Although significant progress has been made in mapping white matter connections in the marmoset brain using ex vivo tracing techniques, the application of in vivo virtual dissection of major white matter fiber tracts has been established by few studies in the marmoset literature. Here, we demonstrate the feasibility of whole-brain diffusion-weighted tractography in anesthetized marmosets at ultrahigh-field MRI (9.4T) and propose protocols for isolating nine major white matter fiber tracts in the marmoset brain.
Keywords: marmoset, fiber tractography, diffusion-weighted imaging
Abstract
The common marmoset (Callithrix jacchus) is a small New World primate that is becoming increasingly popular in the neurosciences as an animal model of preclinical human disease. With several major disorders characterized by alterations in neural white matter (e.g., multiple sclerosis, Alzheimer’s disease, schizophrenia), proposed to be transgenically modeled using marmosets, the ability to isolate and characterize reliably major white matter fiber tracts with MRI will be of use for evaluating structural brain changes related to disease processes and symptomatology. Here, we propose protocols for isolating major white matter fiber tracts in the common marmoset using in vivo ultrahigh-field MRI (9.4T) diffusion-weighted imaging (DWI) data. With the use of a high angular-resolution DWI (256 diffusion-encoding directions) sequence, collected on four anesthetized marmosets, we provide guidelines for manually drawing fiber-tracking regions of interest, based on easily identified anatomical landmarks in DWI native space. These fiber-tract isolation protocols are expected to be experimentally useful for visualization and quantification of individual white matter fiber tracts in both control and experimental groups of marmosets (e.g., transgenic models). As disease models in the marmoset advance, the determination of how macroscopic white matter anatomy is altered as a function of disease state will be relevant in bridging the existing translational gap between preclinical rodent models and human patients.
NEW & NOTEWORTHY Although significant progress has been made in mapping white matter connections in the marmoset brain using ex vivo tracing techniques, the application of in vivo virtual dissection of major white matter fiber tracts has been established by few studies in the marmoset literature. Here, we demonstrate the feasibility of whole-brain diffusion-weighted tractography in anesthetized marmosets at ultrahigh-field MRI (9.4T) and propose protocols for isolating nine major white matter fiber tracts in the marmoset brain.
the common marmoset (Callithrix jacchus) is a small New World primate that is becoming increasingly popular as an animal model of preclinical human disease. As a primate, the marmoset brain is structurally and functionally more similar to the human brain than the rodent brain, with a well-developed frontal cortex (Belcher et al. 2016; Homman-Ludiye and Bourne 2017; Mitchell and Leopold 2015; ’t Hart et al. 2016; Uylings and van Eden 1990; Walker et al. 2017). With the recent advent of transgenic marmosets (Sasaki et al. 2009), the marmoset model is well positioned to inform the translational gap between neurological disease models in rodents and human patients (Oikonomidis et al. 2017). As in vivo models of neurological, neurodegenerative, and psychiatric conditions are developed, the use of MRI to track brain changes noninvasively will be crucial in the execution of these studies.
With several major disorders characterized by alterations in neural white matter (e.g., multiple sclerosis, Alzheimer’s disease, schizophrenia), proposed to be modeled transgenically using marmosets (Okano and Mitra 2015), the ability to isolate and characterize reliably major white matter fiber tracts with MRI will be of use for evaluating structural brain changes related to disease processes and symptomatology. Diffusion-weighted imaging (DWI) is an MRI sequence that is widely applied in human, macaque, and rodent samples that allows for noninvasive characterization of major white matter fibers. This is accomplished by sensitizing the magnetic resonance signal to molecular water diffusivity surrounding white matter structures (Basser et al. 1994). Although significant progress has been made in mapping white matter anatomical connections in the marmoset brain using ex vivo tracing techniques (Burman et al. 2015; Majka et al. 2016; Reser et al. 2013; Roberts et al. 2007; Warner et al. 2015), the application of in vivo DWI has yet to be established in the marmoset literature.
Diffusion tractography (DT) is a type of DWI data analysis that allows for three-dimensional modeling of major white matter structures and quantification of the structural integrity of those tracts. DT provides a means for characterizing structural integrity by modeling water diffusion in brain tissue (Jones and Leemans 2011). With the modeling of translational motion of water molecules, neuroanatomical structure can be inferred. As water is diffusing in the brain, the microstructural environment (e.g., cell membranes, cytoskeletons, and protein structures) (Stejskal and Tanner 1965) influences its path in a predictable way: collisions with microstructure result in changes in the direction of diffusion. In regions where water can move freely, such as in the ventricles or gray matter, diffusion is largely isotopic: the magnitude of diffusion is equal in all directions. In white matter, however, water diffusion is restricted by neuronal structure and thus tends to diffuse anisotropically, with the magnitude of diffusion greatest in the direction that is parallel to axons. DT relies on similarities in the magnitude and direction of water diffusivity across voxels, generating virtual fiber representations across voxels with similar diffusion profiles that ostensibly correspond to white matter structure.
DT has been successfully applied in marmosets to isolate specific fiber pathways in stroke (Bihel et al. 2011) and spinal cord injury (Fujiyoshi et al. 2007, 2013). Additionally, DT has been used to isolate neuronal fiber trajectories of visual tracts in marmosets (Warner et al. 2015; Yamada et al. 2007); importantly, the validity of these DT-based fiber-tract isolation tracts was confirmed with ex vivo fiber tracing (Warner et al. 2015). To date, however, DT studies in marmosets have been driven by tract-specific hypotheses and therefore focused on a constricted area of interest, rather than isolating fiber tracts across the brain. Because the marmoset model is used to study a wide range of disorders—each with its own diverse set of affected brain loci—systematic protocols for isolating and constructing major fiber tracts across the marmoset brain are needed.
Reliable protocols for isolating fiber tracts have been devised for human samples through cleverly placed regions of interest that have a low sensitivity to small variations in user-region placement (Catani et al. 2002; Wakana et al. 2007). These guidelines for reproducing DT-based fiber tracts have been applied in hundreds of studies with both healthy and diseased samples [e.g., Ellison-Wright et al. (2014); Santillo et al. (2016); Surova et al. (2016); Tak et al. (2016)]. Within diseased samples, these protocols are broadly applied to a range of illnesses, from neurodegenerative diseases (Santillo et al. 2016; Surova et al. 2016) to psychiatric disorders, such as schizophrenia (Ellison-Wright et al. 2014). A similar protocol in marmosets would then likely be useful for a broad audience of researchers interested in in vivo virtual dissection of fiber tracts.
The aim of the present study was to demonstrate feasibility of whole-brain tractography in the common marmoset using ultrahigh-field MRI (9.4T) DWI data and to develop a protocol for isolating major white matter fiber tracts based on easily identified anatomical features. We used a high angular-resolution DWI sequence (256 diffusion-encoding directions) in four adult marmoset monkeys and manually isolated nine major white matter fiber tracts using DT. These fiber-tract isolation protocols are expected to be experimentally useful for visualization and quantification of individual white matter fiber tracts in both control and experimental groups of marmosets (e.g., transgenic models). As disease models in the marmoset advance, the determination of how macroscopic white matter anatomy is altered as a function of disease state will be of critical importance for translational research.
METHODS
Image acquisition.
DWI data were acquired on four male common marmoset monkeys (C. jacchus), aged 2.5–4.5 yr and weighing 300–450 g. Before each imaging session, anesthesia was induced in marmosets, with 4% isoflurane in 2 l/min oxygen in a plastic chamber. Isoflurane level was set to 1.5–3% during MRI acquisition and maintained throughout the scan by means of inhalation [see Mundinano et al. (2016) for an overview of anesthetic protocols]. Oxygen flow rate was kept between 1 and 2.5 l/min throughout the scan. Respiration, blood oxygen saturation level, and heart rate were monitored continuously via a pulse oximeter and were observed to be within the normal range throughout the scans. Body temperature was also measured and recorded throughout and maintained using warm water circulating blankets, thermal insulation, and warmed air. The positioning of the animal in the custom-built MRI bed was implemented, similar to the setup presented by Belcher et al. (2013). Experimental procedures were in accordance with the Canadian Council of Animal Care policy, and protocols were approved by the Animal Use Subcommittee of the University of Western Ontario Council on Animal Care.
DWI data were acquired using a 9.4T small animal MRI scanner, equipped with a 12-cm gradient coil set of 400 mT/m strength (Agilent Technologies, Santa Clara, CA). A custom in-house, two-channel transmit coil and adjustable eight-channel receive coil were used to accommodate a range of head sizes (from 37 to 50 mm). DWI data for each animal were acquired with the following parameters: number of shots = 1, repetition time = 5 s, echo time = 26 ms, diffusion time = 12.32 ms, diffusion-encoding directions = 256 (plus 9 b = 0 s/mm2 images), b-value = 1,000 s/mm2, number of averages = 2. Each volume comprised 36 slices with an in-plane resolution of 0.6 × 0.6 mm and a slice thickness of 0.6 mm. The field of view was 48 × 48 mm, and the corresponding matrix size was 80 × 80.
