Abstract
One strategy for improving the throughput of human plasma proteomics discovery analysis while maintaining good depth of analysis is to multiplex using isobaric tags. At present, the greatest multiplexing that is commercially available uses the TMT10plex kit. As an example of this approach, we describe efficient shotgun discovery proteomics of large numbers of human plasma to identify potential biomarkers. In the analysis strategy, a common pooled reference was used to enable comparisons across multiple experiments. Duplicate samples showed excellent overall reproducibility across different TMT experiments. Data filters that improved the quality of individual peptide and protein quantitation included using a filter for purity of the targeted precursor ion in the isolation window and using only unique peptides.
Keywords: Isobaric tag quantitation, TMT10plex, Plasma biomarkers, Proteomics
1. Introduction
Quantitative comparisons of plasma or serum proteomes for discovery of potential clinical biomarkers continues to be of great interest despite substantial challenges in achieving in-depth analysis of samples with adequate throughput. Despite impressive improvements in mass spectrometer performance over the past decade, most plasma discovery strategies require substantial fractionation prior to LC-MS/MS analysis in order to effectively detect low abundance proteins (<100 ng/ml), which is the concentration range of most clinical biomarkers. The major quantification methods applied in shotgun proteomics can be categorized as label-free, metabolic labeling, and isobaric chemical labeling.1 In comparison with label-free quantification, stable isotopic labeling approaches make it possible to multiplex samples: that is, to analyze multiple samples in the same LC–MS/MS run to provide direct comparisons. Metabolic labeling methods, such as stable isotope labeling by amino acids in cell culture (SILAC),2 are very tolerant of variations in any processing steps because samples to be compared can be mixed immediately upon sample collection. A moderate level of multiplexing can be achieved by combining two or three differentially-labeled samples but this also increases the total peptide sample complexity by two- or three-fold respectively, which decreases overall depth of analysis. Regardless, such methods are not feasible for analysis of human plasma. In contrast, chemical labeling methods are sensitive to any variations that may occur prior to and during the labeling step, which is typically performed after protease digestion. The chemical labeling approach that is most widely used is isobaric tags because they have the dual advantage that fractionation and LC-MS/MS analysis can be substantially multiplexed without greatly increasing peptide complexity.
A number of different isobaric labeling reagents have been introduced over the past decade with isobaric tag for relative and absolute quantification (iTRAQ)3 and tandem mass tag (TMT)4 being the most popular ones. These two isobaric labeling reagents have very similar molecular structures which consist of an amine-specific reactive group, a mass reporter group for quantification, and a mass normalizer group to link the reactive and reporter groups and balance the total masses prior to fragmentation. The reactive group employed in these reagents is an N-hydroxysuccinimide ester which reacts with primary amines, i.e., unblocked N-terminals and lysine side chains. The reporter groups are partially fragmented from the peptide during precursor fragmentation in the mass spectrometer. Because each reagent in a multiplex kit has a reporter with a different mass, peptides from different biological samples are readily quantified according to the reporter ion intensities. The mass normalizer group ensures that the peptide complexity in the MS1 spectra does not increase with multiplexing.
Since the initial introduction of TMT reagents, this labeling strategy has undergone modifications to improve accurate quantification and extent of multiplexing capacity. For example, Dayton et al.5 expanded the number of quantification channels to make a 6plex version by incorporating different numbers of 13C atoms in the reporter ion group. Specifically, the 6plex TMT reagent produced a series of six different reporter ions with nominal masses from 126 to 131 Da at 1 Da intervals. Subsequently, McAlister et al.6 and Werner et al.7 both expanded the reagents to 8plex. In their design, they made very similar reporter ions that differed by 0.0063 Da by replacing one 13C with a 15N on the TMT 127 and 129 Da reporter groups. This took advantage of capacities of current high end mass spectrometers that have sufficient resolution and mass accuracy to resolve such small mass differences. Viner et al.8 applied the same 15N replacement strategy to the 128 and 130 Da channels to extend the TMT multiplexing capacity to its current 10plex version, and made this a reliable commercial kit. In addition, Everley et al.9 extended the reagents to 18plex based on the 6plex version. The 18plex reagents included the original 6plex reagents, 6plex medium TMT reagents by inserting an N-Ethylformamide group in the mass normalizer group, and 6plex heavy TMT reagents by inserting an N-propylformamide group in the mass normalizer group. Another type of 18plex method was also proposed by Everley et al.9 by combining TMT6plex with triple labeling SILAC. Furthermore, a 54plex TMT method was designed and demonstrated by combining the proposed two types of 18plex methods.
