Abstract
Bax is known for its pro-apoptotic role within the mitochondrial pathway of apoptosis. However, the mechanism for transitioning Bax from cytosolic to membrane-bound oligomer remain elusive. Previous nuclear magnetic resonance (NMR) and electron paramagnetic resonance (EPR) studies defined monomeric Bax as conformationally homogenous. Yet, it has, recently, been proposed that monomeric Bax exists in equilibrium with a minor state that is distinctly different from its NMR structure. Here, we revisited the structural analysis of Bax using methods uniquely suited for unveiling “invisible” states of proteins, namely, NMR paramagnetic relaxation enhancements (PREs) and EPR double electron-electron resonance (DEER). Additionally, we examined the effect of glycerol, the co-solvent of choice in DEER studies, on the structure of Bax using NMR chemical shift perturbations (CSPs) and residual dipolar couplings (RDCs). Based on our combined NMR and EPR results, Bax is a conformationally homogenous protein prior to its activation.
Keywords: Apoptosis, Bax, NMR, DEER, EPR, residual dipolar coupling
Graphical abstract
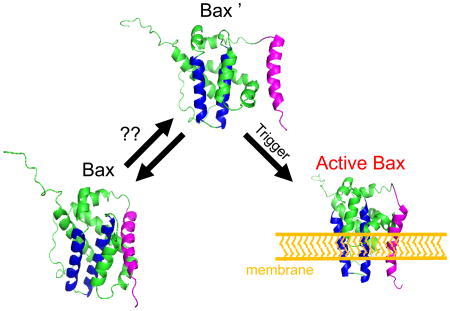
Critical to delineating apoptosis initiation is the structural characterization of monomeric Bax's conformational state or states, if they exist. Barnes et al. thoroughly investigated the conformation ensemble of monomeric Bax using uniquely suited NMR and EPR methodologies. Based on their results, the conformational distribution of monomeric Bax is homogenous.
Introduction
Apoptosis, or programmed cell death, is a naturally occurring process in higher order eukaryotic organisms and is essential for many biological activities including cell turnover, embryonic development, and immune system function (Youle and Strasser, 2008). Dysregulation of apoptosis is at the root of various human conditions such as tumorigenesis, neurodegenerative diseases, autoimmune disorders, and cardiovascular disease (Czabotar et al., 2014; Elmore, 2007). The activation pathway for apoptosis is generally classified as either extrinsic or intrinsic. The former is induced by extracellular ligands while the latter, also known as the mitochondrial pathway, is initiated by intracellular signals. Both pathways lead to caspase activation and can converge at the mitochondrial outer membrane (MOM), the major hallmark of the mitochondrial pathway.
The mitochondrial pathway of apoptosis is regulated by the Bcl-2 family of proteins, generally grouped according to their pro-apoptotic, anti-apoptotic, or sensitizer functions (Shamas-Din et al., 2013; Volkmann et al., 2014; Youle and Strasser, 2008). Prior to apoptosis, this protein family exists in states which favor cell proliferation. However, upon receipt of a death signal, the Bcl-2 proteins will engage in “kiss-and-run” interactions that lead to cell death (Shamas-Din et al., 2014). Bax and Bak are structurally homologous pro-apoptotic members of the Bcl-2 family of proteins and are key protagonists in the irreversible process of mitochondrial outer membrane permeabilization (MOMP) and subsequent release of mitochondrial proteins such as cytochrome c and SMAC to the cytosol which lead to the activation of caspases and other apoptotic factors (Renault and Chipuk, 2014). Thus, Bax/Bak are critical to deciding the life and death fate of cells.
In healthy cells, Bax primarily localizes in the cytosol while Bak is mostly membrane-bound (Volkmann et al., 2014). Following activation of apoptosis, Bax undergoes a series of conformational changes and translocates to the MOM where it becomes anchored and oligomerized (Gahl et al., 2014). The structure of Bax, solved by NMR, is monomeric and defined by a single hydrophobic helix (α5) surrounded by eight amphipathic helices (Suzuki et al., 2000). The membrane-bound structure of Bax is unknown but several studies have provided insights based on sparse structural restraints (Bleicken et al., 2014; Gahl et al., 2014; GroBe et al., 2016; Kuwana et al., 2016; Salvador-Gallego et al., 2016). For instance, a recent study of active Bax at the membrane, using electron paramagnetic resonance (EPR) double electron-electron resonance (DEER), shows significant displacement of helices α6-α9, in comparison to monomeric Bax, with membrane binding helices α5 and α6, assembling into a “clamp-like” structure, while the C-terminal helix (α9) displays conformational dynamics indicative of its participation in dimer formation and oligomerization (Bleicken et al., 2014). Although Bax's monomeric state is well-defined and progress is being made to characterize its membrane-bound state, the mechanism by which Bax is activated, arguably the most important step in apoptosis, remains elusive.
Cytosolic Bax is viewed as a monomeric and conformationally homogeneous protein. (Renault and Chipuk, 2014; Renault and Manon, 2011; Schellenberg et al., 2013; Suzuki et al., 2000; Youle and Strasser, 2008). Recently, however, it was reported, using the mouse homolog of Bax, that monomeric Bax is in equilibrium between two conformationally distinct states, with the previously unobserved state serving as a transitionary state to Bax's membrane-bound form (Tsai et al., 2015; Kao et al., 2017). Using electron paramagnetic resonance double electron-electron resonance at varying temperatures and pressures, Chiang and coworkers “captured” and provided a structural model of this “new” form of monomeric Bax. Although the populations of the two monomeric states are similar at 50 K, it is noted that Bax's conformational ensemble is likely skewed towards the form depicted by NMR at higher temperatures. Hence, the reason for the invisibility of this new form of monomeric Bax in other solution-state studies.
Cytosolic Bax heterogeneity raises intriguing questions about Bax's activation mechanism and potential new therapeutic approaches for regulating its function. Thus, we were stimulated to revisit the structural analysis of Bax in solution using techniques that are sensitive to the presence of “invisible” or lowly populated/minor states. Here we report the results of our search for an “invisible” state of monomeric Bax using NMR paramagnetic relaxation enhancements (PREs) and electron paramagnetic resonance double electron-electron resonance studies as well as complementary chemical shift perturbations (CSPs) and residual dipolar couplings (RDCs) to quantify monomeric Bax's conformational ensemble. At least under the conditions used in our study, monomeric Bax is a conformationally homogenous protein.