Image preprocessing.
Raw diffusion images were converted from Flexible Data Format to Neuroimaging Informatics Technology Initiative format and reoriented from the sphinx position. The gradient components in the b-vector files were also flipped and transposed to correspond to the reorientation from the sphinx position. As an image-quality assessment, temporal signal-to-noise ratio (TSNR) was calculated for each scan, using procedures outline by Roalf et al. (2016). Specifically, TSNR was calculated across diffusion-weighted volumes (nonweighted volumes excluded) before averaging the two diffusion runs. Across the four monkeys, TSNR was found to be sufficiently high (mean = 7.50, SD = 1.89) compared with the quality standards set by Roalf et al. (2016). The images were then processed using the ExploreDTI software package (Leemans et al. 2009). First, the images were converted into the.mat format and then corrected for Eddy current-induced geometric distortions and subject motion (with corresponding b-matrix rotation) (Leemans and Jones 2009). Whole-brain fiber tractography was conducted for each brain using a constrained spherical deconvolution (CSD) algorithm (Jeurissen et al. 2011), with the following parameters: seed-point resolution = 0.6 mm, step size = 0.3 mm, angle threshold = 30°, fiber-length range = 1–100 mm, fiber-orientation distribution = 0.1. The output of CSD was a fiber-orientation distribution for each voxel across the brain from which fibers could be traced between voxels.
Fiber tractography.
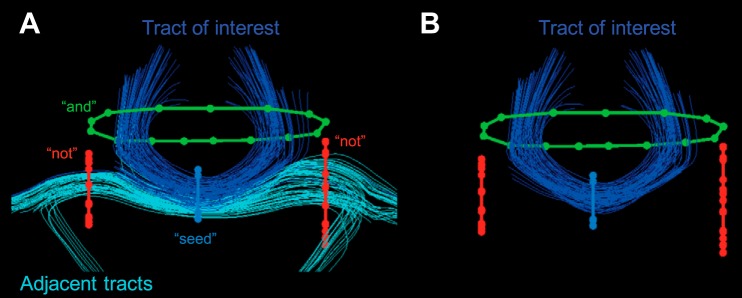
Fiber tractography was performed using the ExploreDTI software package (Leemans et al. 2009). To isolate fibers of interest, two-dimensional regions of interest were manually placed in DWI native space on fractional anisotropy maps, color coded for the principle direction of diffusion. The location of white matter landmarks was based on existing anatomical knowledge in marmosets (Hardman and Ashwell 2012; Paxinos et al. 2012) and loosely guided by existing fiber-isolation protocols that have been shown to be reliable in human samples (Catani and Thiebaut de Schotten 2008; Wakana et al. 2007). To isolate fibers of interest, Boolean operator regions of interest—“seed,” “and,” and “not”—were used (see Fig. 1, for example). The seed regions of interest were drawn first and encompassed obligatory passages of the tract of interest; these obligatory passages are brain structures that all fibers of a tract must pass through to reach their end points (Catani and Thiebaut de Schotten 2008). For example, an obligatory passage of the corticospinal tract is the cerebral peduncle, and thus a seed can be drawn to encompass all of that structure in an axial slice. Next, the and operator was used as a second obligatory passage to isolate further the tract. To continue with the corticospinal tract example, an and region of interest can be drawn in the posterior limb of the internal capsule—a second obligatory passage of the corticospinal tract. Additional and operators were used as needed for further isolation. Finally, not regions were used to exclude fibers that were not part of the tract of interest. With the corticospinal tract, not regions could be drawn to isolate only fibers reaching the motor cortex. Generally, we attempted to draw the seed and and regions to be maximally inclusive, often covering entire lobes on a given slice; this decreases the extent to which variability in user region-of-interest placement affects fiber isolation. Finally, mean values of diffusivity and length were extracted from each tract.
Fig. 1.
A demonstration of fiber-tract isolation using Boolean operator regions of interest. To isolate the blue tracts from the cyan tracts, “seed,” “and,” and “not” region of interests are used (A). First, a seed region is placed around an obligatory passage of the blue tract; i.e., all of the fibers of the blue tract must pass through the seed region. Second, 2 not regions of interest were drawn to exclude fibers of the cyan tract that also pass through the seed region. Finally, an and region is used to isolate further only anteriorly facing fibers of the blue tract. B: resultant fiber traces after applying the region-of-interest operators.
Reproducibility of fiber tracking.
To quantify inter-rater reliability of the nine fiber-tracking protocols, two raters (D. J. Schaeffer and R. Adam), experienced in fiber tractography, preformed the protocols independently for each monkey. Spatial overlap of tract structure between the two raters was then assessed using analysis of spatial matching by Cohen’s kappa (κ) using a procedure described previously by Danielian et al. (2010) and Wakana et al. (2007). First, fiber tracts were converted into a binary format (i.e., pixels containing the tract of interest = 1, and all other pixels = 0) for each rater; then, the two binary maps were superimposed. From this superimposition, four different voxel categories were yielded: 1) voxels that did not contain tracts from either rater (nn), 2) voxels that contained tracts from only the first rater (pn), 3) voxels that contained tracts from only the second rater (np), and 4) voxels that contained tracts from both raters (pp). Expected and observed agreement values were then calculated using Matlab (MathWorks, Natick, MA) [see Wakana et al. (2007) for equations]. Spatial Cohen’s κ was then calculated for each tract across the four monkeys, with κ = (observed agreement − expected agreement)/(100 − expected agreement). As defined by Landis and Koch (1977), a κ value of 0.11–0.20 is considered as “slight,” 0.21–0.40 “fair,” 0.41–0.60 “moderate,” 0.61–0.80 “substantial,” and 0.81–1.00 “almost perfect” agreement.
RESULTS
Fiber tractography.
Nine major fiber tracts were isolated, based on easily identified structures: anterior commissure, cingulum, forceps major, forceps minor, fornix, inferior fronto-occipital fasciculus, inferior longitudinal fasciculus, superior longitudinal fasciculus, and uncinate fasciculus. Figures 2–10 illustrate the placement of the regions of interest for each tract on a directionally encoded fractional anisotropy map. Apart from the bilateral tracts (i.e., anterior commissure and forceps major and minor), all regions were drawn in the left hemisphere, but identical procedures can be used to isolate the above tracts in the right hemisphere.
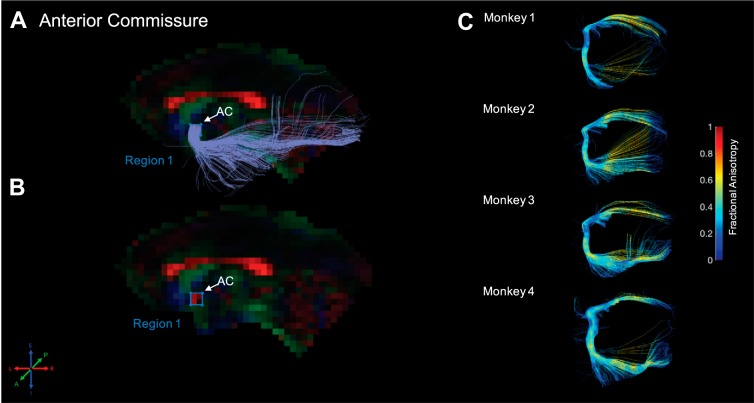
Fig. 2.
Anterior commissure. Fiber traces of anterior commissure (AC) are displayed in light purple (A) and overlaid on fractional anisotropy maps, color coded [red (R), green (G), blue (B)] for the principle direction of diffusion [R = left–right (L–R), G = anterior–posterior (A–P), B = inferior–superior (I–S)]. The region of interest for anterior commissure was drawn in the midsagittal plane. The location for region 1 (“seed”) was identified in the midsagittal slice and drawn around the anterior commissure (A), indicated by red voxels. The resultant fiber traces from this protocol are shown for each monkey and color coded for fractional anisotropy (C). For this figure and Figs. 3–10, left: locations of the regions are shown for monkey 3 (randomly chosen); right: tracts resulting from these region-of-interest protocols are shown for each monkey, color coded for fractional anisotropy.
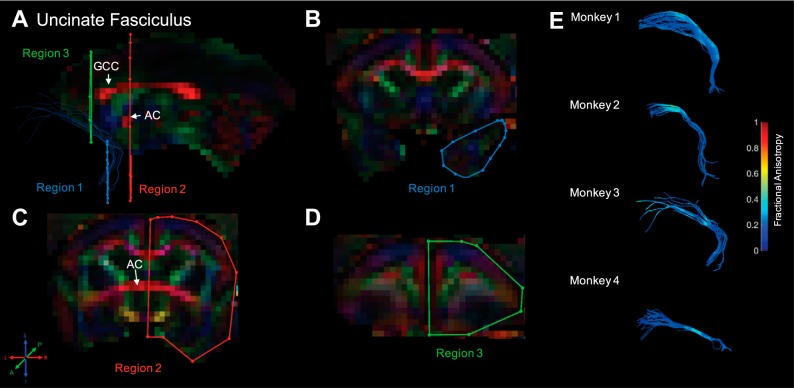
Fig. 10.