The method described in this protocol uses commercial TMT10plex reagent kits to compare a relatively large number of plasma samples that requires multiple sets of 10plex samples with quantitative comparisons across 10plex experiments. As an example, shotgun proteome analysis was conducted of plasma samples from breast cancer patients in efforts to identify potential cardiotoxicity biomarkers caused by therapeutic treatment. Currently, a commonly used and highly effective breast cancer treatment combines doxorubicin and trastuzumab (Herceptin®);10 however, the adverse effects of cardiotoxicity become a major issue as up to 18% of patients develop cardiac dysfunction.11 Since current cardiovascular biomarkers lack sufficient specificity and sensitivity for detection of onset of cardiotoxicity in cancer patients receiving these therapies, it is important to discover better markers both for doctors to make decisions and for researchers to uncover the disease mechanisms.
Our protocol starts with depleting 20 abundant human plasma proteins on an immunoaffinity depletion column. The depleted plasma are then reduced by dithiothretiol, alkylated by iodoacetamide, and in-gel digested by trypsin. The digested peptides are labeled with TMT10plex reagents and combined. In order to increase the depth of analysis, the combined peptides are fractionated by high pH HPLC into 20 fractions. The fractions are analyzed by LC-MS/MS. The raw data files are searched with MaxQuant software.
2. Materials
2.1. FPLC Affinity Depletion of Plasma
2.1.1. Human plasma.
2.1.2. Microcentrifuge tube with 0.22 μm filter.
2.1.3. HPLC or FPLC system capable of operating at low pressure (< 30 psi) with automatic sample collector.
2.1.4. Equilibration buffer: 1× phosphate buffered saline (PBS).
2.1.5. Elution buffer: 0.1 M glycine and 0.1% (w/v) octyl-β-glucopyranoside (OGP) adjusted to pH 2.5 with HCl.
2.1.6. ProteoPrep® 20 Immunodepletion Column (Sigma-Aldrich).
2.2. Ethanol Precipitation
2.2.1. Unbound fraction from ProteoPrep® 20 LC depletion.
2.2.2. Ethanol (200 proof, −20°C).
2.2.3. SpeedVac® centrifuge (Thermo Scientific).
2.3. Reduction and Alkylation of Samples Prior to 1D SDS-PAGE
2.3.1. Depleted and ethanol-precipitated pellet of human plasma.
2.3.2. Protein resuspension buffer: 1% (w/v) SDS buffer solution containing 50 mM Tris-HCl, pH 8.0.
2.3. 1 M aqueous dithiothreitol.
2.4. 0.5 M iodoacetamide in 50 mM Tris-HCl, pH 8.0.
2.5. 37°C thermostatically controlled incubator/shaker.
2.4. 1D Sds-Page
2.4.1. Reduced and alkylated protein sample.
2.4.2. 2 × Protein solubilizing buffer: 0.4 M sucrose, 6% (w/v) SDS, 125 mM Tris-HCl 4 mM Na2EDTA, 2% (v/v) 2-mercaptoethanol, and 2% (v/v) saturated bromophenol blue solution, pH 8.0.
2.4.3. 1-D SDS-PAGE gel (e.g., NuPAGE® Bis-Tris Mini Gels, 1 mm, 10 wells).
2.4.4. Running buffer: 3-(N-morpholino)propanesulfonic acid (MOPS, 50 mM) SDS.
2.4.5. XCell SureLock™ Mini-Cell (Invitrogen).
2.4.6. Heat block set to 90°C.
2.4.7. BenchMark™ (Invitrogen) molecular weight marker.
2.4.8. Novex® Colloidal blue staining kit (Invitrogen) containing Stainer A and Stainer B.
2.4.9. Fixing solution: 50% (v/v) methanol, with 10% (v/v) acetic acid in water.
2.4.10. Staining solution: 20% (v/v) methanol, with 20% (v/v) Stainer A in water.
2.4.11. Staining trays.
2.5. In-Gel Trypsin Digestion
2.5.1. PCR laminar flow hood with a HEPA filter and a lightbox.
2..2. 0.1% (v/v) trifluoroacetic acid (TFA), 50% (v/v) methanol in water.
2.5.3. 96-well V-bottomed pierced plate and storage plates with polystyrene plate covers.
2.5.4. Gel-cutting device: e.g., MEG-1.5 Gel Cutter (The Gel Company) or stainless steel razor blades.
2.5.5. SpeedVac centrifuge equipped with 96-well plate centrifuge rotor.
2.5.6. 37°C thermostatically controlled incubator/shaker.
2.5.7. Destain solution: 50% (v/v) acetonitrile in 50 mM aqueous 4-(2-hydroxyethyl)-1 piperazineethanesulfonic acid (HEPES), pH 8.5.
