Abstract
The mechanics of the protein-lipid interactions of transmembrane proteins are difficult to capture with conventional atomic molecular dynamics, due to the slow lateral diffusion of lipids restricting sampling to states near the initial membrane configuration. The highly mobile membrane mimetic (HMMM) model accelerates lipid dynamics by modeling the acyl tails nearest the membrane center as a fluid organic solvent while maintaining an atomic description of the lipid headgroups and short acyl tails. The HMMM has been applied to many peripheral protein systems, however the organic solvent used to date caused deformations in transmembrane proteins by intercalating into the protein and disrupting interactions between individual side chains.
We ameliorate the effect of the solvent on transmembrane protein structure through the development of two new in silico Lennard-Jones solvents. The parameters for the new solvents were determined through an extensive parameter search in order to match the bulk properties of alkanes in a highly simplified model. Using these new solvents, we substantially improve the insertion free energy profiles of ten protein side chain analogs across the entire bilayer. In addition, we reduce the intercalation of solvent into transmembrane systems, resulting in native-like transmembrane protein structures from five different topological classes within a HMMM bilayer. The parameterization of the solvents, in addition to their computed physical properties are discussed. By combining high lipid lateral diffusion with intact transmembrane proteins, we foresee the developed solvents being useful to efficiently identify membrane composition inhomogeneities and lipid binding caused by the presence of membrane proteins.
Graphical Abstract
Introduction
The biological membranes are an indispensable component of all living cells, playing host to 26% of the human proteome,1 and an estimated 60% of current drug targets.2 Membranes allow gradients to be established, and the exploitation of these gradients is the ultimate cellular energy source. The main players of cellular signaling and transport are also localized within the membrane.3–5 Rather than being a simple host substrate within which membrane-associated proteins act,6 the evolving consensus is that the composition of the local membrane environment can modulate the activity of peripheral and transmembrane proteins.7,8
Studying the interplay of membrane proteins and their environment using molecular dynamics (MD) simulations offers an opportunity to capture specific membrane-protein interactions whose timescale is too fast to probe experimentally, and has been used to great success for a number of membrane associated systems.9–16 However, over the course of a 100 ns MD trajectory, individual lipids may only exchange with their neighbors once or twice, due to a relatively low lateral lipid diffusion constant of ~ 8 × 10−8 cm2s−1 (0.8 Å2ns−1).17,18 The resulting membrane representation is effectively static, with the local environment around the protein biased strongly by its initial configuration. Therefore, sampling specific protein-membrane contacts in a conventional mixed membrane environment require long equilibration times to obtain samples independent from the starting configuration. To accelerate the sampling process, the Highly Mobile Membrane Mimetic (HMMM) representation was developed. HMMM accelerates lateral lipid diffusion and sampling by shortening lipid acyl tails and filling the membrane core with an organic solvent.16,19,20 The model demonstrably expedites membrane insertion of biomolecules,21 increases the lateral lipid diffusion constant by an order of magnitude20 and recapitulates the interaction energetics and membrane pressure profiles on the membrane periphery.22,23 The model has been previously applied to determining the bound conformation distribution and specific membrane-protein contacts for a range of peripheral membrane proteins.19,24–35
Considerable experimental evidence suggests that lipid-specific membrane-protein contacts can modulate the protein function,36–39 with particular emphasis on the lipid head-groups playing an active role in regulating protein activity.40,41 However, unless lipids are co-crystallized with the transmembrane protein42 (a rare phenomenon since the native lipids are often replaced by surfactants in membrane protein crystallization protocols43), the specific lipid binding sites remain unknown. It is inefficient for conventional lipid simulations to probe for these binding sites because of the slow lipid lateral diffusion that also plauged peripheral systems. This need forms the basis for extending the HMMM to transmembrane systems, so that lipids in heterogeneous bilayers will be able to rapidly diffuse and sample their binding sites on much shorter timescales before converting the system to a conventional bilayer representation for further study.
The HMMM was initially designed with a solvent, 1,1-dicholorethane (DCLE), that was meant to represent the entire bilayer.20,44 These model DCLE-based HMMM membranes (DCLE-HMMM) capture the free energy profiles for amino acid analog insertion into a full-length conventional membrane on the membrane interface, while the free energy profiles of DCLE-HMMM membranes deviate substantially from the behavior seen in full membranes in the membrane core.22 This difference at the membrane core is due to the existence of a dipole in DCLE, which in part allows it to be liquid at standard temperatures and pressures. Consequently, interactions between protein side-chains and the solvent were too favorable, leading to spontaneous intercalation of DCLE into transmembrane proteins when this was attempted. Thus, there is need for a new solvent to extend the applicability of the HMMM to transmembrane systems. Here, we develop another solvent that better captures the properties of the membrane interior and improves the behavior of HMMM simulations in transmembrane systems.
Designing the appropriate new solvent to mimic the membrane core is a non-trivial optimization problem, balancing the need for a high lateral lipid diffusion constant to accelerate dynamics against the accuracy in describing the properties of the membrane interior.20 Membrane interiors are most naturally described by linear alkanes or similarly hydrophobic species, however to maximize the lateral lipid diffusion the size of the solvent molecule should be minimized. Small alkanes are gaseous, which renders them unsuitable as a membrane representation, and long alkanes have substantially slower lateral lipid diffusion constants in a HMMM system.20,44 The original solution to this optimization problem was to use a small molecule with a modest dipole that remains liquid, and DCLE was chosen from selection of organic solvents.44 Moving beyond this compromise solution to improve the solvent behavior for transmembrane systems, we have developed a pair of novel solvents to address these competing objectives.
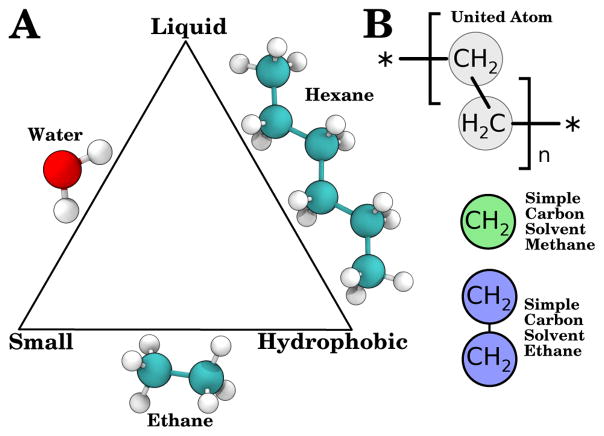
The ideal solvent to use for this application would be small to maximize diffusion, non-polar to reflect the dielectric environment of the membrane, and liquid under physiological conditions. No natural compound can simultaneously satisfy these three constraints, satisfying only two of the three criteria (Fig. 1A). However, with the considerable freedom afforded to us by working in silico, we were not limited to molecules that exist in nature. In the spirit of united-atom lipid force-fields that came before,45–48 we developed liquid small molecule solvents for use in the membrane interior (Fig. 1B). The solvents that emerged were composed of one or two Lennard-Jones particles and termed the Simple Carbon Solvent Methane (SCSM) and Simple Carbon Solvent Ethane (SCSE), designed to mimic the membrane interior by representing a single acyl chain carbon (SCSM) or two connected acyl chain carbons (SCSE). These solvents are non-polar and small by construction, and have effectively four tunable parameters to control their interaction within an MD simulation to create a liquid.
Figure 1.
Optimization triangle for membrane-mimicking solvents and the atomic mapping used here to create Simple Carbon Solvent Methane (SCSM) and Simple Carbon Solvent Ethane (SCSE). (A) Optimization triangle for highly mobile membrane mimics, which should be liquid, hydrophobic, and small. Molecules that satisfy two of the criteria are represented on the edges of the triangle. (B) Atomic mapping used for individual particles in united-atom lipid acyl chains (top, grey), compared with the SCSM (middle, green) and SCSE (bottom, blue) solvents, where each particle represents a distinct methanediyl group.
Through extensive parameter testing, we have arrived at parameters that improve the insertion free energy profile for amino acid analogs into HMMM systems. After testing these parameters with a diverse set of transmembrane HMMM systems, we conclude that when used with appropriate minimal lipid restraints, SCSM and SCSE permit stable simulation of a wide array of transmembrane proteins in a HMMM representation without significantly perturbing the protein structure. The new solvents thereby enable fast equilibration of the membrane environment around transmembrane systems, enabling the identification of lipid-specific binding sites at reduced computational cost while retaining atomic detail.
Methods
There are three steps in our solvent optimization procedure, the initial search for parameters, their validation, and finally testing with five archetypal transmembrane protein systems. The simulations were all carried out with NAMD 2.9,49 using CHARMM36 lipid50 and protein51 parameters, and the CGenFF parameters for DCLE52 where appropriate. Simulation parameters common to all simulations include the non-bonded cutoffs, with a real space cutoff of 12 Å (switching after 10 Å), the inclusion of long-range electrostatics through the use of the particle mesh Ewald (PME) method53,54 with a 1 Å grid spacing, as well as 2 fs timesteps and the requisite use of SETTLE55 to constrain hydrogen bond lengths. All simulations were performed under constant temperature (310 K unless specified otherwise) and pressure (1 atm), maintained by Langevin dynamics and Langevin piston Nosé-Hoover methods.56,57 For simulations with a bilayer, the area of the bilayer was held constant at 75 Å2 per lipid20 (compared with slightly lower ~67 Å2 per lipid for a conventional bilayer58). Maintaining a constant surface area prevents the membrane from shrinking in the event of the surfactant-like short tailed lipids entering into solution, and has been used extensively in peripheral membrane simulations.19,23–35 The lower membrane density also further increases the lateral lipid diffusion by approximately a factor of 2,20 and for this reason is the recommended approach in all HMMM simulations.20,23
To simplify the nomenclature, we introduce the abbreviations DCLE–HMMM, SCSE–HMMM, and SCSM–HMMM, which indicate the solvent used within a specific HMMM system. Conventional bilayers with full-length acyl chains may be labeled as FULL in figures where appropriate.