Uncinate fasciculus. Fiber traces of uncinate fasciculus are displayed in blue (A) and overlaid on fractional anisotropy maps, color coded (RGB) for the principle direction of diffusion (R = left–right, G = anterior–posterior, B = inferior–superior). Regions of interest for uncinate fasciculus were drawn in the coronal plane. The location for region 1 (“seed”) was identified by locating the most posterior coronal slice in which frontal and temporal cortices were separated and covered the entire temporal lobe in that slice (B). The location for region 2 (“not”) was drawn, immediately posterior to the knee of the uncinate fasciculus at the level of the anterior commissure (AC; C); region 2 served to exclude tracts (e.g., inferior frontal-occipital fasciculus) that have adjacent projections to the superior portion of the uncinate fasciculus (i.e., those innervating frontal cortex). Finally, region 3 (“and”) was drawn in a coronal slice, immediately anterior to the genu of the corpus callosum (GCC) and covered the entire hemisphere (D). The resultant fiber traces from this protocol are shown for each monkey and color coded for fractional anisotropy (E).
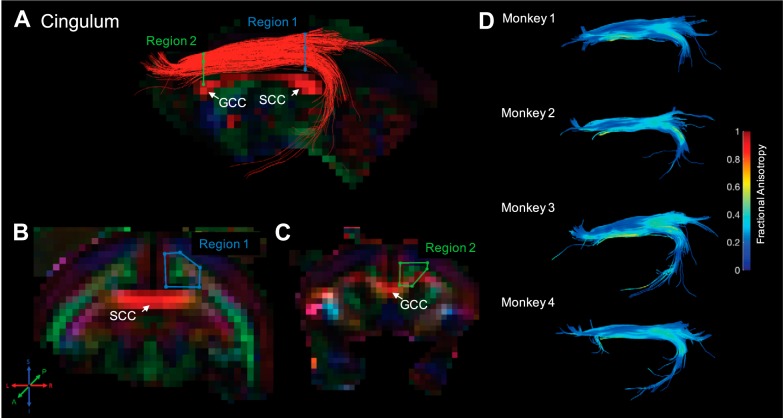
Fig. 3.
Cingulum. Fiber traces of cingulum are displayed in red (A) and overlaid on fractional anisotropy maps, color coded (RGB) for the principle direction of diffusion (R = left–right, G = anterior–posterior, B = inferior–superior). Regions of interest for cingulum were drawn in the coronal plane. The location for region 1 (“seed”) was identified in the midsagittal plane at the center of the splenium of the corpus callosum (SCC) and then drawn in a coronal slice to encompass the anterior–posterior-oriented fibers (indicated by green voxels), just superior to fibers of the corpus callosum (indicated by red voxels; B). Similarly, region 2 (“and”) was selected at the center of the genu of the corpus callosum (GCC) and drawn in the same way as region 1, surrounding anterior–posterior-oriented fibers (C). The resultant fiber traces from this protocol are shown for each monkey and color coded for fractional anisotropy (D).
Fig. 4.
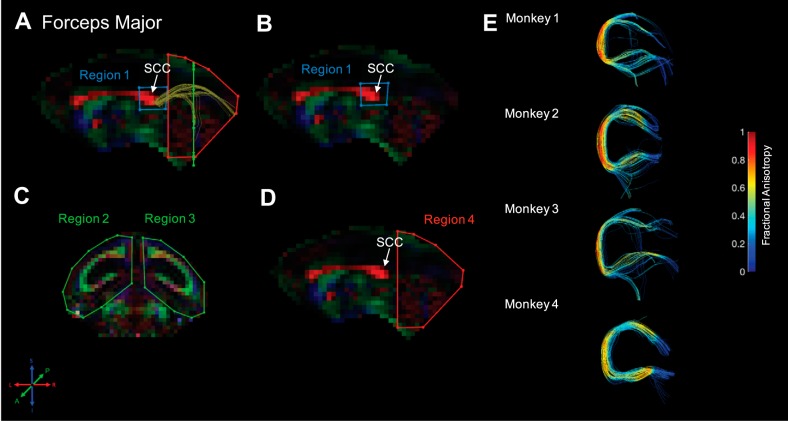
Forceps major. Fiber traces of forceps major are displayed in dark yellow (A) and overlaid on fractional anisotropy maps, color coded (RGB) for the principle direction of diffusion (R = left–right, G = anterior–posterior, B = inferior–superior). Regions of interest for forceps major were drawn in the sagittal and coronal planes. The location for region 1 (“seed”) was identified in the midsagittal plane and drawn around the splenium of the corpus callosum (SCC; B). The location for regions 2 and 3 (“and”) was drawn in a coronal slice, equidistant from the posterior edge of the SCC and the most posterior point in the brain; region 2 covered the entire left hemisphere, and region 3 covered the entire right hemisphere (C). Finally, region 4 (“not”) was drawn in a midsagittal slice and covered the entire slice posterior to the edge of the SCC (D). The resultant fiber traces from this protocol are shown for each monkey and color coded for fractional anisotropy (E).
Fig. 5.
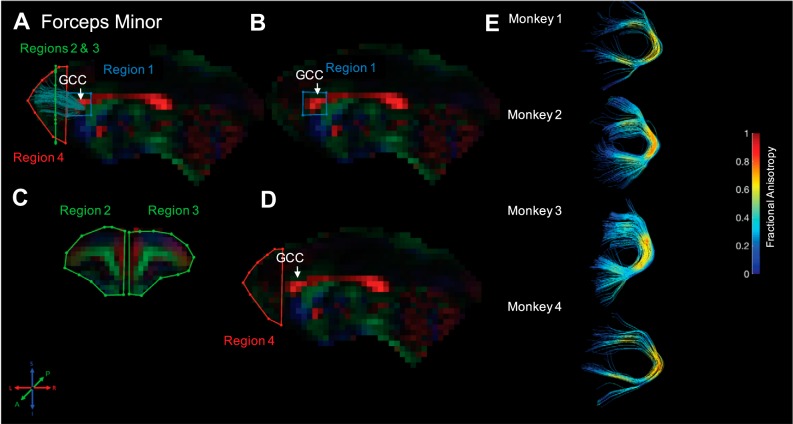
Forceps minor. Fiber traces of forceps minor are displayed in dark green (A) and overlaid on fractional anisotropy maps, color coded (RGB) for the principle direction of diffusion (R = left–right, G = anterior–posterior, B = inferior–superior). Regions of interest for forceps minor were drawn in the sagittal and coronal planes. The location for region 1 (“seed”) was identified in the midsagittal plane and drawn around the genu of the corpus callosum (GCC; B). The location for regions 2 and 3 (“and”) was drawn in a coronal slice, equidistant from the anterior edge of the GCC and the most anterior point in the brain; region 2 covered the entire left hemisphere, and region 3 covered the entire right hemisphere (C). Finally, region 4 (“not”) was drawn in a midsagittal slice and covered the entire slice anterior to the edge of the GCC (D). The resultant fiber traces from this protocol are shown for each monkey and color coded for fractional anisotropy (E).
Fig. 6.
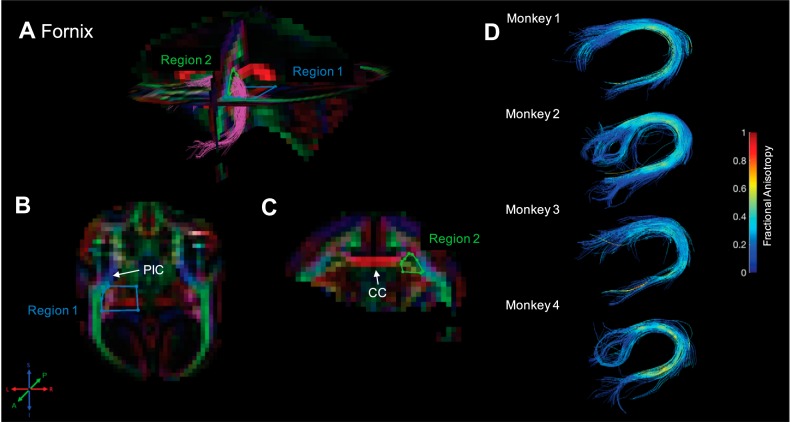
Fornix. Fiber traces of fornix are displayed in magenta (A) and overlaid on fractional anisotropy maps, color coded (RGB) for the principle direction of diffusion (R = left–right, G = anterior–posterior, B = inferior–superior). Regions of interest for fornix were drawn in the axial and coronal planes. The location for region 1 (“seed”) was identified in the axial plane, just posterior to the posterior limb of the internal capsule (PIC; blue voxels), and drawn around the magenta voxels that indicate a mixture of inferior–superior and medial–lateral tract trajectory (i.e., the knee of the fornix; B). The location for region 2 (“and”) was also identified in the axial plane, just posterior to the PIC, but drawn in the coronal plane to surround anterior–posterior-oriented fiber traces (green voxels), just lateral to fiber traces of the corpus callosum (CC; C). The resultant fiber traces from this protocol are shown for each monkey and color coded for fractional anisotropy (D).