2.5.8. Sequencing grade-modified trypsin (Promega)
2.5.9. Trypsin working solution: 0.02 μg/μL trypsin in 50 mM aqueous HEPES, pH 8.
2.5.10. Trypsin wash buffer: 50 mM aqueous HEPES.
2.6. Tryptic Peptides Desalting
2.6.1. SpeedVac® centrifuge.
2.6.2. Spectrafuge™ 16M microcentrifuge (Labnet).
2.6.3. MacroSpin™ column (30 – 300 μg sample capacity, 50 – 150 μL elution volume).
2.6.4. Conditioning solvent: acetonitrile.
2.6.5. Loading buffer: 1% (v/v) aqueous TFA.
2.6.6. Equilibration buffer: 5% (v/v) acetonitrile in 1% (v/v) aqueous TFA.
2.6.7. Releasing buffer A: 50% (v/v) acetonitrile in 0.1% (v/v) aqueous formic acid.
2.6.8. Releasing buffer B: 80% (v/v) acetonitrile in 0.1% (v/v) aqueous formic acid.
2.7. Peptide TMT10plex Labeling
2.7.1. TMT10plex™ isobaric label reagent set (available in either 0.8 mg or 0.2 mg aliquots per label).
2.7.2. Resuspend buffer: 200 mM aqueous HEPES.
2.7.3. Anhydrous acetonitrile.
2.7.4. Quench buffer: 5% (w/v) hydroxylamine.
2.8. Labeled Peptide Mixing Ratio Checking
No additional materials are needed for the corresponding step in methods.
2.9. High pH Reverse Phase HPLC Fractionation
2.9.1. 1100 HPLC platform (Agilent).
2.9.2. 2.1 × 10 mm Xbridge™ C18 guard column (Waters).
2.9.3. 2.1 × 250 mm Xbridge™ BEH300 C18 column (Waters).
2.9.4. Buffer A: 10 mM ammonium formate aqueous solution (pH 10).
2.9.5. Buffer B: 10 mM ammonium formate in 80% (v/v) acetonitrile (pH 10).
2.9.6. 1.5 mL micro centrifuge tube with push cap.
2.10. Lc-Ms/Ms
2.10.1. Q Exactive™ Plus mass spectrometer (Thermo Scientific) coupled to an nanoACQUITY UPLC system (Waters) using a nanospray ion source.
2.10.2. Symmetry trap column (180 μm i.d. × 2 cm packed with 5 μm C18 resin; Waters).
2.10.3. BEH C18 nanocapillary analytical column (75μm i,d, × 25 cm, 1.7 μm particle size, Waters).
2.10.4. Solvent A: 0.1% (v/v) aqueous formic acid.
2.10.5. Solvent B: acetonitrile containing 0.1% (v/v) formic acid.
2.11. Data Processing and Analysis
2.11.1. MaxQuant software.
3. Methods
3.1. FPLC Affinity Depletion of Plasma or Serum
Plasma depletion using a ProteoPrep® 20 column that removes 20 abundant proteins (∼97% of total plasma protein) is described. The ProteoPrep® 20 LC column requires a low pressure HPLC or FPLC system because its packing has a pressure limit of 30 psi, which is much lower than the minimum operating pressure for most regular HPLC systems (typical 100 psi or higher). More or less plasma proteins can be depleted by using alternative immunoaffinity columns (see for example the Chapter by Beer et al. in this volume).
3.1.1. Thaw and filter plasma samples through a 0.22 μm microcentrifuge tube. Keep the filtered plasma on ice before injection onto the depletion column.
3.1.2. Before connecting the depletion column, flush the system with equilibration buffer for 15 min at 3 mL/min to remove any trapped air.
3.1.3. Immediately before usage, take the ProteoPrep® 20 LC column from storage at 2 − 8°C to equilibrate at room temperature for 15 min.
3.1.4. Connect the column to the system while the system flushing with equilibration buffer at 0.5 mL/min. Avoid introducing gas into the column.
3.1.5. It is recommended to run a blank before injection of the first sample of the day.
- 3.1.6. Inject 80 – 100 μL filtered plasma sample at 0.3 mL/min. The 60 min HPLC gradient is as follows:
- 100% equilibration buffer for 30 min at 0.3 mL/min.
- 100% elution buffer for 15 min at 3 mL/min.
- 100% equilibration buffer for 15 min at 0.3 mL/min.
3.1.7. Collect the eluent into pre-cleaned 10 mL polystyrene test tubes using the fraction collector. Switch test tubes every 7.5 min for the first 30 min and then every 2 min thereafter.
3.1.8. Unbound proteins are collected in three test tubes from 7.5 to 30 min. Combine the unbound proteins into a pre-cleaned 50 mL centrifuge tube. Keep the proteins on ice.