Results
Paramagnetic Relaxation Enhancement (PRE) of Monomeric Bax
PREs can be used to identify minor or “invisible” states (with populations as low as <1%) of a macromolecule (Anthis and Clore, 2015). Assuming the exchange between states is rapid (<250-500 μs), then an increase in the transverse relaxation rate (R2) of the major state due to exchange will provide an indirect measurement of the minor state. The presence of a paramagnetic relaxation moiety attached to the molecule of interest will heighten this effect for nearby (<20-35 Å) nuclei via dipolar interactions between the nuclei and unpaired electron(s) of the paramagnetic center.
In consideration of the spin-label location in Bax, we wanted (1) the spin-labeling site to induce minimal perturbations to the structure of Bax using the most conservative site-directed mutations and (2) provide the capability to monitor exchange events for Bax's membrane-binding domain (α5 and α6), which was predicted to be in a different conformation in the minor state. For this purpose, one of the two native cysteines of Bax (Cys62 or Cys126) was mutated to a serine, thereby, leaving the remaining cysteine free for spin-labeling with MTSL and probing of changes in helices α5 and/or α6. Although there were some NMR chemical shift perturbations in the HSQC spectra of the single-cysteine Bax mutants, in comparison to the HSQC spectrum of native Bax, the perturbations were relatively modest (data not shown) and localized only to the mutation site.
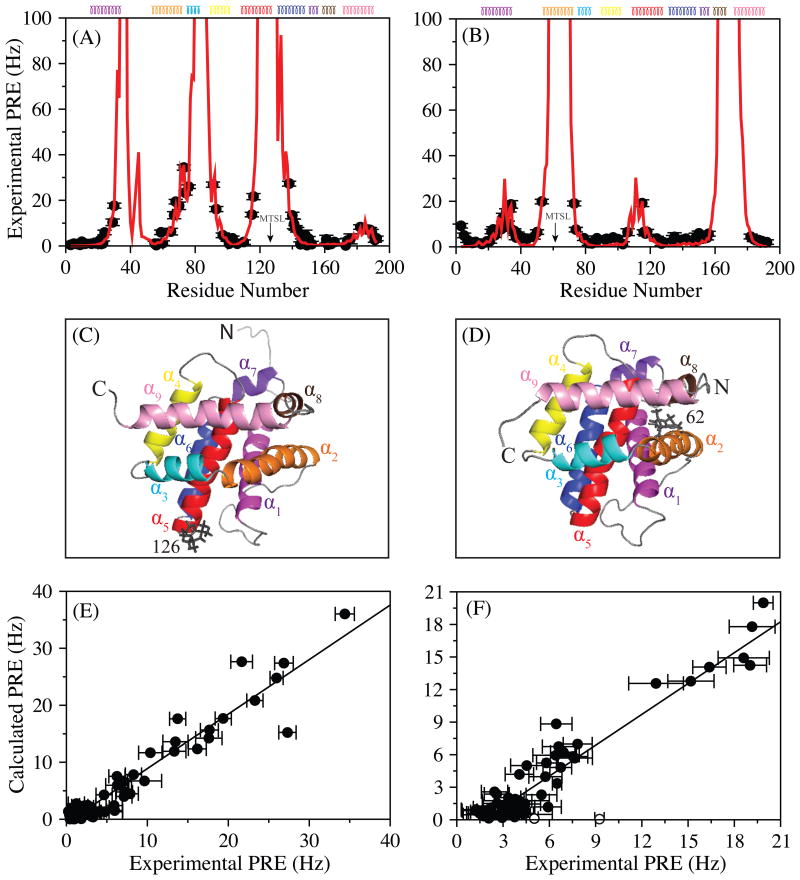
Representative HSQC's of Bax, tagged with the dia- and paramagnetic forms of MTSL at two distinct positions, Cys126 and Cys62, are shown in Figure S1. Experimental PRE's from these two sites of Bax are displayed in Figures 1A and 1B. The PRE values qualitatively agree with Bax's NMR structure. Residues of Bax in the vicinity (<10 Å) of the MTSL tag have high PRE rates or missing resonances due to significant paramagnetic broadening, while those further away display lower PRE rates. To better assess the agreement between the experimental and calculated (using Bax's NMR structure) PREs of Bax, we optimized the position of the MTSL relative to the protein using the measured PREs. To account for the potential flexibility of the MTSL tag, the optimization was carried out using a varying number of MTSL conformers. Three MTSL conformers provided the best agreement. A representative optimized conformation of the MTSL relative to the protein is displayed in Figures 1C and 1D. The calculated PRE values are exhibited as a function of Bax residue number in Figures 1A and 1B and they compare quite well with the experimental values. The most common way to evaluate this agreement is with the PRE Q-factor (Iwahara et al., 2004), whose value ranges from 0 to 1 with a smaller Q-factor implying a better representation of the experimental PRE values with respect to the structure of the protein. The calculated PRE Q-factor for Bax C62S 126-MTSL and Bax C126S 62-MTSL was 0.28 and 0.33, respectively. This agreement between the experimental and calculated PREs of Bax are displayed in correlation plots (Figures 1E and 1F) with correlation factors, R, of 0.96 and 0.94 for C62S 126-MTSL and C126S 62-MTSL, respectively. The excellent Q factors suggest that the experimental PRE values are representative of a single conformation corresponding to the NMR structure of Bax in solution (including helices α5 and α6). If another population were present, with a shorter distance than that reflected in Bax's NMR structure, then residues participating in the exchange between states should have abnormal PREs in comparison to those calculated using the known structure. It should be noted, however, that the first two residues of the N-terminus in Bax C126S 62-MTSL fit poorly to the NMR structure (Figures 1E and 1F), most likely due to a conformation of the highly mobile N-terminal end of Bax not represented by the NMR structure of Bax chosen (lowest energy in the NMR ensemble of structures) in this analysis.
Figure 1.