Fig. 7.
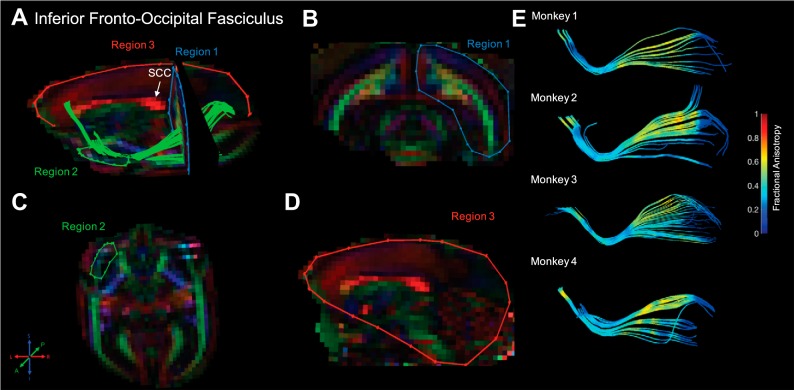
Inferior fronto-occipital fasciculus. Fiber traces of inferior fronto-occipital fasciculus are displayed in green (A) and overlaid on fractional anisotropy maps, color coded (RGB) for the principle direction of diffusion (R = left–right, G = anterior–posterior, B = inferior–superior). Regions of interest for inferior fronto-occipital fasciculus were drawn in 3 planes (coronal, sagittal, and axial). The location for region 1 (“seed”) was identified in the midsagittal plane, immediately posterior to the splenium of the corpus callosum (SCC), and was drawn in the coronal plane to cover the entire hemisphere (B). The location of region 2 (“and”) was identified and drawn in the axial plane around voxels of the external capsule (C). Finally, region 3 (“not”) was drawn in the midsagittal plane to exclude fibers of corpus callosum that mixed with the anterior portion of the inferior front-occipital fasciculus (D). The resultant fiber traces from this protocol are shown for each monkey and color coded for fractional anisotropy (E).
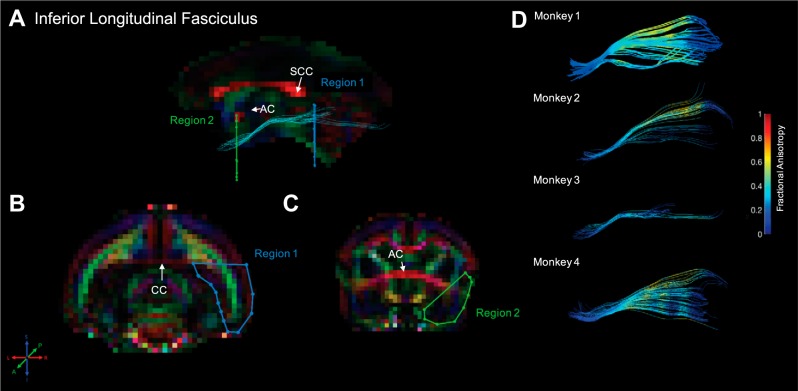
Fig. 8.
Inferior longitudinal fasciculus. Fiber traces of inferior longitudinal fasciculus are displayed in cyan (A) and overlaid on fractional anisotropy maps, color coded (RGB) for the principle direction of diffusion (R = left–right, G = anterior–posterior, B = inferior–superior). Regions of interest for inferior longitudinal fasciculus were drawn in the coronal plane. The location for region 1 (“seed”) was identified in the midsagittal plane, immediately posterior to the splenium of the corpus callosum (SCC), and was drawn in the coronal plane to cover all of the hemisphere inferior of the corpus callosum (CC; B). The location of region 2 (“and”) was identified in the midsagittal plane at the level of the anterior commissure (AC) and drawn in the axial plane to cover the entire temporal cortex (C). The resultant fiber traces from this protocol are shown for each monkey and color coded for fractional anisotropy (D). Note that the fiber traces of inferior longitudinal fasciculus and inferior frontal-occipital fasciculus (Fig. 7) show a large degree of overlap; although anatomically separate, the posterior portion of these fiber tracts cannot be fully delineated at the present resolution.
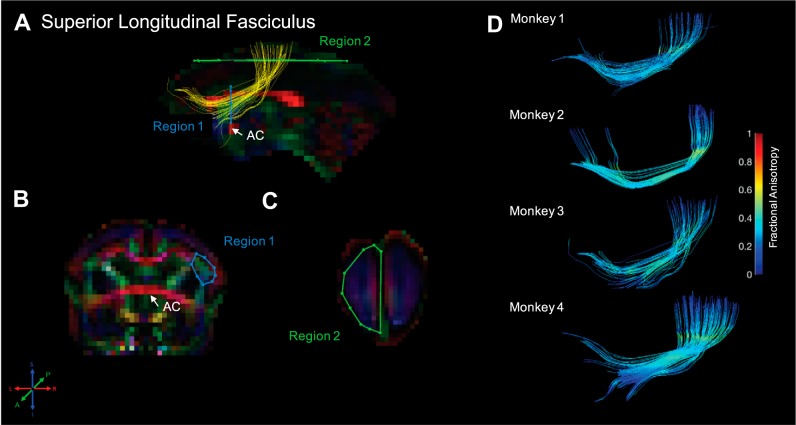
Fig. 9.
Superior longitudinal fasciculus. Fiber traces of superior longitudinal fasciculus are displayed in yellow (A) and overlaid on fractional anisotropy maps, color coded (RGB) for the principle direction of diffusion (R = left–right, G = anterior–posterior, B = inferior–superior). Regions of interest for superior longitudinal fasciculus were drawn in the coronal and axial planes. The location for region 1 (“seed”) was identified in a coronal slice at the level of the anterior commissure (AC; A) and drawn around anterior–posterior-oriented fiber traces of the superior longitudinal fasciculus, as identified by green voxels, immediately lateral to fiber traces of the corpus callosum (red voxels; B). Region 2 (“and”) was drawn in an axial slice, ~1 mm from the most superior point of the brain, and encompassed the entire hemisphere in that slice (C). The resultant fiber traces from this protocol are shown for each monkey and color coded for fractional anisotropy (D).
For each tract, means and SDs were extracted for fractional anisotropy, mean diffusivity, length, axial diffusivity, and radial diffusivity (Table 1). Because all four monkeys were healthy, these values could be useful as a baseline comparison for future studies. Additionally, these values provide guiding parameters for setting minimum and maximum fiber-tracking parameters. For example, the present data suggest that the fractional anisotropy threshold should be set lower for marmoset samples than what is typically used for fiber tracking in humans (e.g., fractional anisotropy = 0.2) (Wakana et al. 2007). Across the brain, diffusivity values showed the following range: fractional anisotropy = 0.17–0.43, mean diffusivity = 6.54–9.46 mm2/s (×10,000), axial diffusivity = 8.60–13.12 mm2/s (×10,000), radial diffusivity = 5.48–8.22 mm2/s (×10,000). Similarly, tract-length tracking parameters should be set lower for marmoset samples, as fiber-tracking software is typically calibrated for human samples (e.g., 50–500 mm). Here, tract length ranged from 13.41 to 33.41 mm.
Table 1.