3.2. Ethanol Precipitation
Ethanol precipitation of proteins is an easy and efficient way to remove salts and detergent. The ethanol used in the precipitation should be of high quality, pre-cooled at −20°C, and added quickly for efficient precipitation.
3.2.1. Quickly add ∼9 fold volumes of ethanol (200 proof, −20°C,) relative to unbound fraction volume and vortex thoroughly.
3.2.2. Incubate at 0°C overnight.
3.2.3. Centrifuge for 25 min at 3500 rpm (g-force 3480) at 4°C.
3.2.4. Remove the ethanol supernatants carefully and leave the intact pellet. Dry the pellet carefully by blowing a gentle stream of argon across the surface.
3.2.5. Store the pellet at −20°C for future use or immediately precede with next steps.
3.3. Reduction and Alkylation of Samples Prior to 1D SDS-PAGE
3.3.1. Thaw ethanol-precipitated depleted plasma pellet.
3.3.2. Resuspend pellet in 100 μL resuspension buffer.
3.3.3. Reduce the proteins by adding dithiothreitol to a final concentration of 20 mM and incubate for one hour at 37°C with shaking.
3.3.4. Alkylate the proteins by adding iodoacetamide to final concentration of 60 mM and incubate for one hour at 37°C in dark with shaking.
3.3.5. Quench the alkylation by adding additional dithiothreitol to the sample solutions with the final concentration reaching 50 mM. Incubate for 15 min at 37°C with shaking.
3.4. 1d Sds-Page
This protocol uses a short SDS gel to clean up and digest samples. An alternative approach is to ethanol precipitate the alkylated protein and perform a solution trypsin digestion (see Note 1).
3.4.1. Mix the reduced and alkylated plasma proteins with 2 × solubilizing buffer.
3.4.2. Heat the mixtures at 90°C for 2 min.
3.4.3. Assemble the gel in the XCell SureLock™ Mini-Cell unit and fill the chambers with running buffer. Load samples into each lane with BenchMark™ molecular weight marker in the first lane.
3.4.4. Run gels at constant 200 V. Stop when the dye front has migrated ∼ 0.5 cm.
3.4.5. Disassemble the gel unit and transfer the gel to a plastic container.
3.4.6. Fix the gel by adding 100 mL fixing solution. Shake gently for 10 min. Discard the fixing solution carefully.
3.4.7. Stain the gel by adding 95 mL staining solution. Shake gently for 10 min. Add 5 mL Stainer B to the staining solution. Shake gently for another 3 – 12 h. Discard the staining solution carefully.
3.4.8. Destain the gel with water till the gel shows a clear background.
3.5. In-Gel Trypsin Digestion
3.5.1. Turn on the fan in the PCR hood at least 15 min before doing any experiment to achieve optimal flow of dust-free air.
3.5.2. Excise the entire stained area (∼ 0.5 cm) from each gel lane of interest and cut it into 6 vertical slices. Transfer the slices into 2 wells of a pre-cleaned, pierced, 96-well plate (3 slices each well to avoid excessive gel volume per reaction).
3.5.3. Destain the gel slices by adding 50 μL destaining solution per well (see Note 2). Incubate for 15 min at 37°C with shaking. Centrifuge the plates for 1 min to remove the buffer. Repeat the destaining step until the gels appear light blue and white.
3.5.4. Dry the gel slices using a SpeedVac® evaporator for at least 30 min.
3.5.5. Re-hydrate gel slices by adding 45 μL trypsin working solution. Incubate at 37°C for 16 – 18 h in a thermostatically controlled incubator and another 15 min at room temperature. Collect the digested protein extract into a clean, 96-well collecting plate by centrifuging for 1 min.
3.5.6. Add 25 μL trypsin wash buffer per well to the 96-well pierced plate. Incubate at 37°C for 30 min and another 15 min at room temperature. Collect the second extract into the same 96-well collecting plate by centrifuging for 1 min.
3.5.7. Transfer digested extracts into pre-cleaned 0.5 mL centrifuge tubes and combine 2 separately digested sample halves.
3.6. Tryptic Peptide Desalting
3.6.1. Lyophilize the tryptic peptides using a SpeedVac® evaporator.
3.6.2. Resuspend the dried tryptic peptides in 100 μL loading buffer.
3.6.3. Condition the MacroSpin™ Column by pipetting 400 μL conditioning solvent into the column and centrifuging it for 1 min at ∼110 × g. Flush the column by pipetting 400 μL water into the column and centrifuging it for 1 min. Repeat the flush once.