Parameterization
For the parameterization of SCSM, a box 1000 SCSM particles was simulated for 400 ps at 298 K using 486 different sets of randomly generated van der Waals parameters ε (which determines the strength of the interaction) and rmin (which determines the resting distance between particles). The volume (V) and isothermal compressibility (κT ) over the last 300 ps of simulation were compared to literature values59,60 for cyclohexane at 298 K of 30.5 Å3 per CH2 group and 1120 TPa−1. The isothermal compressibility was determined as follows:61
The best fitting SCSM parameter set was further refined by testing 20 small adjustments to this parameter set using 2 ns simulations and thereby determining the optimum set of parameters for SCSM bulk properties, which are specified in the final parameters set as an NBFIX term. Interactions between SCSM and other species were tuned to avoid excessive interaction with water, using the radius of a CH2 group to define rmin and an intermediate value for ε that strikes the appropriate balance between interacting too strongly with the surrounding environment at high ε and not interacting at all at low ε.
The parameterization of SCSE began from the bulk values of SCSM, and was manually optimized by a series of 400 ps simulations to a comparable volume by adjusting ε, the depth of the Lennard-Jones potential well. The parameters for the bond between the two Lennard-Jones particles were taken by analogy from the CTL2-CTL2 bond from the CHARMM36 lipid parameter set, which is the term for the bond between adjacent acyl chain carbons in saturated tails.50 Unlike in the case of SCSM, interspecies interaction van der Waals parameters involving SCSE are best described using the sum of the ε values of the constituent carbon and hydrogen atoms, and use a calculated effective radius for a methanediyl group as rmin.
Free Energy Profile and Solvation Free Energy
The membrane insertion free energy profile for 10 amino acid side chain analogs were calculated for both SCSM and SCSE membranes, following the methodology of previous work for conventional membranes62 and DCLE–HMMM membranes,22 and are briefly summarized here. The 10 side chain analogs were chosen from a representative sample of amino acids, with aliphatic residues (Ala and Ile), aromatic residues (Trp, Phe, and Tyr), polar residues (Asn, Cys, and Ser) and charged residues (Asp, and Arg). The analogs were prepared by replacing the α-carbon with a hydrogen, assigning the hydrogen the charge of an aliphatic hydrogen, and transferring the remaining residual α-carbon charge to the β-carbon, consistent with previous atomistic amino acid membrane transfer studies.22,62 To determine the free energy profile, a pair of side chain analogs was placed in a phosphatidylcholine HMMM system with 36 lipids per leaflet and separated by 32.5 Å along the membrane normal. Umbrella sampling63 simulations were performed using 36 windows each separated by 1 Å and simulated for 10.5 ns. During simulation, the positions of the carboxy carbon of each lipid tail were constrained along the membrane normal to limit membrane fluctuations along z to ±3.5 Å, as was done for previous calculations with HMMM membranes.22 The sampling from the last 10 ns of each window was used to determine the symmetric insertion free energy profile using the weighted histogram analysis method (WHAM)64 as implemented by g wham.65
The solvation free energy for SCSM, SCSE, and DCLE, as well as the solvation free energy of water into HMMM solvents were computed using alchemical free energy perturbation.66,67 Each solvation free energy was computed by taking a single molecule of the solute of interest and slowly growing it in a solvent cube of side-length 30 Å. Progressive growth and annihilation of the solute molecule was completed in 20 equal steps. The solute particles made no electrostatic contributions to system forces for the 11 steps nearest to annihilation (alchElecLambdaStart was set to 0.5), and used soft core potentials (as implemented by alchVdwShiftCoeff being set to 5 in the NAMD configuration file) to avoid end-point catastrophes.68,69 Each intermediate state was simulated for 500 ps, with the final 400 ps of sampling being used to calculate the solvation free energy. The calculation and convergence analysis was carried out by the ParseFEP plugin of VMD.70,71
Test Applications
We carried out short, 10 ns simulations on both conventional and HMMM bilayers without protein present to assess the lateral lipid diffusion constant of each membrane type. Additionally, we simulated five representative transmembrane proteins inserted into conventional, DCLE–HMMM, SCSM–HMMM and SCSE–HMMM bilayers, with the initial membrane positioning taken from the Orientations of Proteins in Membranes database.72 The five proteins were chosen for their distinct membrane topologies, representative of different classes of transmembrane proteins. Glycophorin A (PDB: 1AFO73) was chosen as a model small transmembrane α-helix, the heterotrimeric assembly of DAP12-NKG2C (PDB: 2L3574) was chosen to represent a more complex set of small transmembrane α-helices, the potassium channel KcsA (PDB: 1R3J75) was the test monomeric helical transmembrane protein, Aquaporin I (PDB: 1H6I76) was our example of a multimeric helical transmembrane protein, and the mitochondrial ion channel VDAC-1 (PDB: 2K4T77) was our example of a β-barrel. Two simulations were conducted with HMMM systems, one with and one without a 1 kcal/mol/Å2 restraint potential applied to the z-position of the carboxy carbon of each lipid tail, as was performed for the side chain analog insertion profile simulation. Each protein-membrane combination was simulated for 5 ns under exponentially decaying constraints on the protein backbone, followed by at least 20 ns of equilibrium molecular dynamics. These short timescales are typical for previous HMMM insertion simulations, where the fast lipid diffusion of the HMMM is used initially to quickly equilibrate the membrane, then use already developed tools23,25 to start longer simulations in a conventional membrane from a nearly-equilibrated starting point, as was done for membrane binding studies of human cytochrome P450.24,78
Results and Discussion
Solvent Parameterization and Bulk Properties
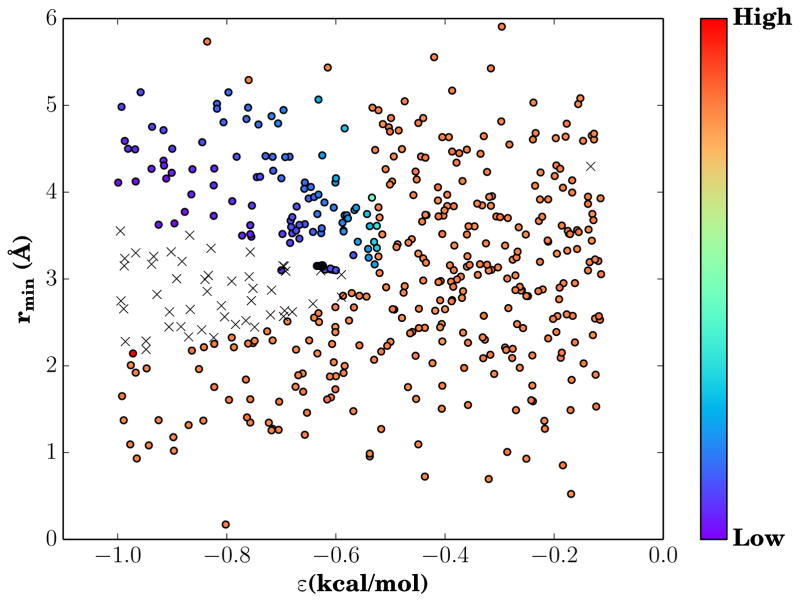
Searching parameter space for a suitable pair of vdW parameters to replicate a membrane interior is subject to the choice of target observables and the target values for those observables. During optimization, the volume per molecule and bulk compressibility of the solvent were the observables chosen to be matched to experimental values for cyclohexane. Volume and compressibility were chosen as the target observables due to their ease of measurement computationally, and their transferability to a simple system of Lennard-Jones particles. Since each Lennard-Jones particle is meant to replace a methanediyl group, choosing a model compound made entirely out of methanediyl groups such as cyclohexane is the logical choice. By testing many different van der Waals parameter values, we construct an effective phase diagram, showing both the accessible values for volume and compressibility (Fig. S1), and the dependence of compressibility on ε and rmin (Fig. 2). From the two distinct populations of points in Figure S1, we observe that through our random sampling of potential van der Waals parameters, we have sampled across a sufficient range to observe liquid-like behavior of low volume and compressibility, as well as gaseous behavior where our Lennard-Jones particles are highly compressible with a high volume per molecule. The gaseous form of SCSM predominates, until ε increases to such a degree that SCSM forms stable interactions. This phase-like behavior is demonstrated by the clear demarcations of low compressibility and high compressibility regions of parameter space in Figure 2, as well as a region that was sampled but did not produce stable simulation conditions. The value for ε required for liquidity is significantly higher than typical ε values seen in conventional force fields by approximately an order of magnitude, and is the fundamental change needed to yield liquid behavior at typical biological temperatures. The final values for the van der Waals parameters for SCSM that recover the bulk properties of cyclohexane are shown in Table 1, and are implemented as an NBFIX term that are specific to SCSM-SCSM contacts.
Figure 2.