Tract diffusivity values and length across 9 fiber tracts
| Anterior Commissure |
Cingulum |
Forceps Major |
Forceps Minor |
Fornix |
Inferior Fronto-Occipital Fasciculus |
Inferior Longitudinal Fasciculus |
Superior Longitudinal Fasciculus |
Uncinate Fasciculus |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| Fractional anisotropy | ||||||||||||||||||
| Monkey 1 | 0.32 | 0.13 | 0.24 | 0.08 | 0.42 | 0.22 | 0.39 | 0.17 | 0.30 | 0.12 | 0.35 | 0.15 | 0.31 | 0.14 | 0.29 | 0.06 | 0.20 | 0.07 |
| Monkey 2 | 0.37 | 0.14 | 0.29 | 0.09 | 0.43 | 0.23 | 0.38 | 0.18 | 0.29 | 0.13 | 0.38 | 0.15 | 0.32 | 0.15 | 0.34 | 0.10 | 0.21 | 0.09 |
| Monkey 3 | 0.38 | 0.13 | 0.28 | 0.10 | 0.37 | 0.14 | 0.31 | 0.12 | 0.31 | 0.12 | 0.37 | 0.11 | 0.32 | 0.14 | 0.31 | 0.09 | 0.19 | 0.07 |
| Monkey 4 | 0.35 | 0.13 | 0.29 | 0.08 | 0.39 | 0.19 | 0.40 | 0.17 | 0.29 | 0.12 | 0.38 | 0.15 | 0.29 | 0.13 | 0.31 | 0.10 | 0.17 | 0.08 |
| Average | 0.36 | 0.13 | 0.28 | 0.09 | 0.40 | 0.20 | 0.37 | 0.16 | 0.30 | 0.12 | 0.37 | 0.14 | 0.31 | 0.14 | 0.31 | 0.09 | 0.19 | 0.08 |
| Mean diffusivity, mm2/s, ×10,000 | ||||||||||||||||||
| Monkey 1 | 7.93 | 0.71 | 7.87 | 0.47 | 7.96 | 0.77 | 8.94 | 1.29 | 9.46 | 1.70 | 7.82 | 0.65 | 7.81 | 0.91 | 7.85 | 0.34 | 8.44 | 0.67 |
| Monkey 2 | 7.47 | 0.82 | 7.63 | 0.82 | 8.12 | 1.84 | 8.51 | 0.86 | 8.87 | 1.78 | 7.58 | 0.63 | 7.66 | 0.54 | 7.48 | 0.55 | 8.28 | 0.52 |
| Monkey 3 | 7.30 | 0.79 | 6.64 | 0.99 | 7.78 | 0.82 | 7.97 | 1.13 | 7.97 | 1.13 | 7.03 | 0.76 | 7.01 | 1.04 | 6.54 | 0.78 | 8.35 | 1.67 |
| Monkey 4 | 7.72 | 0.73 | 7.14 | 0.60 | 7.98 | 1.76 | 8.09 | 1.02 | 9.10 | 2.28 | 7.44 | 0.65 | 7.75 | 0.67 | 7.36 | 0.57 | 9.07 | 1.13 |
| Average | 7.61 | 0.76 | 7.32 | 0.72 | 7.96 | 1.30 | 8.38 | 1.08 | 8.85 | 1.72 | 7.47 | 0.67 | 7.56 | 0.79 | 7.31 | 0.56 | 8.54 | 1.00 |
| Length, mm | ||||||||||||||||||
| Monkey 1 | 32.74 | 9.49 | 18.04 | 2.56 | 26.97 | 4.03 | 17.21 | 1.97 | 19.44 | 3.97 | 27.30 | 4.40 | 23.83 | 2.01 | 13.60 | 5.35 | 15.32 | 3.44 |
| Monkey 2 | 33.18 | 12.19 | 18.77 | 3.31 | 24.99 | 2.74 | 17.12 | 2.28 | 25.18 | 6.21 | 24.25 | 3.14 | 20.62 | 3.20 | 18.29 | 2.22 | 15.18 | 2.20 |
| Monkey 3 | 33.41 | 9.30 | 16.04 | 3.33 | 20.56 | 3.18 | 13.41 | 6.78 | 13.41 | 6.78 | 27.08 | 3.75 | 20.00 | 5.71 | 15.23 | 2.53 | 14.83 | 3.76 |
| Monkey 4 | 29.01 | 10.83 | 18.13 | 3.17 | 22.65 | 1.84 | 18.28 | 0.93 | 23.41 | 7.45 | 21.33 | 2.62 | 17.37 | 2.46 | 15.10 | 1.48 | 19.92 | 1.34 |
| Average | 32.09 | 10.45 | 17.75 | 3.09 | 23.79 | 2.95 | 16.51 | 2.99 | 20.36 | 6.10 | 24.99 | 3.48 | 20.46 | 3.35 | 15.56 | 2.90 | 16.31 | 2.69 |
| Axial diffusivity, mm2/s, ×10,000 | ||||||||||||||||||
| Monkey 1 | 10.79 | 1.33 | 9.80 | 0.86 | 12.14 | 3.10 | 13.12 | 2.87 | 12.67 | 2.40 | 10.97 | 1.47 | 10.50 | 1.21 | 10.20 | 0.83 | 10.27 | 0.85 |
| Monkey 2 | 10.76 | 1.53 | 9.95 | 1.07 | 12.53 | 3.18 | 12.36 | 2.66 | 11.80 | 2.41 | 11.00 | 1.57 | 10.49 | 1.49 | 10.00 | 0.80 | 10.06 | 1.04 |
| Monkey 3 | 10.53 | 1.50 | 8.60 | 1.35 | 11.21 | 2.12 | 10.81 | 2.09 | 10.81 | 2.09 | 9.95 | 1.37 | 9.49 | 1.51 | 8.67 | 1.20 | 9.91 | 2.14 |
| Monkey 4 | 10.79 | 1.32 | 9.22 | 1.01 | 11.87 | 3.34 | 12.05 | 2.76 | 12.05 | 2.81 | 10.77 | 1.77 | 10.31 | 1.45 | 9.76 | 1.09 | 10.76 | 1.49 |
| Average | 10.72 | 1.42 | 9.39 | 1.07 | 11.94 | 2.94 | 12.09 | 2.60 | 11.83 | 2.43 | 10.67 | 1.55 | 10.20 | 1.42 | 9.66 | 0.98 | 10.25 | 1.38 |
| Radial diffusivity, mm2/s, ×10,000 | ||||||||||||||||||
| Monkey 1 | 6.50 | 1.13 | 6.90 | 0.75 | 5.87 | 1.63 | 6.84 | 1.57 | 7.85 | 1.76 | 6.24 | 1.22 | 6.46 | 1.40 | 6.67 | 0.73 | 7.52 | 0.90 |
| Monkey 2 | 5.83 | 1.20 | 6.47 | 1.10 | 5.91 | 2.37 | 6.58 | 1.42 | 7.40 | 1.88 | 5.88 | 1.13 | 6.25 | 1.05 | 6.22 | 1.03 | 7.39 | 0.80 |
| Monkey 3 | 5.68 | 1.09 | 5.66 | 1.16 | 6.06 | 1.10 | 6.56 | 1.19 | 6.56 | 1.19 | 5.58 | 0.97 | 5.77 | 1.35 | 5.48 | 0.96 | 7.57 | 1.60 |
| Monkey 4 | 6.19 | 1.12 | 6.09 | 0.79 | 6.04 | 1.84 | 6.11 | 1.28 | 7.62 | 2.31 | 5.78 | 0.96 | 6.47 | 1.04 | 6.17 | 0.89 | 8.22 | 1.23 |
| Average | 6.05 | 1.14 | 6.28 | 0.95 | 5.97 | 1.74 | 6.52 | 1.37 | 7.36 | 1.79 | 5.87 | 1.07 | 6.24 | 1.21 | 6.14 | 0.90 | 7.68 | 1.13 |
Reproducibility of fiber tracking.
Cohen’s κ values, which quantified spatial agreement of tract shape (i.e., overlap between independent fiber-tracking results from 2 raters) demonstrated almost perfect (κ > 0.80) or substantial (κ = 0.61–0.80) agreement, with an average agreement of κ = 0.88 across tracts (Table 2).
Table 2.
Spatial κ overlap values (inter-rater reliability) across 9 fiber tracts
| Anterior Commissure | Cingulum | Forceps Major | Forceps Minor | Fornix | Inferior Fronto-Occipital Fasciculus | Inferior Longitudinal Fasciculus | Superior Longitudinal Fasciculus | Uncinate Fasciculus | |
|---|---|---|---|---|---|---|---|---|---|
| Monkey 1 | 0.911 | 0.884 | 0.917 | 1.000 | 0.804 | 1.000 | 0.843 | 0.872 | 0.848 |
| Monkey 2 | 0.877 | 0.794 | 0.872 | 0.857 | 0.819 | 0.938 | 0.874 | 0.822 | 0.732 |
| Monkey 3 | 0.761 | 0.851 | 0.937 | 0.957 | 0.846 | 0.926 | 1.000 | 0.920 | 0.820 |
| Monkey 4 | 0.887 | 0.881 | 0.909 | 1.000 | 0.784 | 0.921 | 0.783 | 0.975 | 0.837 |
| Average | 0.859 | 0.853 | 0.909 | 0.954 | 0.813 | 0.946 | 0.875 | 0.897 | 0.809 |
Cohen's kappa (κ) value of 0.11–0.20 is considered as “slight,” 0.21–0.40 “fair,” 0.41–0.60 “moderate,” 0.61–0.80 “substantial,” and 0.81–1.00 “almost perfect” agreement.