3.6.4. Load the resolubilized peptides onto the column. Centrifuge it for 1 min at ∼110 × g. Pipette 200 μL loading buffer into the column and centrifuge it for 1 min. Repeat once (see Note 3).
3.6.5. Pipette 200 μL equilibration buffer into the column and centrifuge it for 1 min. Repeat once (see Note 3).
3.6.6. Replace the collecting tube with another pre-cleaned tube. Release the peptides by pipetting 100 μL releasing buffer A into the column and centrifuge it for 1 min. Pipette 100 μL releasing buffer B into the column and centrifuge it for 1 min. Combine the two elutes.
3.6.7. Lyophilize the combined elutes using a SpeedVac® evaporator.
3.7. Peptide TMT10plex Labeling
Studies that involve analysis of more than 10 samples require the use of a reference sample to compare peptide yields across multiple TMT10plex experimental sets. In the example described here, the reference is a pool of all plasma samples in the study, which is assigned to the same reporter ion channel (e.g. 126 Da) in all the experimental sets. It is recommended that at least some samples be replicated across multiple TMT10plex experimental sets to evaluate reproducibility. In this example, we analyzed 41 different plasma samples that were assigned to six TMT10plex experimental sets with 13 duplicated samples. Each experimental set consist of the reference and 9 different plasma samples. Optimal distribution of samples within and across experiments depends upon goals of the study (see Note 4).
3.7.1. Resuspend desalted peptides with resuspension buffer at an estimated concentration of ∼1 μg/μL (see Note 5).
3.7.2. Pool 3 μL of peptides from all 41 plasma samples to form a reference. Adjust the reference volume to 200 μL.
3.7.3. Equilibrate the TMT label reagents at room temperature for 10-15 min with the lid sealed. Resuspend each 0.8 mg TMT label reagent with 42 μL anhydrous acetonitrile (see Note 6). Vortex briefly to make sure the reagents are fully dissolved.
3.7.4. Label peptides by adding 42 μL TMT label reagent to every 100 μL resuspended peptides. Incubate the reaction for 1 h at room temperature (see Note 7).
3.7.5. Quench the reaction by adding 8 μL quench buffer to the reaction. Incubate for 15 min at room temperature.
3.8. Labeled Peptide Mixing Ratio Checking
After isobaric labeling, similar levels of total peptide per sample should be combined. However, recoveries can be variable when equal volumes of plasma are processed as described above. Typically, protein or peptide assays are used to check yields and to ensure mixing of similar amounts of peptides.12 An alternative quantification method is to perform a pilot mixing experiment followed by a single LC-MS/MS run (without peptide fractionation) to check the ratios of total reporter ion intensity in each channel as described below.
3.8.1. Combine 2 μL of each labeled peptide sample containing different tags to be compared in each experimental set.
3.8.2. Desalt the pooled peptides following the same desalting method described in 3.6.
3.8.3. Resuspend the desalted peptides with 40 μL 0.1% (v/v) aqueous FA.
3.8.4. Inject 4 μL resuspended peptide sample (estimated ∼ 0.9 μg, see Note 5) into the LC-MS/MS system and run a two hour gradient (see 3.10 for LC-MS/MS method details).
3.8.5. Search the resulting LC-MS/MS raw file using MaxQuant (see 3.11 for MaxQuant method details). Sum the total reporter ion intensity per channel, which represents the total amounts of identified peptides in each channel (see Note 8).
3.8.6. Calculate adjusted volumes of labeled peptides by dividing each reporter ion intensity by the average intensity and use these correction factors for a second pilot experiment. Combine 1 – 5 μL of each labeled sample to achieve equal total reporter ion intensities. Repeat the LC-MS/MS analysis following 3.8.2 – 3.8.5. If all reporter ion channels show similar total intensities in the second check, apply these adjusted mixing ratios to the bulk samples. If reporter ion channels continue to still show large variations (> ±30%), repeat this step (see Note 9) prior to preparing the bulk pooled multiplexed sample.
3.9. High pH Reverse Phase HPLC Fractionation
In simple to obtain in depth analysis for the plasma proteome, it is necessary to apply two dimensional HPLC separation. In addition to the low pH reversed phase LC-MS/MS analysis, digested peptides are usually fractionated by another peptide separation method, such as high pH reversed phase HPLC, strong cation exchange (SCX) chromatography, electrostatic repulsion hydrophilic interaction chromatography (ERLIC), etc. We chose high pH reversed phase HPLC as it yields the highest resolution separation of tryptic peptides,13 is easy to perform, and uses MS friendly volatile solutions that eliminate the need for an extra desalting step prior to LC-MS/MS analysis.