Compressibility phase diagram. The color of each point indicates the compressibility for the corresponding ε and rmin pair, with blues being less compressible than red. The sharp color gradient between highly compressible and effectively incompressible states marks the phase transition between gaseous and liquid solvents. The black crosses represent ε and rmin pairs where simulations crashed due to instability prior to completion, and may represent a metastable state.
Table 1.
Optimized parameter table for SCSM and SCSE.
| Interspecies | NBFIX | |||
|---|---|---|---|---|
|
| ||||
| ε (kcal/mol) | (Å) | ε (kcal/mol) | rmin (Å) | |
| SCSM | −0.350 | 2.08 | −0.625 | 3.15 |
| SCSE | −0.112 | 2.08 | −0.274 | 3.15 |
Optimization of contacts between SCSM and other molecular species was treated separately, and contacts were tuned to improve the interface between water and SCSM using standard lipid parameters as the guide. Thus the default is the effective radius for a methanediyl group (2.08 Å), and the first ε tried was the sum of ε from methanediyl constituents (0.112 kcal/mol). With high values for ε, SCSM and water interact too strongly, and SCSM no longer acts hydrophobic enough to be a good membrane mimic. Conversely, when ε is small, as it is in the initial trial, the self-interactions between the two species are too strong, and an interfacial gap forms between SCSM and water. The balanced value chosen was determined to improve the fit between the amino acid analog insertion energy profiles for SCSM-HMMM and conventional membrane representations.
Parameterization of SCSE began from the final bulk parameter values for SCSM, and iterated until the volume per carbon was correct. The final parameters for SCSE are tabulated in Table 1, and are somewhat altered relative to SCSM, despite sharing the same split between self-interaction parameters (given as an NBFIX term) and a set of parameters for interactions with other species as in SCSM. Due to the increased mass of SCSE relative to SCSM, and the self-ordering brought about by pairing particles together, the ε values required to maintain liquidity are lower than those for SCSM. As a consequence, the naive summation of ε for methanediyl constituents was the appropriate balance to generate realistic interfaces between SCSE and other species. By choice, the rmin values are shared with SCSM, and were not further optimized to match the compressibility of cyclohexane.
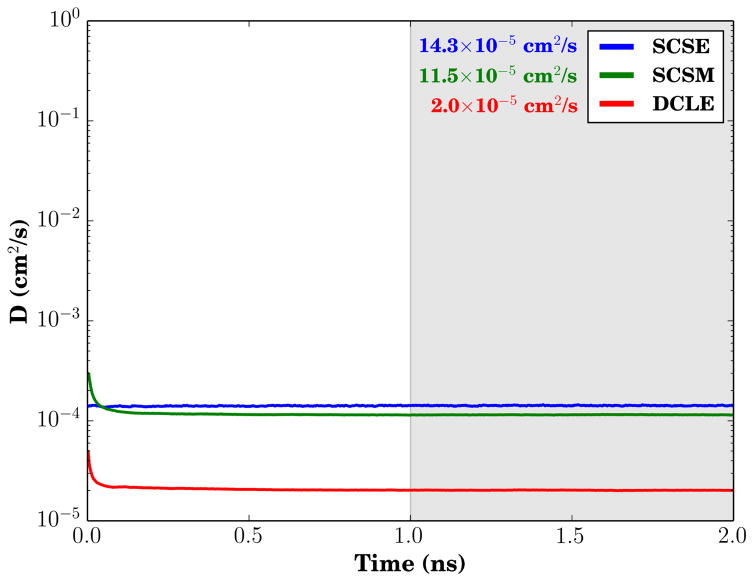
As a result of the parameterization, the bulk properties of SCSM and SCSE are substantial improvements over DCLE for the purposes of facilitating membrane simulations. The summary table of key bulk properties (Table 2) compares the solvents. From a volumetric perspective, the bulky chloride atoms of DCLE increase the average volume per carbon too much, making it difficult to map existing solvent carbons to carbons that would exist in a conventional bilayer, as evidenced by the bilayer shrinking slightly when such a substitution is made.25 Both the volumes of SCSM and SCSE particles per represented CH2 group are comparable to experimentally determined values for alkanes, and thus one can utilize them to regrow the short-tailed HMMM lipids to their full length by using the particle positions of SCSM or SCSE particles as the new position for acyl tail carbons. The bulk compressibility modulus κ of DCLE and SCSM are comparable to the compressibility of alkanes, while SCSE has a compressibility that is substantially higher. We believe that the higher compressibility is actually a useful feature rather than a flaw, as it appears to play a role in making SCSE a better mimic of the membrane interior. One area where this becomes apparent is in comparing the bulk diffusion constants for the solvents (Fig. 3), where the high compressibility of SCSE manifests itself by eliminating transiently high diffusion constant measurements on the picosecond timescale. SCSE also has the highest bulk diffusion constant among the solvents, which increases the lateral lipid diffusion constant for short-tailed lipids on its surface.
Table 2.
Bulk properties comparison between HMMM solvents and cyclohexane.
Figure 3.
Bulk solvent diffusion constant comparison. The mean diffusion constant value over the last 1 ns of trajectory (shaded gray) is provided alongside the infigure legend. The diffusion constant value was calculated using Einstein’s relation for three-dimensional diffusion equation: .
The solvation free energies for SCSM and SCSE are more membrane-like than those of DCLE. Table 3 shows that it is favorable to add water to DCLE, whereas it is energetically unfavorable to add water to SCSM or SCSE. Similarly, DCLE will favorably enter into water, whereas the novel solvents will not. Indeed, the novel solvents appear to follow an established principle where the alkyl chain length linearlly scales with the solvation free energy.80 This diminished hydrophobicity for DCLE relative to the new solvents is a result of favorable interactions between the chlorine atoms of DCLE and water, a structural feature not present in SCSM or SCSE.
Table 3.
Solvation free energy comparison for various HMMM solvents. Solvation free energies are the difference in free energy between a compound in a vacuum and the same compound in solution.79 This is computed in three steps, 1. an annihilation free energy for the compound in vacuum, which is only nonzero for DCLE due to its internal nonbonded interactions, 2. a transition for an annihilated compound from vacuum to solution, which is always zero, and 3. a creation free energy for the compound in solution, with the sum for all three steps reported here.
| ΔG Water as Solute (kcal/mol) | ΔG Water as Solvent (kcal/mol) | |
|---|---|---|
| DCLE | −1.74 ± 0.03 | −0.30 ± 0.04 |
| SCSM | 2.12 ± 0.04 | 1.68 ± 0.03 |
| SCSE | 0.28 ± 0.02 | 3.07 ± 0.03 |
Amino Acid Analog Insertion
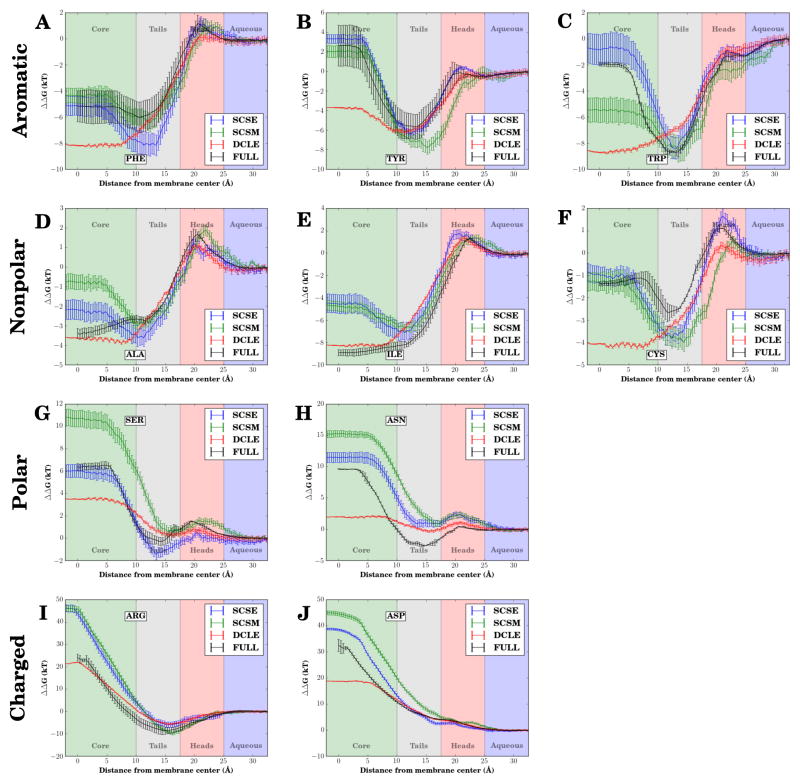
The motivation for developing new solvents such as SCSM and SCSE was to improve the behavior of transmembrane proteins within the HMMM model. While DCLE–HMMM membranes do an adequate job of reproducing insertion free energy profiles on the membrane periphery, the free energy profiles for these membranes did not capture a common theme of profiles from conventional full membranes where there is an “optimum” insertion depth, below which further insertion is energetically less favorable.22,62 Instead, the profile near the membrane center for DCLE–HMMM membranes is frequently an extremum of a profile that is monotonic within the membrane interior. Through the use of designer solvents, these additional features of the free energy landscape are recovered. Each class of amino acids will be presented in turn.