DISCUSSION
The common marmoset is becoming increasingly popular as a research model for studying human brain diseases. Because marmosets can be transgenically modified, they have received considerable attention as a powerful tool for studying the mechanisms of human brain diseases. As primates, marmosets share characteristic features of the human brain (e.g., highly granular frontal lobe, expanded temporal lobe), offering a distinct advantage over rodent models (Homman-Ludiye and Bourne 2017; Izpisua Belmonte et al. 2015; Mitchell and Leopold 2015; Wise 2008). Marmosets also offer advantages over Old World primates, such as macaques (e.g., Macaca mulatta), as diseases can be modeled on a shorter time scale in marmosets with a lower interbirth interval and faster sexual maturation time than macaques (Kishi et al. 2014; Sasaki et al. 2009). Marmosets also have frequent chimeric twinning, which presents tremendous potential for genetic studies (Schiel and Souto 2017; Ward et al. 2014). As researchers begin to use genetically modified marmosets to model human brain diseases, noninvasive techniques, such as MRI, will allow for unprecedented longitudinal tracing of disease-related brain changes without the necessity of killing the animal.
In this study, we developed protocols for isolating nine major white matter fiber tracts in the marmoset brain based on ultrahigh-field MRI data. With the use of fiber tractography, white matter fiber tracts were three dimensionally rendered across the brain, and regions of interest were defined to isolate anterior commissure, cingulum, forceps major, forceps minor, fornix, inferior fronto-occipital fasciculus, inferior longitudinal fasciculus, superior longitudinal fasciculus, and uncinate fasciculus. With the generation of large regions of interest with reference to easily identified structures (e.g., anterior commissure, genu or splenium of corpus callosum), we sought to minimize the effect of variability due to individual differences in region-of-interest drawing (Huang et al. 2004). Indeed, we demonstrated almost perfect inter-rater reliability across the majority of tracts, as assessed by spatial agreement of tract shape using Cohen’s κ (almost perfect agreement = κ > 0.81). These reconstruction procedures are intended to be maximally inclusive and robust. That is, with the use of multiple large regions of interest that included obligatory passages, we were able to isolate the body of a given fiber tract but did not restrict end-point distributions, which vary as a function of individual anatomy and fiber-tracking algorithm stringency (Christidi et al. 2016). As the voxel resolution of DWI in marmosets increases, and a comprehensive white matter atlas is developed (Majka et al. 2016), researchers will be better able to refine the specificity of these protocols, as dictated by their needs.
The present fiber-tracking results highlight the parallels between human and marmoset white matter architecture. The marmoset association tracts (i.e., those connecting different areas of the cortex) shown here bear surprising resemblance to human association tracts [see Catani et al. (2002); Hua et al. (2008); Wakana et al. (2007)]. Recent resting-state functional MRI work in marmosets suggests evolutionary preservation of brain connectivity in marmosets, demonstrating clear homologs in functional connectivity between marmosets and humans (Belcher et al. 2013, 2016; Ghahremani et al. 2017). This can be contrasted to rodent models that show a lower degree of frontal connectivity (Jonckers et al. 2011). Between the homologous parallels in marmoset connectivity and the ability to model diseases in this small nonhuman primate, the marmoset holds tremendous promise for studying neuropsychiatric and neurodegenerative disorders.
Human homologs of the frontally connected tracts shown here (e.g., cingulum, uncinate fasciculus, and superior longitudinal fasciculus) have been implicated in major psychiatric disorders. For example, altered white matter integrity of superior longitudinal fasciculus is associated with autism spectrum disorder (Libero et al. 2016), and altered integrity in uncinate fasciculus and superior longitudinal fasciculus is related to cognitive symptomology in schizophrenia (Karlsgodt et al. 2008; Kubicki et al. 2002; Schaeffer et al. 2015). The ability to track frontally mediated sequelae of neuropsychiatric models in marmosets has the potential to provide insight to the nature of these behavioral abnormalities. Indeed, the pursuit of genetically modified marmosets to model autism spectrum disorder, schizophrenia, and degenerative disorders, such as Alzheimer’s and Parkinson’s diseases, is occurring (Okano et al. 2016; ’t Hart et al. 2016). As one of the first few studies to track these frontally connected white matter fibers in the marmoset using fiber tractography, we expect that the fiber-isolation protocols presented here will be useful for examining the existence of such deficits in marmoset disease models.
Given the small size of the marmoset brain, it is necessary to image at comparatively high spatial resolution (180 times smaller than the human brain) to resolve tract structures (Solomon and Rosa, 2014; Uylings and van Eden 1990). Small-bore animal MRI scanners (i.e., those typically used for rodent studies) offer clear advantages over clinical MRI systems, with purpose-built hardware (e.g., head coils), higher field strengths, and higher signal-to-noise ratios. In rodents, ultrahigh resolution fiber-tracking data have been achieved in vivo (<0.15 mm in-plane) using 9.4T small bore scanners (Cai et al. 2011; Harsan et al. 2010). The attainment of data at such a high resolution, however, is not only equipment dependent but comes at the cost of long scan times (i.e., multishot sequences with long repetition times) that are not optimal for live marmosets. Here, we used a single-shot echo-planar imaging sequence with a reasonably high resolution (0.6 mm isotropic) and a short total scan time of ~22 min (44 min, with 2 averages). With a short repetition time of 5 s, a high angular-resolution sequence was achieved (256 diffusion-encoding directions), which allowed for the use of multifiber/voxel modeling (CSD) (Jeurissen et al. 2011) and thus estimation of multiple intravoxel fiber populations.
These protocols should be used with the following caveats in mind. As an indirect measure of white matter at a macroscopic resolution (0.6 mm isotropic here), DWI is fundamentally limited in the ability to detect true fiber structure, particularly in voxels that include tissues other than white matter (e.g., gray matter, vasculature). Here, we chose to focus on major white matter fiber tracts based on their size and high level of anisotropy, but some other tracts (e.g., thalamic afferents) could not be reliably detected at the present resolution. As such, direct labeling of these tracts [e.g., Roberts et al. (2007); Warner et al. (2015)] remains a superior method for studies that necessitate a high level of anatomical accuracy. Recent two-photon laser microscopy work in mice, however, suggests that the canonical DWI-derived measure of white matter integrity (fractional anisotropy) in major white matter tracts shows a strong correlation with directly labeled axonal elements (Chang et al. 2017). This work suggests that DWI-derived fibers are likely most accurate in areas of coherently organized, densely myelinated structures (e.g., corpus callosum) but less accurate as tract structures branch into smaller fiber bundles. There is also a nontrivial amount of variability in reconstruction results that stem from image quality (e.g., poor SNR, artifacts, motion) and reconstruction patterns (e.g., complexity of fiber-orientation modeling, fiber-tracking parameters) (Huang et al. 2004). In this study, we sought to ameliorate such effects through the use of ultrahigh-field MRI; a custom-built, multichannel head coil; and a high angular-resolution sequence paired with a multifiber/voxel reconstruction protocol (i.e., CSD). Because the aforementioned pitfalls are a function of voxel resolution and SNR, these sources of variance will likely be mitigated as MRI hardware and sequences improve (e.g., higher SNR, higher spatial resolution protocols) (Roebroeck et al. 2008).
In summary, the present manuscript documents protocols for isolating nine major white matter fiber tracts in the common marmoset brain using DWI data and fiber tractography. To our knowledge, this is the first study to provide guidelines for conducting whole-brain fiber tractography in a marmoset sample. The present results corroborate evidence of evolutionary preservation of functional brain connectivity in the marmoset (Ghahremani et al. 2017), albeit some specific connectivity differences do exist [see Burman et al. (2014)], and give further credence to the use of marmosets as nonhuman primate homologs to study human brain disease. As researchers leverage the genetic proximity of marmosets and humans to study intractable brain diseases, we expect that these protocols will be useful for characterizing alterations in neural white matter in both healthy and diseased states.
GRANTS
Support for this work was provided by the Canadian Institutes of Health Research (FRN 148365), Brain Canada, and Canada First Excellence Fund.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
D.J.S., K.M.G., J.S.G., R.S.M., and S.E. conceived and designed research; D.J.S. and A.X.L. performed experiments; D.J.S. and R.A. analyzed data; D.J.S. and S.E. interpreted results of experiments; D.J.S. prepared figures; D.J.S. and S.E. drafted manuscript; D.J.S., R.A., and S.E. edited and revised manuscript; D.J.S., R.A., K.M.G., J.S.G., A.X.L., R.S.M., and S.E. approved final version of manuscript.
ACKNOWLEDGMENTS
The authors thank Nicole Hague, Ashley Kirley, and Lauren Schaeffer for their outstanding technical support.