3.9.1. Desalt the bulk pooled sample following the desalting method described in 3.6 (see Note 10).
3.9.2. Resuspend the peptides with 100 μL buffer A.
3.9.3. Set the flow rate at 0.2 mL/min and equilibrate the column with 5% buffer B.
- 3.9.4. Inject and separate the resuspended 100 μL sample using an HPLC gradient is as follows:
- Hold at 5% buffer B for 8 min.
- 5 – 24% buffer B over 7 min.
- 24 – 50% buffer B over 52 min.
- 50 – 55% buffer B over 7 min.
- 55 – 60% buffer B over 5 min.
- 60 – 90% buffer B over 1 min.
- Hold at 90% buffer B for 15 min.
- Return to 5% buffer B over 0.5 min.
- Re-equilibrate at 5% buffer B for 10 min.
3.9.5. Collect HPLC elutes into 1.5 mL micro centrifuge tubes from 8 to 95 min. Switch collection tube every 1 min. After collection, consolidate the 1 min aliquots into 20 fractions in a checkerboard manner by pooling every 20th fraction; e.g., 1+21+41+61+81; 2+22+42+62+82; etc.
3.9.6. Acidify pooled fractions using FA to a final pH=3. Lyophilize the fractions using a SpeedVac® evaporator.
3.10. Lc-Ms/Ms
Each pooled fraction from the high pH reversed phase HPLC separation is analyzed individually with a 150 min LC-MS/MS gradient. In simple to obtain good quantification results from the reporter ions, high resolution mass spectrometers are necessary to clearly resolve the reporter ions with very similar masses. A fast scan speed is also essential in simple to achieve a good depth of analysis. The study described here was performed using a Thermo Q Exactive™ Plus mass spectrometer.
3.10.1. Set the flow rate at 250 nL/min and equilibrate the column with 5% solvent B.
3.10.2. Resuspend the dried fractions with solvent A. Inject 4 – 8 μL (see Note 5) of each fraction into the LC-MS/MS system. Trap the loaded fractions with the trapping column for 5 min at isocratic 0% solvent B with 6 μL/min flow rate.
- 3.10.3. The 150 min UPLC gradient is as follows:
- 5 – 28% solvent B over 120 min.
- 28 – 40% solvent B over 5 min.
- 40 – 90% solvent B over 10 min.Hold at 90% solvent B for 10 min.
- Return to 5% solvent B over 2 min.
- Re-equilibrate at 5% solvent B for 5 min (see Note 11).
3.10.4. The following parameters are used for MS/MS data acquisition and have been optimized for downstream analysis.
3.10.4.1. Nanospray ion source: 2.5 kV spray voltage and 300°C lens temperature.
3.10.4.2. Scan mode: Full MS / dd-MS2 (TopN).
3.10.4.3. Full scan: 400 – 2000 m/z range with 70,000 resolution, 3×106 automatic gain control (AGC) target and 50 ms maximum injection time (IT).
3.10.4.4. MS/MS scan: top 20 (+1 charge and unassigned ions excluded) selection mode, high-energy collisional dissociation, 1.2 m/z isolation window (see Note 12), 32 normalized collision energy (NCE), first mass fixed at 115 m/z scan range (see Note 13), 35,000 resolution (see Note 14), 1×106 AGC target, 120 ms maximum IT, 5% underfill ratio, and 30 s dynamic exclusion.
3.10.5. Monitor the instrument performance by analyzing standard yeast digests before and after the experiment. For long term experiments also perform a yeast digest QC run approximately every 48 hrs.
3.11. Data Processing and Analysis
A number of different software tools can be used to analyze TMT data. Two programs tested in our lab, are MaxQuant14 and Proteome Discoverer (Thermo Scientific). Proteome Discoverer has a user-friendly visual interface and the latest version of Proteome Discoverer (v2.1) added a number of functions to enhance the TMT data analysis. MaxQuant has the advantage that it is freely available, frequently updated and can use many processors in parallel for fast data-processing. In the example described here, we used MaxQuant for data processing and analysis.
3.11.1. Import all raw mass spectrometric data files into the same MaxQuant session. Define each experimental set and fractions and process them together.
3.11.2. Select the 10plex TMT reporter ion MS2 mode as the searching mode. Set reporter mass tolerance small enough to be able to distinguish the closest reporter ions (see Note 14). Check Filter by PIF and set min. reporter PIF as 0.75 (see Note 12).
- 3.11.3. The following parameters are used for the MaxQuant database search:
- 3.11.3.1. Carbamidomethyl group on cysteine as fixed modification.
- 3.11.3.2. Acetyl group on protein N-terminal and oxidation on methionine as variable modification.
- 3.11.3.3. Trypsin/P as digestion mode with the maximum missed cleavages at 2.