Measured PRE rates are shown, in black circles, as a function of residue number for Bax C62S 126-MTSL (A) and Bax C126S 62-MTSL (B). The secondary structure composition of Bax is indicated at the top of the panels. Helices of Bax are colored as follows: α1 = magenta, α2 = orange, α3 = cyan, α4 = yellow, α5 = red, α6 = blue, α7 = purple blue, α8 = brown, and α9 = pink. The calculated PRE values are displayed in red. Experimental errors were estimated using the noise observed in the 2D 1H-15N transverse relaxation spectra acquired at each time point for both paramagnetic and diamagnetic species (Iwahara et al., 2007). The calculated PREs are obtained from the average of PRE values predicted from ten optimized structures. Three conformers of the MTSL tag were used for the optimizations (Iwahara et al., 2004). A representative optimized conformation of the MTSL tag is displayed for Bax C62S 126-MTSL (C) and Bax C126S 62-MTSL (D). Correlation plots between calculated vs experimental PREs for Bax C62S 126-MTSL (E) and Bax C126S 62-MTSL (F) show very good agreement with a correlation factor (R) of 0.96 and 0.94, respectively. Open circles in the correlation plot of Bax C126S 62-MTSL correspond to the first two residues of the N-terminus. All molecular figures were created using PyMol (Schrodinger, 2015).
Effect of glycerol on the conformation of Monomeric Bax
For DEER studies of soluble proteins at cryogenic temperatures (<100 K), a cryoprotectant/osmolyte, typically glycerol, is added to the protein solution at moderate to high concentrations (>10-50%) to lower the glass transition temperature and, consequently, prevent protein aggregation and ice formation (de Vera et al., 2013). Contrastingly, for membrane proteins, this co-solvent requirement is, normally, eliminated entirely since membrane proteins inherently lower the glass transition temperature of the solvent. Although osmolytes allow the preservation of soluble proteins at cryogenic temperatures, they can perturb a protein's native conformational equilibria (López et al., 2009; Schmidt et al., 2016) and, thus, could allow otherwise energetically unfavorable conformations to be sampled by the protein and/or spin-label. Furthermore, because Bax can be in both membrane and soluble forms, the glycerol percentages (30-50%) used to study Bax with DEER could potentially affect its conformational state. The NMR chemical shift is exquisitely sensitive to changes in the protein local environment and is, therefore, a good reporter for conformational changes (Williamson, 2013).
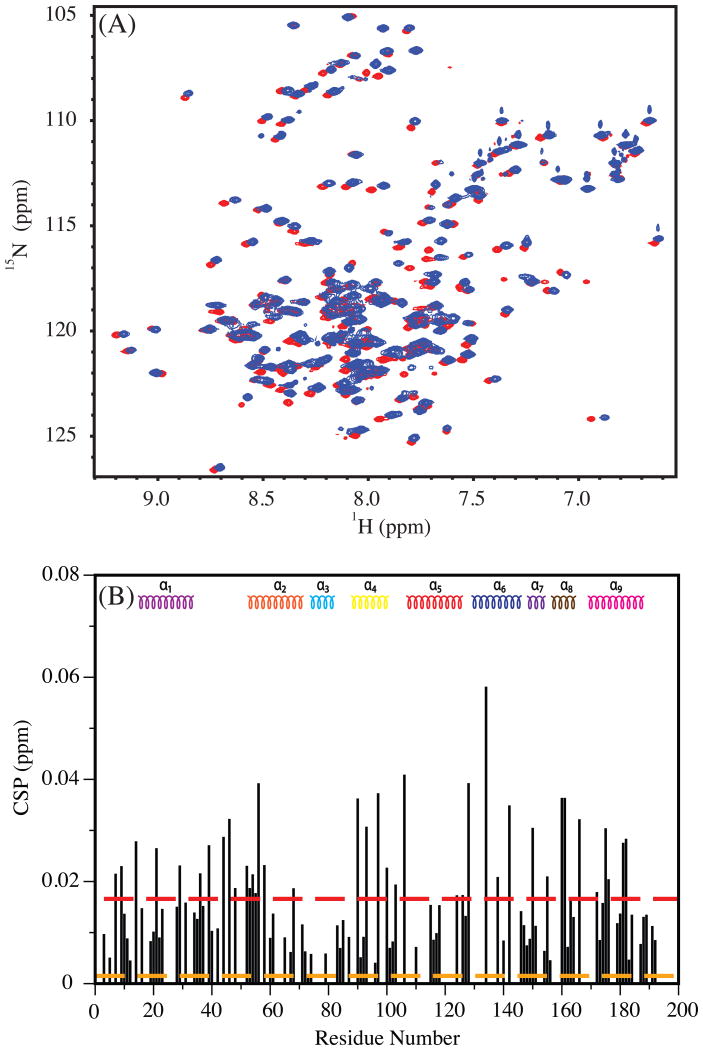
The 1H-15N HSQC spectra of Bax in the absence and presence of glycerol are compared in Figure 2A. As the amount of glycerol increased in the solution, distinct chemical shift changes were observed in the spectrum of Bax. These changes are quantified as chemical shift perturbations, displayed as a function of Bax's residue number in Figure 2B. The CSP's do not appear to be isolated to a specific region of Bax but rather distributed throughout the protein. The CSPs do indicate an interaction between Bax and glycerol. The relatively small magnitude of the CSP (<0.1ppm), however, implies that this interaction is weak and therefore may not lead to significant conformational changes in Bax.
Figure 2.
(A) 1H-15N HSQC of Bax in 0% (red) and 20% (blue) glycerol. (B) A plot displaying the magnitude (in ppm) of the chemical shift changes as a function of residue in Bax due to glycerol. The secondary structure composition of Bax is shown at the top as a reference. The 0% and 20% spectra were used for determining the CSP. Dashed red and orange lines are average CSP (of all observed residues) and the experimental error, respectively.