Aromatic Amino Acids
We calculated the insertion free energy profile for three aromatic amino acids, phenylalanine, tryptohphan, and tyrosine. These amino acids are especially important for transmembrane protein localization, as aromatic amino acids, particularly tryptophan, are noted to be enriched in lipid-exposed locations near the membrane interface.81,82
The free energy profile comparison for phenylalanine (Fig. 4A) is first example of the marked improvement of the insertion free energy profiles of the in silico solvents relative to DCLE–HMMM membranes in recapturing the behavior seen in simulations of conventional membranes. The energies for all systems are similar on the membrane periphery and in solution, with substantial differences between representations developing within 15 Å of the membrane center. Near the membrane center, the insertion free energy for DCLE–HMMM membranes continues to decrease, eventually underestimating the free energy by approximately 3 kT. In contrast, the profiles for both SCSE- and SCSM–HMMM membranes are within a kT of the value for a conventional membrane. The position of the minimum free energy, and thus the most favored location for a phenylalanine side-chain within the membrane, are within 1 Å of one another. The value of the minimum free energy is underestimated by approximately 2 kT in the case of the SCSE–HMMM bilayer, however overall both novel solvents improve upon DCLE–HMMM result for deeply inserted phenylalanine side chains.
Figure 4.
Amino acid-analog insertion free energy profiles along the membrane normal, comparing the new solvents (blue for SCSE-HMMM and green for SCSM-HMMM respectively) to previously published results for DCLE-HMMM22 (red) and conventional bilayers (black).62 The amino acid analogs compared are (A) Phe, (B) Tyr, (C) Trp, (D) Ala, (E) Ile, (F) Cys, (G) Ser, (H) Asn, (I) Arg, and (J) Asp. Membrane regions are demarcated by different background colors. The hydrophobic solvent membrane core is green, the short lipid tails are gray, the headgroups are red, and aqueous solution has a blue background.
The free energy profiles for tyrosine (Fig. 4B) and tryptophan (Fig. 4C) show similar improvements in the insertion free energy profile of the novel solvents relative to DCLE. In both cases, the profiles for the novel solvent membranes have a minimum, indicating a preferred insertion depth for tyrosine and tryptophan side chains, a feature lacking in DCLE–HMMM membranes for tryptophan. The positions of the minima relative to the membrane center for conventional bilayers agree very well with the position of the minima for SCSE–HMMM membranes. However the minimum in SCSM–HMMM membranes in the case of tyrosine is shifted, leading to the conclusion that the use of SCSE as the solvent within the HMMM representation best recovers the behavior of a conventional bilayer for the aromatic amino acid side chain analogs tested.
Non-polar Amino Acids
Another common class of amino acids seen in the membrane interior are the non-polar amino acids, which in our tests are represented by alanine and isoleucine. The profiles in a conventional membrane for these amino acids are simpler than those of the aromatic amino acids, in that the free energy profile monotonically decreases as one approaches the membrane center, implying that the optimal location for these side chains is in the middle of the membrane, rather than in a belt near the membrane interface. The profiles for our designer solvents in the case of both alanine (Fig. 4D) and isoleucine (Fig. 4E) do not show this trend, instead suggesting that there is a most favorable interaction depth. The overall variation at the membrane center is smaller for alanine, where the free energy of insertion at the membrane center is within 2 kT of the conventional membrane value for SCSE–HMMM membranes, and within 3 kT of the conventional membrane value for SCSM–HMMM membranes. The larger isoleucine has a larger deviation, where both novel solvents overestimate the free energy by approximately 5 kT at the membrane center.
The cause for these discrepancies is likely due to the relative strength of the non-bonded interactions between solvent particles and those between solvent particles and other molecules. Due to the large ε values needed to keep the novel solvents liquid, and the epsilon-mixing rules of CHARMM-compatible force fields, there is an energetic penalty to be paid for inserting any foreign substance into these novel solvents. As a foreign substance is inserted into the novel solvents, highly-favorable non-bonded interactions between solvent particles are replaced by less favorable non-bonded interactions. Due to this built-in energetic penalty, mixing of solvent and protein is diminished, as will be discussed later in greater detail, and the resulting reduced solvent intercalation should be considered as a useful feature of the model. Thus while using DCLE as a solvent for this class of amino acids best captures the behavior of a conventional bilayer, we note that the novel solvents do an adequate job of describing the energetics.
Amino Acids Containing Sulfur
The remaining typical group of amino acids to be discussed is that of sulfur-containing amino acids. The profiles for cysteine were compared across the three tested HMMM solvents as well as for the conventional bilayer (Fig. 4F). Echoing behavior seen in the aromatic amino acids, both SCSE–HMMM and SCSM–HMMM were able to capture the overall partitioning between solution and the membrane center, arriving at a value within a kT while also capturing the presence of a membrane depth that is especially favorable to cysteine. Membranes containing SCSE were found to better reproduce the membrane-entry barrier.
In general, using SCSE instead of DCLE yields the most improvement on the side chain insertion free energy profiles in the membrane interior, while retaining the good agreement with conventional bilayers on the membrane periphery and in solution. Utilizing the novel solvents, we capture the shape of the side chain insertion free energy profiles for a majority of the amino acids tested. By capturing the profile lineshapes, our improved HMMM model reproduces favored insertion depths for particular side chains, suggesting that these models are suitable for use in studies with larger transmembrane assemblies when a high lateral lipid diffusion constant is required.
Polar Amino Acids
While polar amino acids are rare within the membrane interior, previously calculated energetics for DCLE–HMMM membranes were substantially less unfavorable than was to be expected from prior studies of conventional bilayers.22,62 Using serine (Fig. 4G) and as-paragine (Fig. 4H) as representative amino acids from the group, we see that SCSE–HMMM membranes best reproduce the shape and values of the free energy insertion profile of a conventional full bilayer. Serine especially shows remarkable agreement between the conventional and SCSE profiles, which may reflect the relative importance of the dipole of the hydroxyl group in the insertion of the amino acid side chain analog (methanol). The dipole present within DCLE stabilizes the insertion to the membrane center, resulting in the DCLE–HMMM representation suggesting that polar residues are more favorable in the middle of membranes than they actually are.
No HMMM solvent is able to exactly capture the stability of asparagine on the membrane periphery. However the shape of the insertion profile in solution for SCSE and SCSM are different, in that they are above zero well into solution. Since the reference point for comparing the profiles was arbitrarily chosen to be in solution, and since free energy profiles calculated by WHAM are cumulative, we can choose a different reference point to compare profiles. Using the location of the local maxima as the side chain enters the membrane as our reference, we see that the profile for a SCSE–HMMM membrane tracks well with a conventional bilayer, maintaining a constant separation within 20 Å of the membrane center. Thus SCSE is the the prefered solvent for describing the side-chain energetics of polar amino acids within the HMMM bilayer.
Charged Amino Acids
Amino acids carrying a naked charge are not anticipated to ever be present within a typical bilayer without forming a water defect.83,84 Spontaneous formation of water defects have been observed previously for both charged residues studied, arginine and aspartate,22 and are observed again in SCSM–HMMM and SCSE–HMMM membranes (Fig. S2). The insertion free energy profile for arginine (Fig. 4I) are quite accurate for DCLE–HMMM membranes overall. However SCSE–HMMM membranes also perform well, replicating behavior in the membrane interior by matching the slope of the profile for a conventional membrane, with the difference in the membrane interior a result of discrepancies at the membrane-water interface. Membrane water defects were found to be formed through all simulations involving arginine.
The free energy profile comparison for aspartate (Fig. 4J) is more accurate for SCSE–HMMM. The profile for inserting aspartate remains steady at approximately 18 kT near the membrane core for DCLE–HMMM systems, whereas the free energy profile for the novel solvents increases as the membrane center is approached, a behavior consistent with conventional membranes. Water defects were less prevalent for aspartate, only occurring when the side chain analog was constrained to be near the membrane-water interface. The lack of water defects in the case of aspartate highlights the role of the HMMM organic solvent in stabilizing charged residues relative to a conventional membrane. While HMMM-models with SCSM and SCSE correctly predict an increasing energetic penalty for charged side chains in the membrane, DCLE–HMMM models feature favorable interactions between the dipole inherent to DCLE and the charge of the aspartate.
SCSM and SCSE in Membrane and Membrane-Protein Systems
The improvement of the insertion free energy profiles of side chains into SCSE- and SCSM–HMMM membranes suggests that these membranes are superior to DCLE for simulating transmembrane systems, but we must establish the suitability of novel solvent HMMM membranes for simulation of transmembrane proteins. We have tested our models with five different transmembrane proteins, each representative of a particular class of membrane proteins. In addition, simulations of pure bilayer systems were also conducted.