- 3.11.3.4. Uniprot human database appended with common expected contaminants including keratins and trypsin.
- 3.11.3.5. False discovery rate (FDR) set to 0.01 for proteins and peptides.
- 3.11.4. After the search is complete, filter out contaminants, reverse hits, and proteins only identified by site (see Note 15).
- 3.11.5. Based on the assumption that each sample should have equal amounts of total protein, the data is then normalized based on the total reporter ion intensity in each channel of each experimental set to correct for variations in total yield. For each channel, the total reporter ion intensities are divided by the total number of identified proteins to get average intensities for that channel. To normalize, each protein reporter ion intensity is divided by the average protein intensity of that channel (see Note 16).
- 3.11.6. Individual protein values are then normalized across experimental sets by dividing each protein reporter ion intensity by the reference reporter intensity in that experimental set (see Note 17).
4. Notes
4.1. To perform the in-solution digestion, depleted plasma is resuspended in 120 μL 8 M urea aqueous solution containing 50 mM HEPES, pH 8.5. Add 6.3 μL 1 M DTT aqueous solution and incubate 30 min at 37°C for reduction. Add 40 μL 0.5 M iodoacetamide aqueous solution containing 50 mM HEPES and incubate 1 h at 37°C in dark for alkylation. Add 2 μL 1 M DTT aqueous solution and incubate 15 min at 37°C to quench the alkylation. Add 800 μL 50 mM HEPES aqueous solution to dilute the urea to ∼ 1M concentration. Add 1:50 (w/w, E:S) trypsin and incubate overnight to digest the sample. After digestion, the samples are desalted, TMT labeled, combined, fractionated, and LC-MS analyzed using the same procedure as described.
4.2. In TMT labeling experiments, 50 mM HEPES is used as the digestion buffer rather than the commonly used ammonium bicarbonate because the TMT reagent reacts with primary amines such as ammonium ion. Although a desalting step is performed before the TMT labeling, the buffer is not typically completely removed. HEPES is also preferred over the triethylammonium bicarbonate buffer recommended by the vendor because the residual HEPES causes less interference in the subsequent high pH reverse phase separation than the residual triethylammonium bicarbonate.
4.3. Collect and save the eluent and wash fractions during loading and desalting of peptides, so that the peptides can be recovered if they are not well retained on the column for any reason.
4.4. A good experiment design is to randomly assign samples to reporter ion channels and to use different reporter ions for duplicates in different TMT experimental sample sets. Also, if some direct comparisons are more important than other, e.g., a case and matched control, these should be placed in the same experimental set. This is because low abundance proteins may be inconsistently detected and quantitated across different TMT experimental sets due to the somewhat stochastic detection of low abundance peptides in very complex samples. Another consideration is to place similar numbers of cases and controls in each experimental set.
4.5. An optimal peptide load on 75 μm columns should be about 1 – 2 μg. For depleted plasma we estimate the recovery of tryptic peptides as follows: 1) a BCA protein assay is used to quantitate the unbound fraction of a ProteoPrep® 20 LC column (typically approximately 400 μg recovered protein/80 μl human plasma); 2) a 50% recovery from the in-gel digest is assumed (this is based on recoveries we typically observe in digestions of standard proteins); 3) losses during TMT labeling, sample cleanup and high pH reverse phase separation are assumed to be low and are not corrected, and 4) the distribution of total peptides among fractions from the high pH separation is assumed to be equal.
4.6. TMT reagents are water-sensitive and have limited stability in solution. Therefore it is important to dissolve the reagents with dehydrated acetonitrile to avoid degrading the reagent before labeling. Because of the water-sensitivity, it is also better to use the entire vial of reagent at one time. If the entire vial is not needed, lyophilize the rest of the reagent and store at −20°C. For smaller scale TMT experiments (6-25 μg peptides per reaction), one can now purchase the reagents in 0.2 mg quantities.
4.7. The TMT reagent from a 0.8 mg vial (42 μL of solution in acetonitrile) can label up to 100 μg of total peptides (100 μL at about 1 μg/μL). Because current mass spectrometers are very sensitive and only about 1 μg of multiplexed peptides is injected per LC-MS/MS run, a 0.8 mg vial can be used to label 2, 3, or 4 different samples. In the experiments described here, we divided the 42 μL reagent solutions into 3 × 14 μL and labeled three different plasma samples (33 μg each). For the pooled reference, two vials for a total of 82 μL of TMT-126 label reagent was used to label 200 μL pooled reference solution. In future experiments we will label 20-25 μg per sample using the new smaller aliquots of reagent (see note 5).