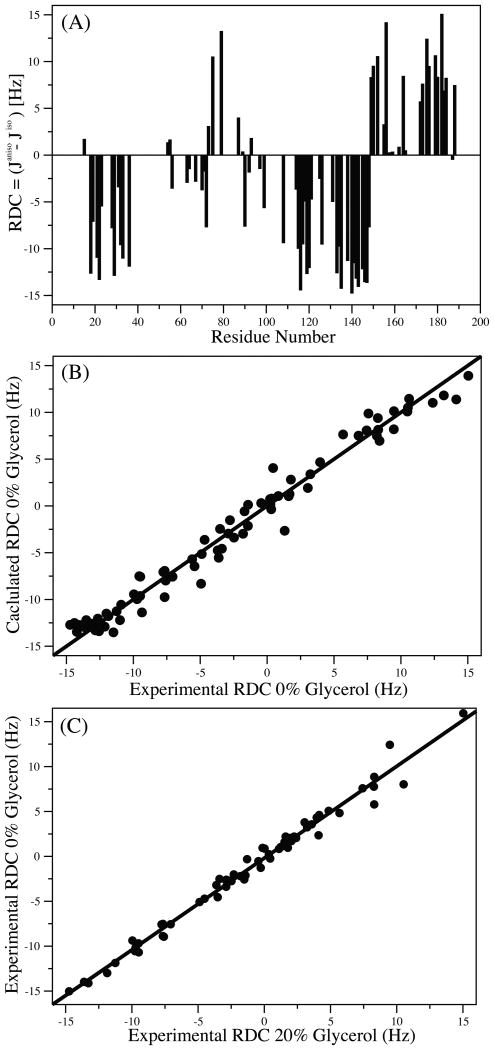
Residual dipolar couplings are excellent probes for minute changes in the structure of a protein (Chen and Tjandra, 2012; Tjandra et al., 1997). Since the NH RDC reports on the orientation of the NH bond relative to a common reference frame (alignment frame) of the protein, even variations of a few degrees would result in a measurable change in the NH RDC. If multiple conformations of a protein in dynamic exchange are present in solution, the NH RDC will be a collective contribution from all conformers. To check whether the changes in chemical shifts are associated with conformational changes in Bax, the NH RDC's of Bax were measured in filamentous phage, pf1, in the presence and absence of 20% glycerol. The measured NH RDC of Bax without glycerol is shown in Figure 3A. Residues with overlapping resonances and those in the unstructured loops of Bax have been omitted from the analysis. The agreement between the experimental NH RDC without glycerol to the NMR structure is shown in Figure 3B with a correlation coefficient, R, of 0.99. Due to severe overlap of resonances in the HSQC spectrum of Bax in 20% glycerol, the number of reliable NH RDCs that could be obtained was fewer than in the absence of glycerol. The comparison between 0% and 20% glycerol NH RDC values is shown in Figure 3C. An excellent agreement with a correlation factor, R, of 0.99 was found between the two NH RDC data sets, reflecting that the same overall conformation of Bax is present in the 0% and 20% glycerol buffers. Therefore, even though glycerol does interact with Bax in a non-specific manner, conformational changes were not detected, at least up to a glycerol concentration of 20%.
Figure 3.
1H-15N Residual dipolar couplings (RDC) of 15N Bax in 0% glycerol (A). A comparison of experimental and calculated 1H-15N RDC's of Bax (using the NMR structure of Bax PDB ID: 1F16) are displayed in (B), with a correlation coefficient R of 0.99. Correlation of the experimental 15N Bax in 0% and 20% glycerol solutions (C). An excellent agreement between the two experimental RDC data sets was observed with R = 0.99. Residues from the loops of Bax were removed in (A) and (B).
Double Electron-Electron Resonance (DEER) of Monomeric Bax
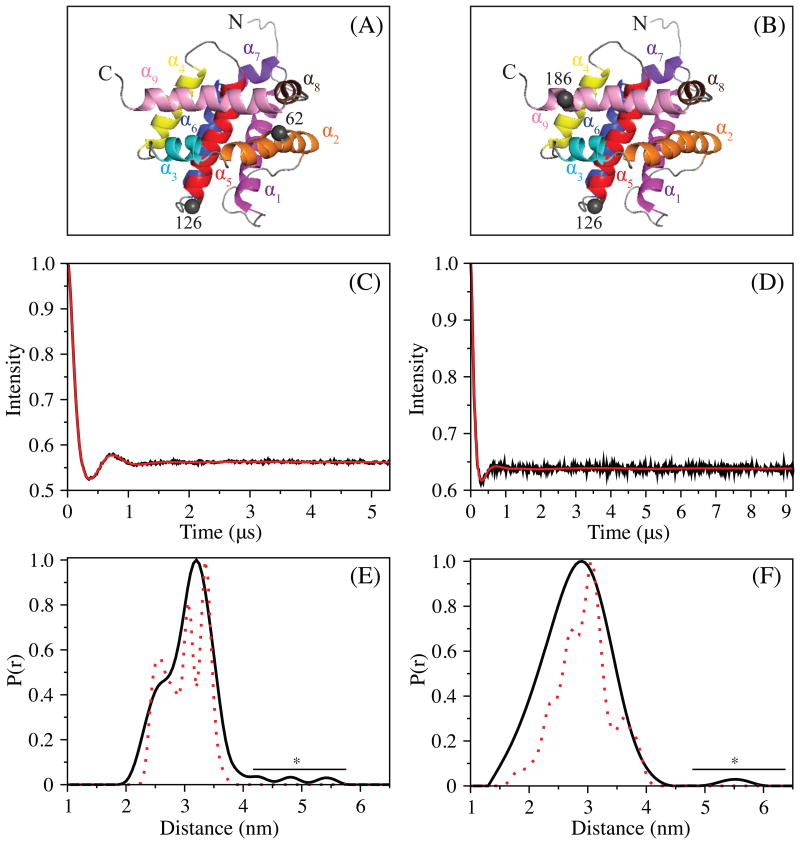
DEER in conjunction with site-directed mutagenesis is a powerful technique that can measure distances (>1.5-10 nm) between two paramagnetic sites of a protein (Larsen and Singel, 1993; Milov et al., 1981; Milov et al., 1984; Pannier et al., 2000). If the spin-labeling sites are chosen carefully, then a conformational change of a protein can be detected in the DEER distance distribution. To probe for the possibility of an equilibrium between two or more conformationally distinct states of monomeric Bax, which may only exist at cryogenic temperatures, we utilized DEER at 50 K in combination with spin-labeling of Bax's native cysteines (i.e., C62 and C126) as well as one engineered at T186. Thus, two different Bax proteins containing a pair of cysteines (Cys62-Cys126 or Cys126-Cys186) were labeled with nitroxide spin label, MTSL. Specifically, 62-126MTSL was used to investigate the possible conformational heterogeneity in the core of the Bax's structure, while that of 126-186MTSL was designed to monitor the conformation of Bax's C-terminal helix (α9). The positions of these tags relative to the structure of Bax are shown in Figure 4A and 4B. Since improper background correction and/or low signal to noise ratios can lead to spurious results in the DEER distance distribution and because our interests lie in the unveiling of lowly populated states, all measurements were conducted at Q-band with perdeuterated Bax. Perdeuteration increases the phase memory time and, resultantly, the signal-to-noise ratio (Baber et al., 2015; Ward et al., 2010). As well, Q-band DEER measurements provide an order of magnitude greater gain in sensitivity versus those performed at X-band (Polyhach et al., 2012). Therefore, if another conformational analog of Bax were present, and at the relative populations (∼20% or greater) reported previously (Tsai et al., 2015), then it should be observed in our DEER distance distributions.
