclerosis: a serial MRI study. Brain 1996;119(pt 6):2009–19 [DOI] [PubMed] [Google Scholar]
- 18. Keshavan A, Paul F, Beyer MK, et al. Power estimation for non-standardized multisite studies. Neuroimage 2016;134:281–94 10.1016/j.neuroimage.2016.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bakshi R, Thompson AJ, Rocca MA, et al. MRI in multiple sclerosis: current status and future prospects. Lancet Neurol 2008;7:615–25 10.1016/S1474-4422(08)70137-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Agartz I, Okuguwa G, Nordström M, et al. Reliability and reproducibility of brain tissue volumetry from segmented MR scans. Eur Arch Psychiatry Clin Neurosci 2001;251:255–61 10.1007/PL00007542 [DOI] [PubMed] [Google Scholar]
- 21. Bartzokis G, Mintz J, Marx P, et al. Reliability of in vivo volume measures of hippocampus and other brain structures using MRI. Magn Reson Imaging 1993;11:993–1006 10.1016/0730-725X(93)90218-3 [DOI] [PubMed] [Google Scholar]
- 22. Maclaren J, Han Z, Vos SB, et al. Reliability of brain volume measurements: a test-retest dataset. Sci Data 2014;1:140037 10.1038/sdata.2014.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Morey RA, Selgrade ES, Wagner HR 2nd, et al. Scan-rescan reliability of subcortical brain volumes derived from automated segmentation. Hum Brain Mapp 2010;31:1751–62 10.1002/hbm.2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schnack HG, van Haren NE, Hulshoff Pol HE, et al. Reliability of brain volumes from multicenter MRI acquisition: a calibration study. Hum Brain Mapp 2004;22:312–20 10.1002/hbm.20040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jovicich J, Marizzoni M, Sala-Llonch R, et al. ; PharmaCog Consortium. Brain morphometry reproducibility in multi-center 3T MRI studies: a comparison of cross-sectional and longitudinal segmentations. Neuroimage 2013;83:472–84 10.1016/j.neuroimage.2013.05.007 [DOI] [PubMed] [Google Scholar]
- 26. Schnack HG, van Haren NE, Brouwer RM, et al. Mapping reliability in multicenter MRI: voxel-based morphometry and cortical thickness. Hum Brain Mapp 2010;31:1967–82 10.1002/hbm.20991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cannon TD, Sun F, McEwen SJ, et al. Reliability of neuroanatomical measurements in a multisite longitudinal study of youth at risk for psychosis. Hum Brain Mapp 2014;35:2424–34 10.1002/hbm.22338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Eloyan A, Shou H, Shinohara R, et al. Health effects of lesion localization in multiple sclerosis: spatial registration and confounding adjustment. PLoS One 2014;9:e107263 10.1371/journal.pone.0107263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shiee N, Bazin PL, Ozturk A, et al. A topology-preserving approach to the segmentation of brain images with multiple sclerosis lesions. Neuroimage 2010;49:1524–35 10.1016/j.neuroimage.2009.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sweeney EM, Shinohara RT, Shiee N, et al. OASIS is Automated Statistical Inference for Segmentation, with applications to multiple sclerosis lesion segmentation in MRI. Neuroimage Clin 2013;2:402–13 10.1016/j.nicl.2013.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Roy S, He Q, Sweeney E, et al. Subject-Specific Sparse Dictionary Learning for atlas-based brain MRI segmentation. IEEE J Biomed Health Inform 2015;19:1598–609 10.1109/JBHI.2015.2439242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lao Z, Shen D, Liu D, et al. Computer-assisted segmentation of white matter lesions in 3D MR images using support vector machine. Acad Radiol 2008;15:300–13 10.1016/j.acra.2007.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Patenaude B, Smith SM, Kennedy DN, et al. A Bayesian model of shape and appearance for subcortical brain segmentation. Neuroimage 2011;56:907–22 10.1016/j.neuroimage.2011.02.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huo Y, Carass A, Resnick SM, et al. Combining multi-atlas segmentation with brain surface estimation. Proc SPIE Int Soc Opt Eng 2016;9784. pii: 97840E 10.1117/12.2216604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Doshi J, Erus G, Ou Y, et al. ; Alzheimer's Neuroimaging Initiative. MUSE: MUlti-atlas region Segmentation utilizing Ensembles of registration algorithms and parameters, and locally optimal atlas selection. Neuroimage 2016;127:186–95 10.1016/j.neuroimage.2015.11.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2016 [Google Scholar]
- 37. Sicotte NL, Voskuhl RR, Bouvier S, et al. Comparison of multiple sclerosis lesions at 1.5 and 3.0 Tesla. Invest Radiol 2003;38:423–27 10.1097/01.RLI.0000065426.07178.f1 [DOI] [PubMed] [Google Scholar]