REFERENCES
- Basser PJ, Mattiello J, LeBihan D. MR diffusion tensor spectroscopy and imaging. Biophys J 66: 259–267, 1994. doi: 10.1016/S0006-3495(94)80775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belcher AM, Yen CC, Notardonato L, Ross TJ, Volkow ND, Yang Y, Stein EA, Silva AC, Tomasi D. Functional connectivity hubs and networks in the awake marmoset brain. Front Integr Nuerosci 10: 9, 2016. doi: 10.3389/fnint.2016.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belcher AM, Yen CC, Stepp H, Gu H, Lu H, Yang Y, Silva AC, Stein EA. Large-scale brain networks in the awake, truly resting marmoset monkey. J Neurosci 33: 16796–16804, 2013. doi: 10.1523/JNEUROSCI.3146-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bihel E, Roussel S, Toutain J, Bernaudin M, Touzani O. Diffusion tensor MRI reveals chronic alterations in white matter despite the absence of a visible ischemic lesion on conventional MRI: a nonhuman primate study. Stroke 42: 1412–1419, 2011. doi: 10.1161/STROKEAHA.110.596650. [DOI] [PubMed] [Google Scholar]
- Burman KJ, Bakola S, Richardson KE, Reser DH, Rosa MG. Patterns of cortical input to the primary motor area in the marmoset monkey. J Comp Neurol 522: 811–843, 2014. doi: 10.1002/cne.23447. [DOI] [PubMed] [Google Scholar]
- Burman KJ, Bakola S, Richardson KE, Yu H-H, Reser DH, Rosa MG. Cortical and thalamic projections to cytoarchitectural areas 6Va and 8C of the marmoset monkey: connectionally distinct subdivisions of the lateral premotor cortex. J Comp Neurol 523: 1222–1247, 2015. doi: 10.1002/cne.23734. [DOI] [PubMed] [Google Scholar]
- Cai Y, McMurray MS, Oguz I, Yuan H, Styner MA, Lin W, Johns JM, An H. Use of high resolution 3D diffusion tensor imaging to study brain white matter development in live neonatal rats. Front Psychiatry 2: 54, 2011. doi: 10.3389/fpsyt.2011.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catani M, Howard RJ, Pajevic S, Jones DK. Virtual in vivo interactive dissection of white matter fasciculi in the human brain. Neuroimage 17: 77–94, 2002. doi: 10.1006/nimg.2002.1136. [DOI] [PubMed] [Google Scholar]
- Catani M, Thiebaut de Schotten M. A diffusion tensor imaging tractography atlas for virtual in vivo dissections. Cortex 44: 1105–1132, 2008. doi: 10.1016/j.cortex.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Chang EH, Argyelan M, Aggarwal M, Chandon T-S, Karlsgodt KH, Mori S, Malhotra AK. The role of myelination in measures of white matter integrity: combination of diffusion tensor imaging and two-photon microscopy of CLARITY intact brains. Neuroimage 147: 253–261, 2017. doi: 10.1016/j.neuroimage.2016.11.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christidi F, Karavasilis E, Samiotis K, Bisdas S, Papanikolaou N. Fiber tracking: a qualitative and quantitative comparison between four different software tools on the reconstruction of major white matter tracts. Eur J Radiol Open 3: 153–161, 2016. doi: 10.1016/j.ejro.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielian LE, Iwata NK, Thomasson DM, Floeter MK. Reliability of fiber tracking measurements in diffusion imaging for longitudinal study. Neuroimage 49: 1572–1580, 2010. doi: 10.1016/j.neuroimage.2009.08.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison-Wright I, Nathan PJ, Bullmore ET, Zaman R, Dudas RB, Agius M, Fernandez-Egea E, Müller U, Dodds CM, Forde NJ, Scanlon C, Leemans A, McDonald C, Cannon DM. Distribution of tract deficits in schizophrenia. BMC Psychiatry 14: 99, 2014. doi: 10.1186/1471-244X-14-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiyoshi K, Konomi T, Yamada M, Hikishima K, Tsuji O, Komaki Y, Momoshima S, Toyama Y, Nakamura M, Okano H. Diffusion tensor imaging and tractography of the spinal cord: from experimental studies to clinical application. Exp Neurol 242: 74–82, 2013. doi: 10.1016/j.expneurol.2012.07.015. [DOI] [PubMed] [Google Scholar]
- Fujiyoshi K, Yamada M, Nakamura M, Yamane J, Katoh H, Kitamura K, Kawai K, Okada S, Momoshima S, Toyama Y, Okano H. In vivo tracing of neural tracts in the intact and injured spinal cord of marmosets by diffusion tensor tractography. J Neurosci 27: 11991–11998, 2007. doi: 10.1523/JNEUROSCI.3354-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghahremani M, Hutchison RM, Menon RS, Everling S. Frontoparietal functional connectivity in the common marmoset. Cereb Cortex 27: 3890–3905, 2017. doi: 10.1093/cercor/bhw198. [DOI] [PubMed] [Google Scholar]
- Hardman CD, Ashwell KW. Stereotaxic and Chemoarchitectural Atlas of the Brain of the Common Marmoset (Callithrix jacchus). Boca Raton, FL: CRC, 2012. doi: 10.1201/b11635. [DOI] [Google Scholar]
- Harsan L-A, Paul D, Schnell S, Kreher BW, Hennig J, Staiger JF, von Elverfeldt D. In vivo diffusion tensor magnetic resonance imaging and fiber tracking of the mouse brain. NMR Biomed 23: 884–896, 2010. doi: 10.1002/nbm.1496. [DOI] [PubMed] [Google Scholar]
- Homman-Ludiye J, Bourne JA. The marmoset: an emerging model to unravel the evolution and development of the primate neocortex. Dev Neurobiol 77: 263–272, 2017. doi: 10.1002/dneu.22425. [DOI] [PubMed] [Google Scholar]
- Hua K, Zhang J, Wakana S, Jiang H, Li X, Reich DS, Calabresi PA, Pekar JJ, van Zijl PC, Mori S. Tract probability maps in stereotaxic spaces: analyses of white matter anatomy and tract-specific quantification. Neuroimage 39: 336–347, 2008. doi: 10.1016/j.neuroimage.2007.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Zhang J, van Zijl PC, Mori S. Analysis of noise effects on DTI-based tractography using the brute-force and multi-ROI approach. Magn Reson Med 52: 559–565, 2004. doi: 10.1002/mrm.20147. [DOI] [PubMed] [Google Scholar]
- Izpisua Belmonte JC, Callaway EM, Caddick SJ, Churchland P, Feng G, Homanics GE, Lee KF, Leopold DA, Miller CT, Mitchell JF, Mitalipov S, Moutri AR, Movshon JA, Okano H, Reynolds JH, Ringach D, Sejnowski TJ, Silva AC, Strick PL, Wu J, Zhang F. Brains, genes, and primates. Neuron 86: 617–631, 2015. doi: 10.1016/j.neuron.2015.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeurissen B, Leemans A, Jones DK, Tournier J-D, Sijbers J. Probabilistic fiber tracking using the residual bootstrap with constrained spherical deconvolution. Hum Brain Mapp 32: 461–479, 2011. doi: 10.1002/hbm.21032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonckers E, Van Audekerke J, De Visscher G, Van der Linden A, Verhoye M. Functional connectivity fMRI of the rodent brain: comparison of functional connectivity networks in rat and mouse. PLoS One 6: e18876, 2011. doi: 10.1371/journal.pone.0018876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D, Leemans A. Diffusion tensor imaging. In: Magnetic Resonance Neuroimaging, edited by Modo M and Bulte J. New York: Springer Science+Business Media, 2011, p. 127–144. doi: 10.1007/978-1-61737-992-5_6. [DOI] [Google Scholar]
- Karlsgodt KH, van Erp TG, Poldrack RA, Bearden CE, Nuechterlein KH, Cannon TD. Diffusion tensor imaging of the superior longitudinal fasciculus and working memory in recent-onset schizophrenia. Biol Psychiatry 63: 512–518, 2008. doi: 10.1016/j.biopsych.2007.06.017. [DOI] [PubMed] [Google Scholar]
- Kishi N, Sato K, Sasaki E, Okano H. Common marmoset as a new model animal for neuroscience research and genome editing technology. Dev Growth Differ 56: 53–62, 2014. doi: 10.1111/dgd.12109. [DOI] [PubMed] [Google Scholar]
- Kubicki M, Westin C-F, Maier SE, Frumin M, Nestor PG, Salisbury DF, Kikinis R, Jolesz FA, McCarley RW, Shenton ME. Uncinate fasciculus findings in schizophrenia: a magnetic resonance diffusion tensor imaging study. Am J Psychiatry 159: 813–820, 2002. doi: 10.1176/appi.ajp.159.5.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics 33: 159–174, 1977. doi: 10.2307/2529310. [DOI] [PubMed] [Google Scholar]