SCSM and SCSE Accelerate Lipid and Protein Sampling
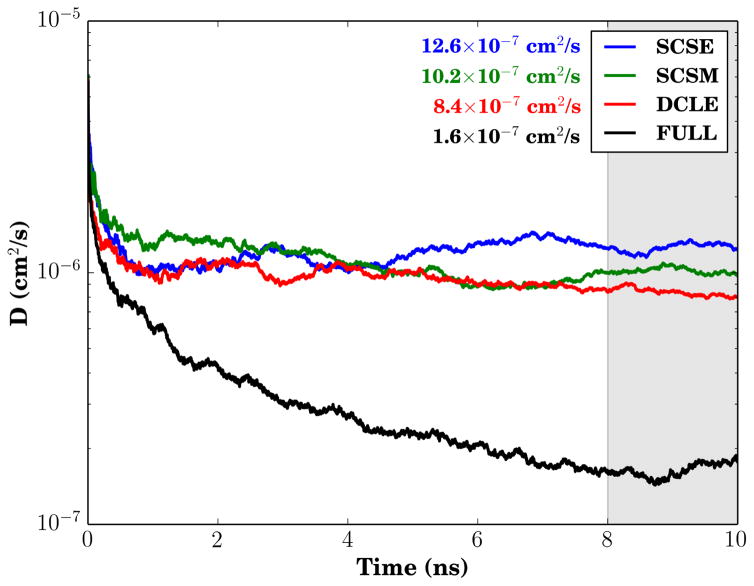
One of the notable features of the HMMM representation is its high lateral lipid diffusion constant. As was already suggested by the higher solvent bulk diffusion constants of SCSE and SCSM shown in Fig. 3, the progression of the lateral lipid diffusion (Fig. 5) shows that the lipid lateral diffusion accelerates by up to 50% for the new solvents relative to DCLE, outside the individual uncertainties of ±1.5 × 10−7cm2/s. The working hypothesis for the origin of the lateral diffusion difference depends on “flows” of the solvent in HMMM systems acting on the lipid tails to move the entire short tailed-lipid floating on the surface. Thus the increased bulk diffusion brought about by the reduced attractive forces between molecules acts also to increase the lateral lipid diffusion relative to the baseline acceleration inherent to HMMM representations.
Figure 5.
Lipid lateral diffusion constant comparison. The mean diffusion constant value over the last 2 ns of trajectory (shaded gray) is provided alongside the infigure legend. The diffusion constant value was calculated using Einstein’s relation for two-dimensional diffusion equation: .
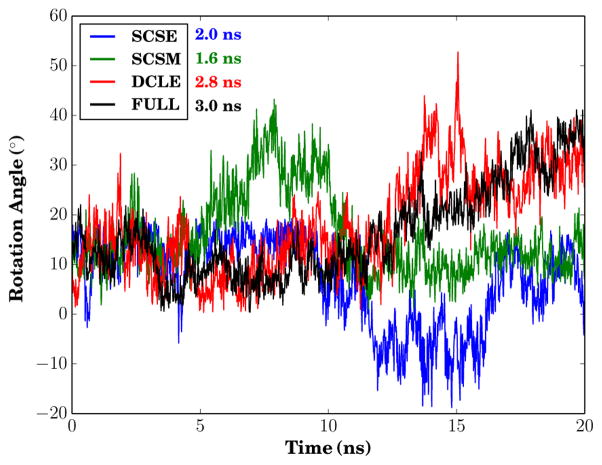
Not only do the lipids sample more rapidly in the HMMM representation, but also certain aspects of protein motion are accelerated, and this acceleration is greater for the new solvents. In particular, we measured differences in protein rotation rate within the different membrane representations. Figure 6 measures the rotation of the glycophorin A dimer over time, and includes an estimated autocorrelation time. The range of observed rotation over the 20 ns simulations are approximately similar, however the integrated autocorrelation times differ substantially, with a factor of two separating the fastest diffusing case (SCSM) from the slowest (conventional bilayer). Stated another way, the effective number independent samples of the dimer rotation vary between the chosen membrane representation. There is a minimal difference between DCLE and a conventional bilayer, and this difference increases by moving to simpler SCSE and SCSM solvents.
Figure 6.
Rotation trace of glycophorin relative to an arbitrary orientation in the four membranes. The integrated autocorrelation times for the rotation angle are displayed beside the infigure legend.
Impact of Lipid Z Constraints
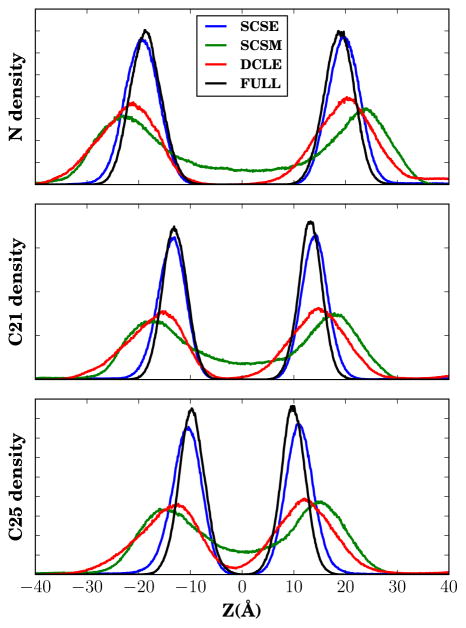
During initial development of the HMMM, it was noted that the membrane density profiles of membrane substituents were broader than those seen in simulations with a conventional membrane.19 This suggested that the short tails were not sufficiently attracted to the solvent base, and as a result, individual short-tailed lipids would diffuse into solution. However, comparing the membrane density profiles for different HMMM solvents (Fig. 7) indicates that the membrane density profiles are dependent on solvent, with SCSE–HMMM membranes displaying nearly-native membrane density profiles in the absence of constraints, while SCSM–HMMM membranes show the wide distribution seen in DCLE–HMMM membranes. Thus while it is seen as essential to apply vertical constraints to correct the membrane density profiles for DCLE- and SCSM–HMMM membranes,20 SCSE–HMMM membranes need no such correction. The success of SCSE relative to SCSM is unexpected, given that the lipid tails have stronger interactions with SCSM, and should in principle maintain the planarity of the bilayer. Measuring the hydrogen order parameter (SCH ) for each carbon in the lipid tails shows that the interaction between SCSM and the tails may be too strong, as the uniformly low order present in SCSM membranes in Figure S3 suggests that the lipid tails lie flatter against the lipid-SCSM interface to increase the number of lipid-SCSM contacts.
Figure 7.
Membrane density profiles for lipid components without constraints. From top to bottom, the densities correspond to the choline nitrogen, the first carbon in one of the acyl tails (C21 in CHARMM nomenclature), and the last carbon in one of the acyl tails (C25).
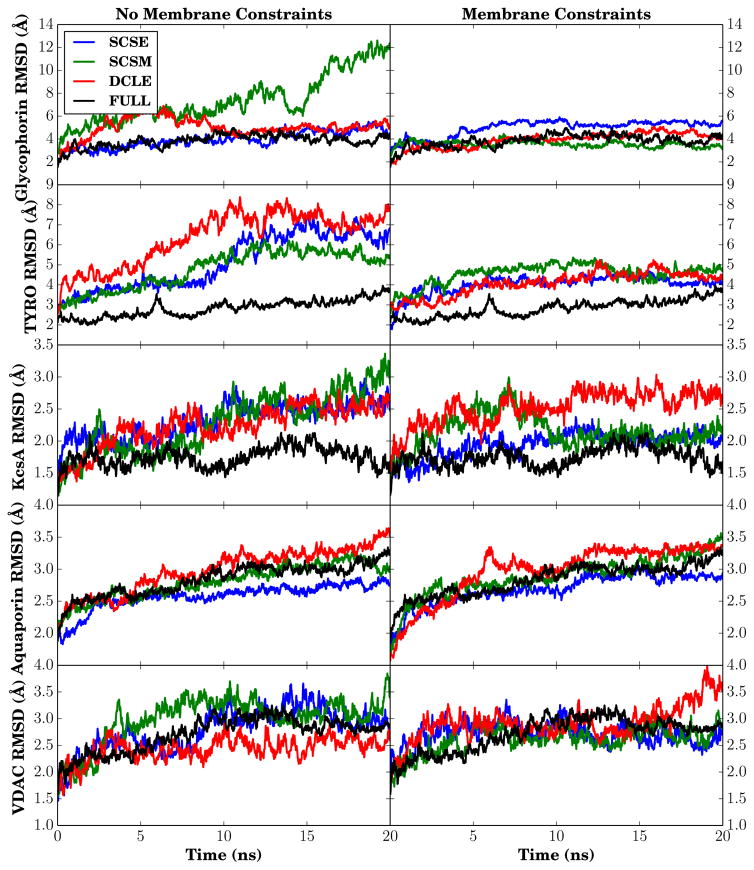
The acid test for suitability is of course to determine whether there are major conformational changes that take place as a result of insertion of membrane proteins into these novel solvents. The simplest measure of conformational change is to monitor the RMSD of membrane-embedded backbone segments for each of the five membrane proteins tested for each HMMM solvent model with and without lipid constraints, and compare against simulations conducted when the protein is placed in a full membrane (Fig. 8). Overall, the change in RMSD for proteins in HMMM membranes over the measured timescale is often within 1.0 Å of the value for a full membrane, and on occasion shows smaller deviations from the structure than was seen in the full membrane case. The case of an unconstrained SCSM–HMMM membrane resulted in the only truly large change in RMSD, and was caused by the separation of the dimer present in the PDB structure. The application of lipid constraints has the largest impact on the smallest systems, with markedly reduced RMSD in both glycophorin A, where the dimers stay together with constraints, and TYRO, where vertical lipid constraints tend to keep the constituent helices straighter as they were in the PDB structure.
Figure 8.