Figure 4.
DEER of 2H, 15N Bax mutants at 50 K. The location of the MTSL tags in Bax 62-126 and Bax 126-186 are shown in (A) and (B), respectively, as gray spheres. The experimental, background subtracted, DEER echo curves and P(r) distance distributions, normalized to 1, for Bax 62-126MTSL and 126-186MTSL are displayed in (C, D) and (E, F), respectively. The P(r) distance distributions were produced using Tikhonov regularization of the echo curves in DeerAnalysis 2015. The simulated distance distributions (dotted red line) were acquired from MMM 2013.2, using model 8 of Bax (PDB ID: 1F16), assuming an MTSL rotamer distribution at 298 K. Peaks resulting from noise artifacts are indicated with a bar and *.
The DEER signals acquired for the two mutants of Bax are displayed in Figures 4C, 4D, and S2. The calculated DEER distance distributions from the acquired dipolar evolution curves are presented in Figures 4E and 4F. The distance distribution of Bax 126-186MTSL is monomodal while that of 62-126MTSL is bimodal. As displayed in the simulated distance distribution of Bax 62-126MTSL, the shoulder at ∼2.3 nm arises from a subpopulation of MTSL rotamers and not changes in protein conformation. Thus, both Bax variants display a single conformation matching the NMR structure of Bax with an intact core and closely packed helix α9, occupying its hydrophobic pocket. These results are in agreement with recent work from Bordignon and coworkers, in which another conformation of monomeric Bax was not observed (Bleicken et al.). Similar to their work, model 8 of the NMR structural ensemble of Bax provided a good fit of the data using MMM2013.2 (Polyhach et al., 2011). In addition, the 25 distances acquired in their study were used as restraints for in silico folding of Bax to the NMR structure (Fischer et al., 2016).
Conclusion
Bax is a central, and arguably the most important, entity in the mitochondrial pathway of apoptosis and the study of its conformational dynamics and structural features as it proceeds from its monomeric to oligomeric and membrane-bound state is critical to not only unraveling its activation mechanism but also in providing insights into new therapeutic treatments which can influence its function. Most studies to date have defined Bax as a conformationally homogeneous protein in its monomeric state. Recently, the existence of a conformationally different form of monomeric Bax has been proposed as an important feature in Bax's activation mechanism. Motivated by this report and the potential implications for developing better therapeutic approaches for modulating Bax's function, we revisited the structural analysis of monomeric Bax using established NMR and EPR methodologies, specifically geared to revealing “invisible” or minor states of a protein. By using wild type and carefully chosen Bax mutants, we performed NMR PRE, NMR RDC, and EPR DEER experiments to search for structurally different forms of monomeric Bax, while considering the sensitivity of those methodologies to detect minor species of a protein. However, at least under the conditions used in our study, another conformational analog of monomeric Bax was not observed.
Contact for Reagent and Resource Sharing
Further information and requests for reagents may be directed to and will be fulfilled by the Lead Contact, Nico Tjandra (tjandran@nhlbi.nih.gov)
Experimental Model and Subject Details
Bax plasmid DNA's were cultured and amplified in Escherichia coli XL2Blue cells (Novagen) in LB at 37 °C. Recombinant proteins were overexpressed in Escherichia coli BL21(DE3) cells (Novagen) in minimal media at 18 °C overnight.
Method Details
Protein Expression, Purification, Spin-Labeling
The human variant of Bax was used throughout this study. The expression vector used was previously described (Suzuki et al., 2000). Single-site point mutations (C62S, C126S, and T186) in the cDNA of Bax were generated using the QuikChange site-directed mutagenesis kit (Agilent Technologies). The protein expression was performed as previously reported (Suzuki et al., 2000) except for implementation of fermentation (>10 L) and overnight expression at 18 °C. Briefly, Bax was overexpressed in Escherichia coli BL21 gold (DE3) cells grown in minimal media with >99% 15NH4Cl as the sole source of nitrogen. For 13C-labeling, >99% 13C-D-glucose was used in place of naturally abundant D-glucose. Perdeuterated Bax was acquired by using deuterium oxide (D2O) instead of H2O in the expression medium. Isotope labels were acquired from Cambridge Isotope Laboratories. Immediately following cell harvesting, cell pellets were frozen in liquid nitrogen and stored at −80 °C. For purification of Bax, frozen cell pellets were thawed and resuspended in 20 mM Tris-HCl, 100 mM NaCl, 1 mM EDTA, pH=8 (Buffer A) containing Roche complete protease inhibitor (Roche Applied Science) and passed twice through an EmulsiFlex-C3 (AVESTIN). The resulting cell lysate was centrifuged at 40000 rpm for 1 hr at 4 °C in a Beckman Type 45 Ti rotor and the supernatant was passed over a chitin affinity column pre-equilibrated with Buffer A (also at 4 °C). The column was then washed with >15 column volumes (CV) of 20 mM Tris-HCl, 2M NaCl, 1 mM EDTA, pH=8, followed by ∼10 CV of Buffer A. The on-column resin was next stirred with 3 CV of Buffer A + 60 mM β-mecaptoethanol and allowed to incubate for ∼48 hrs at 4 °C before elution. Bax protein was eluted with ∼4-5 CVs of Buffer A, concentrated with Amicon Filters (>10 kDa cutoff), and buffer-exchanged into 20 mM Tris-HCl pH=8 (Buffer C). Further purification via mono-Q anion-exchange chromatography was performed with a linear salt gradient using Buffer C and Buffer C + 1M NaCl. The purity of the protein was analyzed at each step of purification using SDS-PAGE. Following the mono-Q purification step, liquid chromatography-mass spectrometry (LC-MS) using an Agilent 6224 spectrometer equipped with an electrospray ionization (ESI) source and time-of-flight (TOF) analyzer was used to confirm the identity and purity of the isotopically (15N, 2H, and/or 13C) labeled recombinant Bax protein.