- Leemans A, Jeurissen B, Sijbers J, Jones D.. ExploreDTI: a graphical toolbox for processing, analyzing, and visualizing diffusion MR data. Proc Intl Soc Mag Reson Med 17: 3537, 2009. [Google Scholar]
- Leemans A, Jones DK. The B-matrix must be rotated when correcting for subject motion in DTI data. Magn Reson Med 61: 1336–1349, 2009. doi: 10.1002/mrm.21890. [DOI] [PubMed] [Google Scholar]
- Libero LE, Burge WK, Deshpande HD, Pestilli F, Kana RK. White matter diffusion of major fiber tracts implicated in Autism Spectrum Disorder. Brain Connect 6: 691–699, 2016. doi: 10.1089/brain.2016.0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majka P, Chaplin TA, Yu H-H, Tolpygo A, Mitra PP, Wójcik DK, Rosa MG. Towards a comprehensive atlas of cortical connections in a primate brain: mapping tracer injection studies of the common marmoset into a reference digital template. J Comp Neurol 524: 2161–2181, 2016. doi: 10.1002/cne.24023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JF, Leopold DA. The marmoset monkey as a model for visual neuroscience. Neurosci Res 93: 20–46, 2015. doi: 10.1016/j.neures.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundinano I-C, Flecknell PA, Bourne JA. MRI-guided stereotaxic brain surgery in the infant and adult common marmoset. Nat Protoc 11: 1299–1308, 2016. doi: 10.1038/nprot.2016.076. [DOI] [PubMed] [Google Scholar]
- Oikonomidis L, Santangelo AM, Shiba Y, Clarke FH, Robbins TW, Roberts AC. A dimensional approach to modeling symptoms of neuropsychiatric disorders in the marmoset monkey. Dev Neurobiol 77: 328–353, 2017. doi: 10.1002/dneu.22446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano H, Mitra P. Brain-mapping projects using the common marmoset. Neurosci Res 93: 3–7, 2015. doi: 10.1016/j.neures.2014.08.014. [DOI] [PubMed] [Google Scholar]
- Okano H, Sasaki E, Yamamori T, Iriki A, Shimogori T, Yamaguchi Y, Kasai K, Miyawaki A. Brain/MINDS: a Japanese national brain project for marmoset neuroscience. Neuron 92: 582–590, 2016. doi: 10.1016/j.neuron.2016.10.018. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Watson C, Petrides M, Rosa M, Tokuno H. The Marmoset Brain in Stereotaxic Coordinates. San Diego: Elsevier Academic, 2012. [Google Scholar]
- Reser DH, Burman KJ, Yu H-H, Chaplin TA, Richardson KE, Worthy KH, Rosa MG. Contrasting patterns of cortical input to architectural subdivisions of the area 8 complex: a retrograde tracing study in marmoset monkeys. Cereb Cortex 23: 1901–1922, 2013. doi: 10.1093/cercor/bhs177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roalf DR, Quarmley M, Elliott MA, Satterthwaite TD, Vandekar SN, Ruparel K, Gennatas ED, Calkins ME, Moore TM, Hopson R, Prabhakaran K, Jackson CT, Verma R, Hakonarson H, Gur RC, Gur RE. The impact of quality assurance assessment on diffusion tensor imaging outcomes in a large-scale population-based cohort. Neuroimage 125: 903–919, 2016. doi: 10.1016/j.neuroimage.2015.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AC, Tomic DL, Parkinson CH, Roeling TA, Cutter DJ, Robbins TW, Everitt BJ. Forebrain connectivity of the prefrontal cortex in the marmoset monkey (Callithrix jacchus): an anterograde and retrograde tract-tracing study. J Comp Neurol 502: 86–112, 2007. doi: 10.1002/cne.21300. [DOI] [PubMed] [Google Scholar]
- Roebroeck A, Galuske R, Formisano E, Chiry O, Bratzke H, Ronen I, Kim DS, Goebel R. High-resolution diffusion tensor imaging and tractography of the human optic chiasm at 9.4 T. Neuroimage 39: 157–168, 2008. doi: 10.1016/j.neuroimage.2007.08.015. [DOI] [PubMed] [Google Scholar]
- Santillo AF, Lundblad K, Nilsson M, Landqvist Waldö M, van Westen D, Lätt J, Blennow Nordström E, Vestberg S, Lindberg O, Nilsson C. Grey and white matter clinico-anatomical correlates of disinhibition in neurodegenerative disease. PLoS One 11: e0164122, 2016. doi: 10.1371/journal.pone.0164122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki E, Suemizu H, Shimada A, Hanazawa K, Oiwa R, Kamioka M, Tomioka I, Sotomaru Y, Hirakawa R, Eto T, Shiozawa S, Maeda T, Ito M, Ito R, Kito C, Yagihashi C, Kawai K, Miyoshi H, Tanioka Y, Tamaoki N, Habu S, Okano H, Nomura T. Generation of transgenic non-human primates with germline transmission. Nature 459: 523–527, 2009. doi: 10.1038/nature08090. [DOI] [PubMed] [Google Scholar]
- Schaeffer DJ, Rodrigue AL, Burton CR, Pierce JE, Unsworth N, Clementz BA, McDowell JE. White matter structural integrity differs between people with schizophrenia and healthy groups as a function of cognitive control. Schizophr Res 169: 62–68, 2015. doi: 10.1016/j.schres.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Schiel N, Souto A. The common marmoset: an overview of its natural history, ecology and behavior. Dev Neurobiol 77: 244–262, 2017. doi: 10.1002/dneu.22458. [DOI] [PubMed] [Google Scholar]
- Solomon SG, Rosa MG. A simpler primate brain: the visual system of the marmoset monkey. Front Neural Circuits 8: 96, 2014. doi: 10.3389/fncir.2014.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stejskal EO, Tanner JE. Spin diffusion measurements: spin echoes in the presence of a time-dependent field gradient. J Chem Phys 42: 288–292, 1965. doi: 10.1063/1.1695690. [DOI] [Google Scholar]
- Surova Y, Lampinen B, Nilsson M, Lätt J, Hall S, Widner H, van Westen D, Hansson O; Swedish BioFINDER study . Alterations of diffusion kurtosis and neurite density measures in deep grey matter and white matter in Parkinson’s disease. PLoS One 11: e0157755, 2016. doi: 10.1371/journal.pone.0157755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ’t Hart BA, Dunham J, Jagessar SA, Kap YS. The common marmoset (Callithrix jacchus): a relevant preclinical model of human (auto)immune-mediated inflammatory disease of the brain. Primate Biol 3: 9–22, 2016. doi: 10.5194/pb-3-9-2016. [DOI] [Google Scholar]
- Tak HJ, Kim JH, Son SM. Developmental process of the arcuate fasciculus from infancy to adolescence: a diffusion tensor imaging study. Neural Regen Res 11: 937–943, 2016. doi: 10.4103/1673-5374.184492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uylings HB, van Eden CG. Qualitative and quantitative comparison of the prefrontal cortex in rat and in primates, including humans. Prog Brain Res 85: 31–62, 1990. doi: 10.1016/S0079-6123(08)62675-8. [DOI] [PubMed] [Google Scholar]
- Wakana S, Caprihan A, Panzenboeck MM, Fallon JH, Perry M, Gollub RL, Hua K, Zhang J, Jiang H, Dubey P, Blitz A, van Zijl P, Mori S. Reproducibility of quantitative tractography methods applied to cerebral white matter. Neuroimage 36: 630–644, 2007. doi: 10.1016/j.neuroimage.2007.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J, MacLean J, Hatsopoulos NG. The marmoset as a model system for studying voluntary motor control. Dev Neurobiol 77: 273–285, 2017. doi: 10.1002/dneu.22461. [DOI] [PubMed] [Google Scholar]
- Ward JM, Buslov AM, Vallender EJ. Twinning and survivorship of captive common marmosets (Callithrix jacchus) and cotton-top tamarins (Saguinus oedipus). J Am Assoc Lab Anim Sci 53: 7–11, 2014. [PMC free article] [PubMed] [Google Scholar]
- Warner CE, Kwan WC, Wright D, Johnston LA, Egan GF, Bourne JA. Preservation of vision by the pulvinar following early-life primary visual cortex lesions. Curr Biol 25: 424–434, 2015. doi: 10.1016/j.cub.2014.12.028. [DOI] [PubMed] [Google Scholar]
- Wise SP. Forward frontal fields: phylogeny and fundamental function. Trends Neurosci 31: 599–608, 2008. doi: 10.1016/j.tins.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Nagakane Y, Yoshikawa K, Kizu O, Ito H, Kubota T, Akazawa K, Oouchi H, Matsushima S, Nakagawa M, Nishimura T. Somatotopic organization of thalamocortical projection fibers as assessed with MR tractography. Radiology 242: 840–845, 2007. doi: 10.1148/radiol.2423060297. [DOI] [PubMed] [Google Scholar]