Backbone RMSD of transmembrane segments for five membrane-embedded proteins. On the left are the RMSD traces for unconstrained HMMM lipids, while on the right the HMMM lipids have been constrained as described in methods. Residues were considered to be membrane embedded if the residue was part of a helix or sheet in the original PDB structure, as determined by STRIDE.85
In larger systems, the success of constraints in recovering native-like structural changes is mixed. In KcsA, conventional membranes always show the smallest change from the crystal structure; however, with constraints SCSM and SCSE are within 0.5 Å of the conventional membrane result, while DCLE shows no improvement over the unconstrained scenario. For the simulations carried out with Aquaporin, HMMM systems with SCSE show the smallest deviations from the crystallographic structure, and there is no obvious difference between conventional membranes and SCSM and DCLE. The β-barrel VDAC is an odd case altogether, and provides the clearest warning against over-interpretation of the resulting RMSD, which appear approximately uniform across conditions. Despite this uniformity, without lipid constraints, SCSE–HMMM membranes are not stable near VDAC. Instead, as evidenced by the snapshots in Figure 9, water forms a pore in the membrane. In these simulations, E73 is deprotonated, attracting water from solution and forming a water defect that expands in the SCSE membrane. Thinning of the membrane around VDAC with a deprotonated E73 has been previously reported,86 however membrane rupture was not expected in the absence of constraints.
Figure 9.
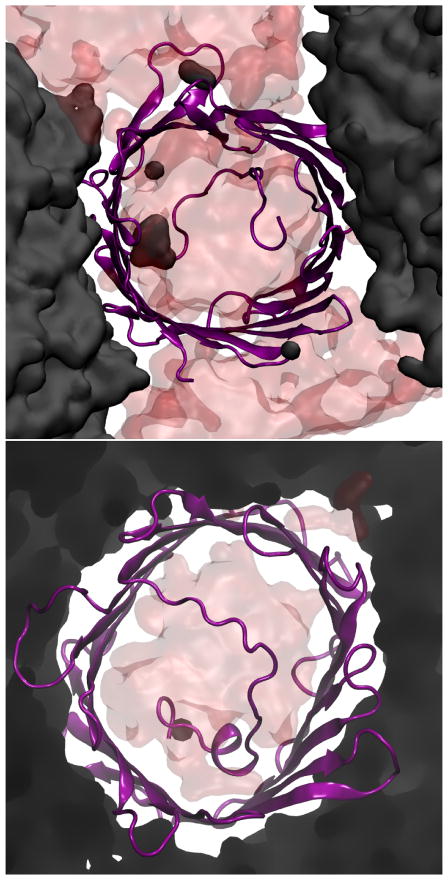
Snapshot of VDAC in SCSE after 20 ns for both unconstrained (top) and constrained (bottom) HMMM lipids. The protein is shown in purple, water in red, and SCSE in black.
Reducing Solvent Intercalation
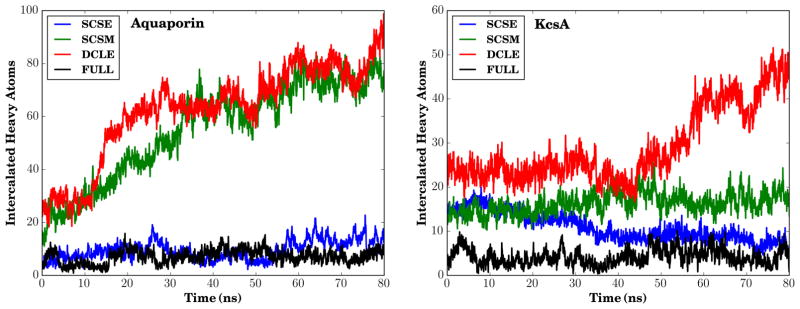
Solvent intercalation, where single molecules of organic solvent of the HMMM representation persisted for long periods of time within the interior of large transmembrane proteins, was one of the primary difficulties in past attempts in using the HMMM representation to model transmembrane systems. Due to the increased favorability for the novel solvents to interact with themselves through their high ε values, we suspected that the new solvents would preferentially interact with one another, and be less likely than DCLE to intercalate into transmembrane proteins. By counting carbons that fall within a surface drawn over the extent of KcsA and Aquaporin in our simulations (the details are given in the Supporting Information, however the method relies on the concepts behind the Quicksurf representation87), we quantitatively assess this hypothesis. For both KcsA and Aquaporin in constrained lipid membranes (Fig. 10), the number of intercalated carbons decreases substantially when SCSE is used rather than DCLE. The large reduction in the number of intercalated carbons agrees well with overall improvements in protein behavior when comparing simulations carried out with SCSE as the solvent relative to DCLE, and is the strongest indicator of the suitability of SCSE–HMMM for use when simulating transmembrane proteins.
Figure 10.
Measured intercalation of solvent into membrane constrained simulations of Aquaporin (left) and KcsA (right). More information on the definition of intercalation used can be found in the Supporting Information.
Conclusion
While it must be stated that the HMMM could never fully replace conventional membrane simulations, both of the new solvents developed here represent a substantial accuracy improvement over DCLE for quickly equilibrating mixed membrane systems around a protein of interest. Viewed from an energetics perspective, HMMM systems containing SCSM or SCSE more accurately reflect the amino acid insertion profiles of a conventional bilayer relative to DCLE. Furthermore, the new solvents intercalate less into transmembrane proteins, resulting in substantially less structural perturbation. Protein structural changes are further reduced when a weak harmonic potential is used to constrain the position along the membrane normal of the short-tailed phospholipids of the HMMM representation, however these constraints are not required for some systems, particularly when using SCSE as the solvent.
When choosing between these two in silico solvents for use in HMMM simulations of transmembrane proteins, we recommend starting with SCSE. In general, the insertion free energy profiles for SCSE agreed the best with those for a conventional bilayer, suggesting that the two-particle solvent strikes the best overall balance between the competing entropic and enthalpic free energy contributions. This was reflected by the near-native density profiles for HMMM lipid components in the absence of constraints, as well as by the further reduced intercalation observed for SCSE into large transmembrane proteins relative to SCSM.
By expanding the HMMM model to transmembrane systems, we are opening new avenues of investigation into delineating the mechanisms by which lipids modulate protein activity.36–39 Through these new solvent models, it is now possible to search for specific lipid or cholesterol binding sites which may be missed by crystallography with an unbiased and non-perturbative procedure by adding the species of interest to a transmembrane HMMM system and allowing lipid diffusion to locally enrich the concentration of specific components near the protein. The all atom nature of the HMMM then enables the straightforward conversion to a conventional full tail bilayer once the binding site is identified. Alternatively stated, the new solvents ability to leave embedded proteins unperturbed combined with the fluidity of the HMMM enables the collection of statistically meaningful sampling in mixed bi-layers at modest computational cost without a coarse grained representation masking specific atomic interactions.
Supplementary Material
Acknowledgments
The research presented here was supported by the grants from NIH to E.T. (R01–GM086749, R01–GM067887, U54–GM087519, and P41–RR05969), and the DOE CSGF Fellowship to J.V.V. (DE-FG02-97ER25308) This research used resources of the National Energy Research Scientific Computing Center (NERSC), which is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Footnotes
Supporting Information Available
Disscussion of the algorithm to calculate intercalated proteins is included in the supplement, as well as CHARMM-compatible topology and parameter files for SCSE and SCSM and the aligned free energy profiles. This material is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Fagerberg L, Jonasson K, von Heijne G, Uhlén M, Berglund L. Prediction of the Human Membrane Proteome. Proteomics. 2010;10:1141–1149. doi: 10.1002/pmic.200900258. [DOI] [PubMed] [Google Scholar]
- 2.Yildirim MA, Goh KI, Cusick ME, Barabasi AL, Vidal M. Drug–Target Network. Nat Biotechnol. 2007;25:1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- 3.Whited A, Johs A. The Interactions of Peripheral Membrane Proteins with Biological Membranes. Chem Phys of Lipids. 2015;192:51–59. doi: 10.1016/j.chemphyslip.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 4.Kholodenko BN, Hancock JF, Kolch W. Signalling Ballet in Space and Time. Nat Rev Mol Cell Biol. 2010;11:414–426. doi: 10.1038/nrm2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laganowsky A, Reading E, Allison TM, Ulmschneider MB, Degiacomi MT, Baldwin AJ, Robinson CV. Membrane Proteins Bind Lipids Selectively to Modulate Their Structure and Function. Nature. 2014;510:172–175. doi: 10.1038/nature13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singer S, Nicolson G. The Fluid Mosaic Model of the Structure of Cell Membranes. Science. 1972;173:720–731. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- 7.van Meer G, Voelker DR, Feigenson GW. Membrane Lipids: Where They Are and How They Behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vereb G, Szollosi J, Matko J, Nagy P, Farkas T, Vigh L, Matyus L, Waldmann TA, Damjanovich S. Dynamic, Yet Structured: The Cell Membrane Three Decades After the Singer-Nicolson Model. Proc Natl Acad Sci USA. 2003;100:8053–8058. doi: 10.1073/pnas.1332550100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kopeć W, Telenius J, Khandelia H. Molecular Dynamics Simulations of the Interactions of Medicinal Plant Extracts and Drugs with Lipid Bilayer Membranes. FEBS J. 2013;280:2785–2805. doi: 10.1111/febs.12286. [DOI] [PubMed] [Google Scholar]