For nitroxide spin-labeling, single- and double-cysteine Bax proteins were reduced with >10-fold molar excess of DTT for at least 2 hrs and, then, buffer exchanged into 100 mM Tris-HCl pH=7.5, 100 mM NaCl, and 1 mM EDTA. An excess (20-fold) of 1-Oxyl-2,2,5,5-tetramethyl-Δ 3-pyrroline-3-methyl methanethiosulfonate (MTSL; Toronto Research Company) was then added to the protein solution and the reaction mixture was incubated in the dark at room temperature for ∼18 hrs. Termination of the reaction and removal of unreacted spin-label was performed by a PD10 buffer exchange column and size-exclusion chromatography (Superdex 16/60 G75). LC-MS, as described above, was used to determine the labeling efficiency. Unlabeled protein was not observed in this analysis.
NMR Spectroscopy
All NMR spectra were acquired with Bruker Advance-III spectrometers using triple resonance z-axis gradient cryogenic probes tuned to 1H Larmor frequencies at 900.27, 800.35, or 600.33 MHz. Carrier frequencies were generally set at ∼4.70 ppm and ∼118.0 ppm for the proton and nitrogen, respectively. The 1H-15N HSQC spectra consisted of 2048(t2) × 256(t1) real data points. HNCACB and CBCA(CO)NH were used to confirm resonance assignments of the backbone for the single-cysteine Bax mutants. All NMR samples were prepared in 20 mM sodium acetate-d3, pH=6, 2 mM dithiothreitol (DTT), and 2 mM ethylenediaminetetraacetic acid (EDTA) in 90% H2O/10% D2O. The reducing agent (DTT) was excluded from spin-labeled samples. All samples were placed in Shigemi NMR tubes and measurements were performed at 305 K. Residue resonances that were missing and/or overlapped were not used in the data analysis. All data was processed using NMRPipe (Delaglio et al., 1995). Data analysis and resonance assignment were performed in SPARKY (Goddard and Kneller, UCSF) and CARA (Goddard and Kneller; Keller, 2004).
Paramagnetic Relaxation Enhancement
Residue specific transverse 1H PRE (1H-Γ2) rates of Bax were determined by the difference between the 1H-R2 rates of the oxidized and reduced (via 10× molar excess of sodium ascorbate) forms of MTSL using the two-time point method (Iwahara et al., 2007). The errors in 1H-Γ2 were calculated using error propagation and the standard deviation in the spectral noise. PRE rates and errors were plotted with respect to residue number and compared to those calculated from Bax's NMR structure (1F16) using XPLOR-NIH (Schwieters et al., 2006; Schwieters et al., 2003).
Bax Chemical Shift Perturbation by Glycerol
Based on its density (1.26 g/mL) glycerol-d8 (Cambridge Isotope Laboratories) was added by mass to achieve the necessary volumes to produce Bax solutions of 0-20% glycerol-d8. For each protein-glycerol solution, the ionic strength and pH were kept constant between samples. The chemical shift perturbation was calculated using the following equation (Ishima, 2015)
where δ and γ are the chemical shift and gyromagnetic ratio of the nuclei, respectively. Perturbations were calculated for each residue and plotted against their residue numbers (i).
Residual Dipolar Coupling
15N-1H scalar (J) couplings were measured by J-modulation experiments (Tjandra et al., 1996). In brief, ten 1JHN-modulated two-dimensional 1H-15N HSQC spectra were acquired with dephasing delays of 23.91, 24.71, 26.11, 27.51, 28.91, 35.11, 36.01, 36.91, 38.51, and 39.61 ms. To determine the NH residual dipolar couplings (RDCs) of Bax, JNH couplings in the presence of filamentous phage (pf1, ∼12 mg/mL) were subtracted by those without. RDC measurements of Bax were performed in 0% glycerol and 20% glycerol. The buffer conditions (i.e. 20 mM sodium acetate-d3, 2 mM EDTA, 2 mM DTT, pH=6) were the same for each solution.
Double Electron-Electron Resonance
The DEER experiments were performed as in Baber, et. al., 2015 using a Bruker E-580 spectrometer operating at Q-band (∼33.8 GHz) and fitted with an ER5107D2 probe and 150 W traveling-wave tube amplifier. Four-pulse DEER was conducted with observe π/2 and π pulses of 12 ns and 24 ns, respectively, and a pump (ELDOR) π pulse of 8 ns. The pump pulse was placed at the center of the most intense peak of the echo-detected field-swept-spectrum. A frequency difference of ∼95 MHz between the pump and observe pulse was maintained for all experiments. Deuterium modulation was averaged by incrementing the first echo period (τ1 = 400 ns) eight times in 16 ns steps. Spin-labeling of double-cysteine Bax proteins was performed as described above. Each protein sample (∼50 μM) for DEER was prepared in 30% d8-glycerol/70% D2O and flash frozen (with liquid N2) in Vitrocom, 1.1 mm internal diameter quartz tubes. The sample temperature for all measurements was 50 K. To increase the phase memory time (Tm) and, consequently, the dipolar evolution time, Bax protein samples for DEER were perdeuterated. A total dipolar evolution time of 10 μs and 6 μs was used for 2H15N Bax 126-186MTSL and 2H15N Bax 62-126MTSL, respectively. Due to artifacts from the “2+1” echo perturbation at the end of the second echo period, a cutoff of 9.3 μs and 5.3 μs was used for processing the data in DeerAnalysis (Jeschke et al., 2006). Each curve was background corrected using a 3D homogeneous model and fit to the optimum Tikhonov regulation parameter (α), 1000 and 10 for Bax 126-186MTSL and Bax 62-126MTSL, respectively, acquired from L-curve analysis (de Vera et al., 2013). Simulated DEER distance distributions were calculated with MMM2013.2 (Polyhach et al., 2011).