- 38. Stankiewicz JM, Glanz BI, Healy BC, et al. Brain MRI lesion load at 1.5T and 3T versus clinical status in multiple sclerosis. J Neuroimaging 2011;21:e50–56 10.1111/j.1552-6569.2009.00449.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chu R, Tauhid S, Glanz BI, et al. Whole-brain volume measured from 1.5T versus 3T MRI in healthy subjects and patients with multiple sclerosis. J Neuroimaging 2016;26:62–67 10.1111/jon.12271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chu R, Hurwitz S, Tauhid S, et al. Deep gray matter segmentation from 1.5T vs. 3T MRI in normal controls and patients with multiple sclerosis. Neurology 2016;86(16 suppl):P4.171 [Google Scholar]
- 41. Gunter JL, Bernstein MA, Borowski BJ, et al. Measurement of MRI scanner performance with the ADNI phantom. Med Phys 2009;36:2193–205 10.1118/1.3116776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Duning T, Kloska S, Steinsträter O, et al. Dehydration confounds the assessment of brain atrophy. Neurology 2005;64:548–50 10.1212/01.WNL.0000150542.16969.CC [DOI] [PubMed] [Google Scholar]
- 43. Sampat MP, Healy BC, Meier DS, et al. Disease modeling in multiple sclerosis: assessment and quantification of sources of variability in brain parenchymal fraction measurements. Neuroimage 2010;52:1367–73 10.1016/j.neuroimage.2010.03.075 [DOI] [PubMed] [Google Scholar]
- 44. Nakamura K, Brown RA, Araujo D, et al. Correlation between brain volume change and T2 relaxation time induced by dehydration and rehydration: implications for monitoring atrophy in clinical studies. Neuroimage Clin 2014;6:166–70 10.1016/j.nicl.2014.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nakamura K, Brown RA, Narayanan S, et al. ; Alzheimer's Disease Neuroimaging Initiative. Diurnal fluctuations in brain volume: statistical analyses of MRI from large populations. Neuroimage 2015;118:126–32 10.1016/j.neuroimage.2015.05.077 [DOI] [PubMed] [Google Scholar]
- 46. Jovicich J, Czanner S, Han X, et al. MRI-derived measurements of human subcortical, ventricular and intracranial brain volumes: reliability effects of scan sessions, acquisition sequences, data analyses, scanner upgrade, scanner vendors and field strengths. Neuroimage 2009;46:177–92 10.1016/j.neuroimage.2009.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kruggel F, Turner J, Muftuler LT; Alzheimer's Disease Neuroimaging Initiative. Impact of scanner hardware and imaging protocol on image quality and compartment volume precision in the ADNI cohort. Neuroimage 2010;49:2123–33 10.1016/j.neuroimage.2009.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Filippi M, Wolinsky JS, Comi G. Effects of oral glatiramer acetate on clinical and MRI-monitored disease activity in patients with relapsing multiple sclerosis: a multicentre, double-blind, randomised, placebo-controlled study. Lancet Neurol 2006;5:213–20 10.1016/S1474-4422(06)70327-1 [DOI] [PubMed] [Google Scholar]
- 49. Shinohara RT, Sweeney EM, Goldsmith J, et al. Statistical normalization techniques for magnetic resonance imaging. Neuroimage Clin 2014;6:9–19 10.1016/j.nicl.2014.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nyúl LG, Udupa JJ, Zhang X. New variants of a method of MRI scale standardization. Med Imaging IEEE Trans 2000;19:143–50 [DOI] [PubMed] [Google Scholar]
- 51. Ghassemi R, Brown R, Narayanan S, et al. Normalization of white matter intensity on T1-weighted images of patients with acquired central nervous system demyelination. J Neuroimaging 2015;25:184–90 10.1111/jon.12129 [DOI] [PubMed] [Google Scholar]
- 52. Fortin JP, Sweeney EM, Muschelli J, et al. ; Alzheimer's Disease Neuroimaging Initiative. Removing inter-subject technical variability in magnetic resonance imaging studies. Neuroimage 2016;132:198–212 10.1016/j.neuroimage.2016.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Madabhushi A, Udupa JK. New methods of MR image intensity standardization via generalized scale. Med Phys 2006;33:3426–34 10.1118/1.2335487 [DOI] [PubMed] [Google Scholar]
- 54. Nyúl LG, Udupa JK. On standardizing the MR image intensity scale. Magn Reson Med 1999;42:1072–81 [DOI] [PubMed] [Google Scholar]
- 55. Chua AS, Egorova S, Anderson MC, et al. Handling changes in MRI acquisition parameters in modeling whole brain lesion volume and atrophy data in multiple sclerosis subjects: comparison of linear mixed-effect models. Neuroimage Clin 2015;8:606–10 10.1016/j.nicl.2015.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Leek JT, Scharpf RB, Bravo HC, et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet 2010;11:733–39 10.1038/nrg2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fennema-Notestine C, Gamst AC, Quinn BT, et al. Feasibility of multi-site clinical structural neuroimaging studies of aging using legacy data. Neuroinformatics 2007;5:235–45 10.1007/s12021-007-9003-9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.