- 10.Lindahl E, Sansom MSP. Membrane Proteins: Molecular Dynamics Simulations. Curr Opin Struct Biol. 2008;18:425–431. doi: 10.1016/j.sbi.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 11.Khalid S, Bond PJ. Multiscale Molecular Dynamics Simulations of Membrane Proteins. Methods Mol Biol. 2013;924:635–657. doi: 10.1007/978-1-62703-017-5_25. [DOI] [PubMed] [Google Scholar]
- 12.Braun AR, Lacy MM, Ducas VC, Rhoades E, Sachs JN. α-Synuclein-Induced Membrane Remodeling Is Driven by Binding Affinity, Partition Depth, and Interleaflet Order Asymmetry. J Am Chem Soc. 2014;136:9962–72. doi: 10.1021/ja5016958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai CL, Landgraf KE, Voth GA, Falke JJ. Membrane Docking Geometry and Target Lipid Stoichiometry of Membrane-Bound PKC C2 Domain: A Combined Molecular Dynamics and Experimental Study. J Mol Biol. 2010;402:301–310. doi: 10.1016/j.jmb.2010.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bucher D, Hsu YH, Mouchlis VD, Dennis EA, McCammon JA. Insertion of the Ca2+-Independent Phospholipase A2 into a Phospholipid Bilayer Via Coarse-Grained and Atomistic Molecular Dynamics Simulations. PLoS Comput Biol. 2013;9:e1003156. doi: 10.1371/journal.pcbi.1003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohkubo YZ, Tajkhorshid E. Distinct Structural and Adhesive Roles of Ca2+ in Membrane Binding of Blood Coagulation Factors. Structure. 2008;16:72–81. doi: 10.1016/j.str.2007.10.021. [DOI] [PubMed] [Google Scholar]
- 16.Baylon JL, Vermaas JV, Muller MP, Arcario MJ, Pogorelov TV, Tajkhorshid E. Atomic-Level Description of Protein-Lipid Interactions Using an Accelerated Membrane Model. Biochim Biophys Acta Biomembr. 2016;1858:1573–1583. doi: 10.1016/j.bbamem.2016.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klauda JB, Brooks BR, Pastor RW. Dynamical Motions of Lipids and a Finite Size Effect in Simulations of Bilayers. J Chem Phys. 2006;125:144710. doi: 10.1063/1.2354486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wohlert J, Edholm O. Dynamics in Atomistic Simulations of Phospholipid Membranes: Nuclear Magnetic Resonance Relaxation Rates and Lateral Diffusion. J Chem Phys. 2006;125:204703. doi: 10.1063/1.2393240. [DOI] [PubMed] [Google Scholar]
- 19.Ohkubo YZ, Pogorelov TV, Arcario MJ, Christensen GA, Tajkhorshid E. Accelerating Membrane Insertion of Peripheral Proteins with a Novel Membrane Mimetic Model. Biophys Biophys J. 2012;102:2130–2139. doi: 10.1016/j.bpj.2012.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vermaas JV, Baylon JL, Arcario MJ, Muller MP, Wu Z, Pogorelov TV, Tajkhorshid E. Efficient Exploration of Membrane-Associated Phenomena at Atomic Resolution. J Membr Biol. 2015;248:563–582. doi: 10.1007/s00232-015-9806-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vermaas JV, Tajkhorshid E. A Microscopic View of Phospholipid Insertion into Biological Membranes. J Phys Chem B. 2014;118:1754–1764. doi: 10.1021/jp409854w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pogorelov TV, Vermaas JV, Arcario MJ, Tajkhorshid E. Partitioning of Amino Acids into a Model Membrane: Capturing the Interface. J Phys Chem B. 2014;118:1481–1492. doi: 10.1021/jp4089113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi Y, Cheng X, Lee J, Vermaas JV, Pogorelov TV, Tajkhorshid E, Park S, Klauda JB, Im W. CHARMM-GUI HMMM Builder for Membrane Simulations with the Highly Mobile Membrane-Mimetic Model. Biophys J. 2015;109:2012–2022. doi: 10.1016/j.bpj.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baylon JL, Lenov IL, Sligar SG, Tajkhorshid E. Characterizing the Membrane-Bound State of Cytochrome P450 3A4: Structure, Depth of Insertion, and Orientation. J Am Chem Soc. 2013;135:8542–8551. doi: 10.1021/ja4003525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vermaas JV, Tajkhorshid E. Conformational Heterogeneity of α-Synuclein in Membrane. Biochim Biophys Acta Biomembr. 2014;1838:3107–3117. doi: 10.1016/j.bbamem.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blanchard AE, Arcario MJ, Schulten K, Tajkhorshid E. A Highly Tilted Membrane Configuration for the Pre-Fusion State of Synaptobrevin. Biophys J. 2014;107:2112–2121. doi: 10.1016/j.bpj.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arcario MJ, Tajkhorshid E. Membrane-Induced Structural Rearrangement and Identification of a Novel Membrane Anchor in Talin F2F3. Biophys J. 2014;107:2059–2069. doi: 10.1016/j.bpj.2014.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baylon JL, Tajkhorshid E. Capturing Spontaneous Membrane Insertion of the Influenza Virus Hemagglutinin Fusion Peptide. J Phys Chem B. 2015;119:7882–7893. doi: 10.1021/acs.jpcb.5b02135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDougle DR, Baylon JL, Meling DD, Kambalyal A, Grinkova YV, Ham-mernik J, Tajkhorshid E, Das A. Incorporation of Charged Residues in the CYP2J2 F-G Loop Disrupts CYP2J2-Lipid Bilayer Interactions. Biochim Biophys Acta. 2015;1848:2460–2470. doi: 10.1016/j.bbamem.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madsen JJ, Ohkubo YZ, Peters GH, Faber JH, Tajkhorshid E, Olsen OH. Membrane Interaction of the Factor VIIIa Discoidin Domains in Atomistic Detail. Bio-chemistry. 2015;54:6123–6131. doi: 10.1021/acs.biochem.5b00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu Z, Schulten K. Synaptotagmin’s Role in Neurotransmitter Release Likely Involves Ca2+-Induced Conformational Transition. Biophys J. 2014;107:1156–1166. doi: 10.1016/j.bpj.2014.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rhéault JF, Gagné E, Guertin M, Lamoureux G, Auger M, Lagüe P. Molecular Model of Hemoglobin N from Mycobacterium Tuberculosis Bound to Lipid Bilayers: A Combined Spectroscopic and Computational Study. Biochemistry. 2015;54:2073–2084. doi: 10.1021/bi5010624. [DOI] [PubMed] [Google Scholar]
- 33.Zhang L, Rajendram M, Weibel DB, Yethiraj A, Cui Q. Ionic Hydrogen Bonds and Lipid Packing Defects Determine the Binding Orientation and Insertion Depth of Reca on Multicomponent Lipid Bilayers. J Phys Chem B. 2016;120:8424–8437. doi: 10.1021/acs.jpcb.6b02164. [DOI] [PubMed] [Google Scholar]
- 34.Fajardo-Sánchez E, Galiano V, Villalan J. Molecular Dynamics Study of the Membrane Interaction of a Membranotropic Dengue Virus C Protein-Derived Peptide. J Biomol Struct Dyn. 2016:1–12. doi: 10.1080/07391102.2016.1179595. [DOI] [PubMed] [Google Scholar]
- 35.Vermaas JV, Tajkhorshid E. Differential Membrane Binding Mechanics of Synaptotagmin Isoforms Observed at Atomic Detail. Biochemistry. 2017;56:281–293. doi: 10.1021/acs.biochem.6b00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeagle PL. Non-Covalent Binding of Membrane Lipids to Membrane Proteins. Biochim Biophys Acta Biomembr. 2014;1838:1548–1559. doi: 10.1016/j.bbamem.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 37.Arnarez C, Mazat JP, Elezgaray J, Marrink SJ, Periole X. Evidence for Cardiolipin Binding Sites on the Membrane-Exposed Surface of the Cytochrome Bc1. J Am Chem Soc. 2013;135:3112–20. doi: 10.1021/ja310577u. [DOI] [PubMed] [Google Scholar]
- 38.Martens C, Stein RA, Masureel M, Roth A, Mishra S, Dawaliby R, Konijnenberg A, Sobott F, Govaerts C, Mchaourab HS. Lipids Modulate the Conformational Dynamics of a Secondary Multidrug Transporter. Nat Struct Mol Biol. 2016;23:744–751. doi: 10.1038/nsmb.3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akyuz N, Georgieva ER, Zhou Z, Stolzenberg S, Cuendet MA, Khelashvili G, Altman RB, Terry DS, Freed JH, Weinstein H, et al. Transport Domain Unlocking Sets the Uptake Rate of an Aspartate Transporter. Nature. 2015;518:68–73. doi: 10.1038/nature14158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee AG. How Lipids Affect the Activities of Integral Membrane Proteins. Biochim Biophys Acta Biomembr. 2004;1666:62–67. doi: 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 41.Li J, James ZM, Dong X, Karim CB, Thomas DD. Structural and Functional Dynamics of an Integral Membrane Protein Complex Modulated by Lipid Headgroup Charge. J Mol Biol. 2012;418:379–389. doi: 10.1016/j.jmb.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palsdottir H, Hunte C. Lipids in Membrane Protein Structures. Biochim Biophys Acta. 2004;1666:2–18. doi: 10.1016/j.bbamem.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 43.Moraes I, Evans G, Sanches-Weatherby J, Newstead S, Stewart PDS. Membrane Protein Structure Determiniation – the Next Generation. Biochim Biophys Acta Biomembr. 2014;1838:78–87. doi: 10.1016/j.bbamem.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arcario MJ, Ohkubo YZ, Tajkhorshid E. Capturing Spontaneous Partitioning of Peripheral Proteins Using a Biphasic Membrane-Mimetic Model. J Phys Chem B. 2011;115:7029–7037. doi: 10.1021/jp109631y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hénin J, Shinoda W, Klein ML. United-Atom Acyl Chains for CHARMM Phospholipids. J Phys Chem B. 2008;112:7008–7015. doi: 10.1021/jp800687p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ulmschneider JP, Ulmschneider MB. United Atom Lipid Parameters for Combination with the Optimized Potentials for Liquid Simulations All-Atom Force Field. J Chem Theory Comput. 2009;5:1803–1813. doi: 10.1021/ct900086b. [DOI] [PubMed] [Google Scholar]