Supplementary Material
Highlights.
NMR and DEER studies defined a monomodal conformational distribution of monomeric Bax.
Although NMR CSP's were observed, Bax's structure is preserved in glycerol.
Monomeric Bax is conformationally homogeneous prior to its activation.
Acknowledgments
We would like to thank Yi He for providing training and use of the fermentation facility (NHLBI) for preparation of the proteins. Also, we thank Dr. Duck-Yeon Lee of the Biochemistry Core at NHLBI for giving us access to the LC-MS instrument as well as his helpful comments and advice regarding our results. We also thank Dr. Thomas Schmidt of NIDDK for helpful comments with regards to our EPR experiments. Finally, we would like to thank Dr. James A. Ferretti for his helpful comments and reading of the manuscript. This work was supported by the Intramural Research Program of the NIH, National Heart, Lung, and Blood Institute.
Footnotes
Author Contributions: C. A. B and N. T. conceptualized the project. C. A. B. performed all sample preparations, experiments, and data analysis as well as manuscript composition and preparation. P. M. assisted in NMR data analysis, while M-P. S. created mutant plasmids of Bax. J. L. B. assisted in performing the DEER experiment as well as data analysis. N. T. assisted in project administration.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anthis NJ, Clore GM. Visualizing transient dark states by NMR spectroscopy. Quarterly Reviews of Biophysics. 2015;48:35–116. doi: 10.1017/S0033583514000122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baber JL, Louis JM, Clore GM. Dependence of Distance Distributions Derived from Double Electron Electron Resonance Pulsed EPR Spectroscopy on Pulse-Sequence Time. Angewandte Chemie International Edition. 2015;54:5336–5339. doi: 10.1002/anie.201500640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleicken S, Jeschke G, Stegmueller C, Salvador-Gallego R, García-Sáez AJ, Bordignon E. Structural Model of Active Bax at the Membrane. Molecular Cell. 2014;56:496–505. doi: 10.1016/j.molcel.2014.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Tjandra N. In: The Use of Residual Dipolar Coupling in Studying Proteins by NMR In NMR of Proteins and Small Biomolecules. Zhu G, editor. Berlin, Heidelberg: Springer Berlin Heidelberg; 2012. pp. 47–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nature Reviews Molecular Cell Biology. 2014;15:49–63. doi: 10.1038/nrm3722. [DOI] [PubMed] [Google Scholar]
- de Vera IMS, Blackburn ME, Galiano L, Fanucci GE. Pulsed EPR Distance Measurements in Soluble Proteins by Site-directed Spin-labeling (SDSL) Current protocols in protein science / editorial board. 2013;74(Unit-17):17. doi: 10.1002/0471140864.ps1717s74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. Journal of Biomolecular NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Elmore S. Apoptosis: A Review of Programmed Cell Death. Toxicologic Pathology. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer AW, Bordignon E, Bleicken S, García-Saez AJ, Jeschke G, Meiler J. Pushing the size limit of de novo structure ensemble prediction guided by sparse SDSL-EPR restraints to 200 residues: The monomeric and homodimeric forms of BAX. Journal of Structural Biology. 2016;195:62–71. doi: 10.1016/j.jsb.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahl RF, He Y, Yu S, Tjandra N. Conformational Rearrangements in the Proapoptotic Protein, Bax, as It Inserts into Mitochondria: A CELLULAR DEATH SWITCH. Journal of Biological Chemistry. 2014;289:32871–32882. doi: 10.1074/jbc.M114.593897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Kneller DG. SPARKY 3. University of California; San Francisco: [Google Scholar]
- GroBe L, Wurm CA, Brüser C, Neumann D, Jans DC, Jakobs S. Bax assembles into large ring-like structures remodeling the mitochondrial outer membrane in apoptosis. The EMBO Journal. 2016;35:402–413. doi: 10.15252/embj.201592789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishima R. Protein-Inhibitor Interaction Studies Using NMR. Applications of NMR spectroscopy. 2015;1:143–181. doi: 10.2174/9781608059621115010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwahara J, Schwieters CD, Clore GM. Ensemble Approach for NMR Structure Refinement against 1H Paramagnetic Relaxation Enhancement Data Arising from a Flexible Paramagnetic Group Attached to a Macromolecule. Journal of the American Chemical Society. 2004;126:5879–5896. doi: 10.1021/ja031580d. [DOI] [PubMed] [Google Scholar]
- Iwahara J, Tang C, Clore GM. Practical Aspects of (1)H Transverse Paramagnetic Relaxation Enhancement Measurements on Macromolecules. Journal of Magnetic Resonance. 2007;184:185–195. doi: 10.1016/j.jmr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao TY, Tsai CJ, Lan YJ, Chiang YW. The role of conformational heterogeneity in regulating the apoptotic activity of BAX protein. Physical Chemistry Chemical Physics. 2017;19:9584–9591. doi: 10.1039/c7cp00401j. [DOI] [PubMed] [Google Scholar]
- Keller R. CARA: computer aided resonance assignment. 2004 http://cara.nmr.ch/
- Kuwana T, Olson NH, Kiosses WB, Peters B, Newmeyer DD. Pro-apoptotic Bax molecules densely populate the edges of membrane pores. Scientific Reports. 2016;6:27299. doi: 10.1038/srep27299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen R, Singel D. Double electron–electron resonance spin–echo modulation: Spectroscopic measurement of electron spin pair separations in orientationally disordered solids. The Journal of Chemical Physics. 1993;98:5134–5146. [Google Scholar]