- 47.Lee S, Tran A, Allsopp M, Lim JB, Hénin J, Klauda JB. CHARMM36 United Atom Chain Model for Lipids and Surfactants. J Phys Chem B. 2014;118:547–556. doi: 10.1021/jp410344g. [DOI] [PubMed] [Google Scholar]
- 48.Tjörnhammar R, Edholm O. Reparameterized United Atom Model for Molecular Dynamics Simulations of Gel and Fluid Phosphatidylcholine Bilayers. J Chem Theory Comput. 2014;10:5706–5715. doi: 10.1021/ct500589z. [DOI] [PubMed] [Google Scholar]
- 49.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable Molecular Dynamics with NAMD. J Comp Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klauda JB, Venable RM, Freites JA, O’Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD, Jr, Pastor RW. Update of the CHARMM All-Atom Additive Force Field for Lipids: Validation on Six Lipid Types. J Phys Chem B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Best RB, Zhu X, Shim J, Lopes PEM, Mittal J, Feig M, MacKerell AD. Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone ϕ, ψ and Side-Chain χ1 and χ2 Dihedral Angles. J Chem Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, et al. CHARMM General Force Field: A Force Field for Drug-like Molecules Compatible with the CHARMM All-Atom Additive Biological Force Fields. J Comp Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Darden T, York D, Pedersen LG. Particle Mesh Ewald: An N ·log(N ) Method for Ewald Sums in Large Systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 54.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A Smooth Particle Mesh Ewald Method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 55.Miyamoto S, Kollman PA. SETTLE: An Analytical Version of the SHAKE and RATTLE Algorithm for Rigid Water Molecules. J Comp Chem. 1992;13:952–962. [Google Scholar]
- 56.Martyna GJ, Tobias DJ, Klein ML. Constant Pressure Molecular Dynamics Algorithms. J Chem Phys. 1994;101:4177–4189. [Google Scholar]
- 57.Feller SE, Zhang Y, Pastor RW, Brooks BR. Constant Pressure Molecular Dynamics Simulation: The Langevin Piston Method. J Chem Phys. 1995;103:4613–4621. [Google Scholar]
- 58.Fogarty JC, Arjunwadkar M, Pandit SA, Pan J. Atomically Detailed Lipid Bilayer Models for the Interpretation of Small Angle Neutron and X-Ray Scattering Data. Biochim Biophys Acta Biomembr. 2015;1848:662–672. doi: 10.1016/j.bbamem.2014.10.041. [DOI] [PubMed] [Google Scholar]
- 59.Haynes W, Lide D, Bruno T. CRC Handbook of Chemistry and Physics. 93. CRC Press; Boca Raton, Florida: 2012. CRC Handbook of Chemistry and Physics. [Google Scholar]
- 60.Aicart E, Tardajos G, Diaz Pena M. Isothermal Compressibility of Cyclohexane + N-Hexane, Cyclohexane + N-Heptane, Cyclohexane + N-Octane, and Cyclohexane + N-Nonane. J Chem Eng Data. 1980;25:140–145. [Google Scholar]
- 61.Landau L, Lifshitz E. Statistical Physics. Elsevier Butterworth-Heinemann; 1980. [Google Scholar]
- 62.MacCallum JL, Bennett WFD, Tieleman DP. Distribution of Amino Acids in a Lipid Bilayer from Computer Simulations. Biophys J. 2008;94:3393–3404. doi: 10.1529/biophysj.107.112805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Torrie GM, Valleau JP. Nonphysical Sampling Distributions in Monte Carlo Free-Energy Estimation: Umbrella Sampling. J Comp Phys. 1977;23:187–199. [Google Scholar]
- 64.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. The Weighted Histogram Analysis Method for Free-Energy Calculations on Biomolecules. I. The Method. J Comp Chem. 1992;13:1011–1021. [Google Scholar]
- 65.Hub JS, de Groot BL, van der Spoel D. g_wham-a Free Weighted Histogram Analysis Implementation Including Robust Error and Autocorrelation Estimates. J Chem Theory Comput. 2010;6:3713–3720. [Google Scholar]
- 66.Zwanzig RW. High-Temperature Equation of State by a Perturbation Method. I. Nonpolar Gases. J Chem Phys. 1954;22:1420–1426. [Google Scholar]
- 67.Chipot C, Pohorille A. Free Energy Calculations: Theory and Applications in Chemistry and Biology. Springer; 2007. [Google Scholar]
- 68.Zacharias M, Straatsma TP, McCammon JA. Separation-Shifted Scaling, a New Scaling Method for Lennard-Jones Interactions in Thermodynamic Integration. J Chem Phys. 1994;100:9025–9031. [Google Scholar]
- 69.Beutler TC, Mark AE, van Schalk RC, Gerber PR, van Gunsteren WF. Avoiding Singularities and Numerical Instabilities in Free Energy Calculations Based on Molecular Simulations. Chem Phys Lett. 1994;222:529–539. [Google Scholar]
- 70.Humphrey W, Dalke A, Schulten K. VMD – Visual Molecular Dynamics. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 71.Liu P, Dehez F, Cai W, Chipot C. A Toolkit for the Analysis of Free-Energy Perturbation Calculations. J Chem Theory Comput. 2012;8:2606–2616. doi: 10.1021/ct300242f. [DOI] [PubMed] [Google Scholar]
- 72.Lomize MA, Lomize AL, Pogozheva LD, Mosberg HI. OPM: Orientations of Proteins in Membranes Database. Bioinformatics. 2006;22:623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 73.MacKenzie KR, Prestegard JH, Engelman DM. A Transmembrane Helix Dimer: Structure and Implications. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 74.Call ME, Wucherpfennig KW, Chou JJ. The Structural Basis for Intramembrane Assembly of an Activating Immunoreceptor Complex. Nat Immunol. 2010;11:1023–1029. doi: 10.1038/ni.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou Y, MacKinnon R. The Occupancy of Ions in the K+ Selectivity Filter: Charge Balance and Coupling of Ion Binding to a Protein Conformational Change Underlie High Conduction Rates. J Mol Biol. 2003;333:965–975. doi: 10.1016/j.jmb.2003.09.022. [DOI] [PubMed] [Google Scholar]
- 76.de Groot BL, Engel A, Grubmüller H. A Refined Structure of Human Aquaporin-1. FEBS Lett. 2001;504:206–211. doi: 10.1016/s0014-5793(01)02743-0. [DOI] [PubMed] [Google Scholar]
- 77.Hiller S, Garces RG, Malia TJ, Orekhov VY, Colombini M, Wagner G. Solution Structure of the Integral Human Membrane Protein VDAC-1 in Detergent Micelles. Science. 2008;321:1206–1210. doi: 10.1126/science.1161302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Denisov IG, Grinkova YV, Baylon JL, Tajkhorshid E, Sligar SG. Mechanism of Drug-Drug Interactions Mediated by Human Cytochrome P450 CYP3A4 Monomer. Biochemistry. 2015;54:2227–2239. doi: 10.1021/acs.biochem.5b00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Limovich PV, Shirts MR, Mobley DL. Guidelines for the Analysis of Free Energy Calculations. Curr Comput Aided Drug Des. 2015;29:397–411. doi: 10.1007/s10822-015-9840-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Smith R, Tanford C. Hydrophobicity of Long Chain N-Alkyl Carboxylic Acids, As Measured by Their Distribution Between Heptane and Aqueous Solutions. Proc Natl Acad Sci USA. 1973;70:289–293. doi: 10.1073/pnas.70.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Killian JA, von Heijne G. How Proteins Adapt to a Membrane-Water Interface. Trends Biochem Sci. 2000;25:429–434. doi: 10.1016/s0968-0004(00)01626-1. [DOI] [PubMed] [Google Scholar]
- 82.Ulmschneider MB, Sansom MSP. Amino Acid Distributions in Integral Membrane Protein Structures. Biochim Biophys Acta Biomembr. 2001;1512:1–14. doi: 10.1016/s0005-2736(01)00299-1. [DOI] [PubMed] [Google Scholar]
- 83.Gumbart J, Roux B. Determination of Membrane-Insertion Free Energies by Molecular Dynamics Simulations. Biophys J. 2012;104:795–801. doi: 10.1016/j.bpj.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bennett WD, Tieleman DP. Water Defect and Pore Formation in Atomistic and Coarse-Grained Lipid Membranes: Pushing the Limits of Coarse Graining. J Chem Theory Comput. 2011;7:2981–2988. doi: 10.1021/ct200291v. [DOI] [PubMed] [Google Scholar]
- 85.Frishman D, Argos P. Knowledge-Based Secondary Structure Assignment. Proteins. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 86.Villinger S, Briones R, Giller K, Zachariae U, Lange A, de Groot BL, Griesinger C, Becker S, Zweckstetter M. Functional Dynamics in the Voltage-Dependent Anion Channel. Proc Natl Acad Sci USA. 2010;107:22546–22551. doi: 10.1073/pnas.1012310108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Krone M, Stone JE, Ertl T, Schulten K. Fast Visualization of Gaussian Density Surfaces for Molecular Dynamics and Particle System Trajectories. EuroVis - Short Papers. 2012;2012:67–71. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.