- López CJ, Fleissner MR, Guo Z, Kusnetzow AK, Hubbell WL. Osmolyte perturbation reveals conformational equilibria in spin-labeled proteins. Protein Science. 2009;18:1637–1652. doi: 10.1002/pro.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milov A, Salikhov K, Shirov M. Use of the double resonance in electron spin echo method for the study of paramagnetic center spatial distribution in solids. Fiz Tverd Tela. 1981;23:975–982. [Google Scholar]
- Milov AD, Ponomarev AB, Tsvetkov YD. Electron-electron double resonance in electron spin echo: Model biradical systems and the sensitized photolysis of decalin. Chemical Physics Letters. 1984;110:67–72. [Google Scholar]
- Pannier M, Veit S, Godt A, Jeschke G, Spiess HW. Dead-Time Free Measurement of Dipole-Dipole Interactions between Electron Spins. Journal of Magnetic Resonance. 2000;142:331–340. doi: 10.1006/jmre.1999.1944. [DOI] [PubMed] [Google Scholar]
- Polyhach Y, Bordignon E, Jeschke G. Rotamer libraries of spin labelled cysteines for protein studies. Physical Chemistry Chemical Physics. 2011;13:2356–2366. doi: 10.1039/c0cp01865a. [DOI] [PubMed] [Google Scholar]
- Polyhach Y, Bordignon E, Tschaggelar R, Gandra S, Godt A, Jeschke G. High sensitivity and versatility of the DEER experiment on nitroxide radical pairs at Q-band frequencies. Physical Chemistry Chemical Physics. 2012;14:10762–10773. doi: 10.1039/c2cp41520h. [DOI] [PubMed] [Google Scholar]
- Renault TT, Chipuk JE. Death upon a Kiss: Mitochondrial Outer Membrane Composition and Organelle Communication Govern Sensitivity to BAK/BAX-Dependent Apoptosis. Chemistry & Biology. 2014;21:114–123. doi: 10.1016/j.chembiol.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault TT, Manon S. Bax: Addressed to kill. Biochimie. 2011;93:1379–1391. doi: 10.1016/j.biochi.2011.05.013. [DOI] [PubMed] [Google Scholar]
- Salvador-Gallego R, Mund M, Cosentino K, Schneider J, Unsay J, Schraermeyer U, Engelhardt J, Ries J, García-Saez AJ. Bax assembly into rings and arcs in apoptotic mitochondria is linked to membrane pores. The EMBO Journal. 2016;35:389–401. doi: 10.15252/embj.201593384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberg B, Wang P, Keeble Já, Rodriguez-Enriquez R, Walker S, Owens TW, Foster F, Tanianis-Hughes J, Brennan K, Streuli Cá, et al. Bax Exists in a Dynamic Equilibrium between the Cytosol and Mitochondria to Control Apoptotic Priming. Molecular Cell. 2013;49:959–971. doi: 10.1016/j.molcel.2012.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T, Ghirlando R, Baber J, Clore GM. Quantitative Resolution of Monomer-Dimer Populations by Inversion Modulated DEER EPR Spectroscopy. ChemPhysChem. 2016;17:2987–2991. doi: 10.1002/cphc.201600726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodinger L. The PyMOL Molecular Graphics System. Version 1.8 2015 [Google Scholar]
- Schwieters CD, Kuszewski JJ, Marius Clore G. Using Xplor-NIH for NMR molecular structure determination. Progress in Nuclear Magnetic Resonance Spectroscopy. 2006;48:47–62. [Google Scholar]
- Schwieters CD, Kuszewski JJ, Tjandra N, Marius Clore G. The Xplor-NIH NMR molecular structure determination package. Journal of Magnetic Resonance. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- Shamas-Din A, Kale J, Leber B, Andrews DW. Mechanisms of Action of Bcl-2 Family Proteins. Cold Spring Harbor Perspectives in Biology. 2013;5 doi: 10.1101/cshperspect.a008714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamas-Din A, Satsoura D, Khan O, Zhu W, Leber B, Fradin C, Andrews DW. Multiple partners can kiss-and-run: Bax transfers between multiple membranes and permeabilizes those primed by tBid. Cell Death and Disease. 2014;5:e1277. doi: 10.1038/cddis.2014.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Youle RJ, Tjandra N. Structure of Bax: Coregulation of Dimer Formation and Intracellular Localization. Cell. 2000;103:645–654. doi: 10.1016/s0092-8674(00)00167-7. [DOI] [PubMed] [Google Scholar]
- Tjandra N, Grzesiek S, Bax A. Magnetic Field Dependence of Nitrogen–Proton J Splittings in 15N-Enriched Human Ubiquitin Resulting from Relaxation Interference and Residual Dipolar Coupling. Journal of the American Chemical Society. 1996;118:6264–6272. [Google Scholar]
- Tjandra N, Omichinski JG, Gronenborn AM, Clore GM, Bax A. Use of dipolar 1H-15N and 1H-13C couplings in the structure determination of magnetically oriented macromolecules in solution. Nature Structural and Molecular Biology. 1997;4:732–738. doi: 10.1038/nsb0997-732. [DOI] [PubMed] [Google Scholar]
- Tsai CJ, Liu S, Hung CL, Jhong SR, Sung TC, Chiang YW. BAX-Induced Apoptosis Can Be Initiated through a Conformational Selection Mechanism. Structure. 2015;23:139–148. doi: 10.1016/j.str.2014.10.016. [DOI] [PubMed] [Google Scholar]
- Volkmann N, Marassi FM, Newmeyer DD, Hanein D. The rheostat in the membrane: BCL-2 family proteins and apoptosis. Cell Death and Differentiation. 2014;21:206–215. doi: 10.1038/cdd.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward R, Bowman A, Sozudogru E, El-Mkami H, Owen-Hughes T, Norman DG. EPR Distance Measurements in Deuterated Proteins. Journal of Magnetic Resonance. 2010;207:164–167. doi: 10.1016/j.jmr.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson MP. Using chemical shift perturbation to characterise ligand binding. Progress in Nuclear Magnetic Resonance Spectroscopy. 2013;73:1–16. doi: 10.1016/j.pnmrs.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nature Reviews Molecular Cell Biology. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.