-
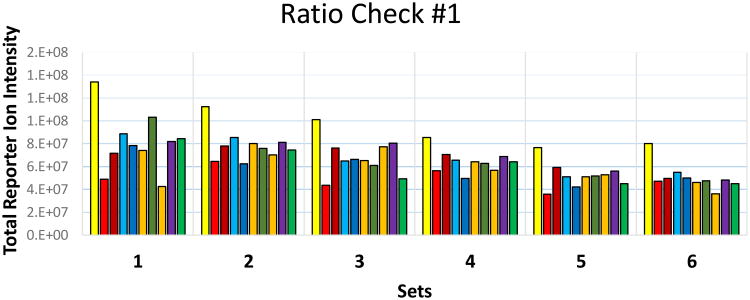
4.8. When a study involves a large number of samples and multiple TMT10plex experimental sets, the first ratio check and adjustment usually does not fully correct for variable yields within and across experimental sets. Figure 1 shows the first ratio check results for the illustrated experiment using 6 TMT10plex experimental sets. It clearly shows large reporter ion intensity variations between different reporter ion channels in the same experimental set and the reference channel (yellow bars) varies substantially across experiments. This illustrates the importance of performing a pilot mixing experiment before committing the bulk samples.
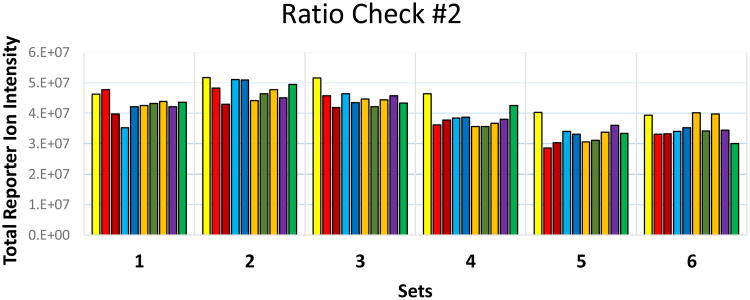
4.9. The second ratio check (Figure 2) showed similar report intensities across all samples, so no further pilots were performed. Specifically, in each experiment set, the lowest yielding reporter ion channel and highest yielding reporter ion channel differ by less than 30%. However, the volumes used to pool the bulk samples were further adjusted based on the relative yields observed in the second ratio check.
4.10. It is recommended that samples should always be desalted after combining the TMT tagged samples. Although high pH reversed phase HPLC should be able to separate the excess TMT reagents and salts/buffers from the labeled peptides, we have observed interference from reagents and buffers with the peptide separation and the subsequent LC-MS/MS runs.
4.11. It is recommended that a blank be run between each sample to minimize carryover between samples. We use a rapid blank gradient that takes a total of ∼ 25 min.
4.12. In large scale or highly complex shotgun proteomics analysis, a general challenge is that the co-elution of peptides with similar mass are co-isolated and co-fragmented.15 This type of co-elution is most detrimental for isobaric tag quantitations that rely on reporter ions detected in MS2 spectra because most peptides in an experiment will be present at similar levels in all samples. Hence, most commonly, a peptide that is differentially abundant in different samples will be co-isolated with a peptide that is similar across all samples. This skews and compresses the calculated ratios on which quantitations are based. In simple to reduce the co-isolation interference, Ting et al. recommended that an additional isolation and fragmentation of a selected high-intensity MS2 fragment ion be performed. This will usually circumvent the co-isolation problem in the MS3 spectra and therefore quantitation is based on the ratios of reporter ions in this spectra which efficiently ignores the co-fragmented peaks in the MS2.16Alternatively, Wenger et al. recommended using proton-transfer ion-ion reactions (PTRs) to reduce the charge states of the isolated ions, which might separate the precursor species from other co-isolated peptides within a fairly large m/z range and provide higher purity isolation and cleaner fragmentation in further MS3 analysis.17 Although these reported novel methods usually efficiently reduce the co-isolation interference, they require a mass spectrometer capable of performing high resolution MS3 measurements. Many currently available instruments, including the Q Exactive Plus mass spectrometer that was used in our study, cannot perform MS2 isolation and fragmentation. Hence, in our case, two other approaches were applied to reduce the co-isolation interference. First, a narrow isolation window of 1.2 m/z was set in MS/MS experiments compared with the 2.0 m/z isolation window that we would typically use. It has also been reported that further narrowing the isolation window from 2.0 to 0.5 m/z could further reduce the interference effect;16 however, the narrow isolation would also result in fewer identifications. Therefore, the 1.2 m/z isolation window is a reasonable compromise. Second, a post-acquisition filter of precursor ion fraction (PIF) of 75% was set, as has been previously recommended.17 PIF is defined as the fraction of the total ion intensity within the isolation window that is contributed by the targeted precursor ion with the range from 0 to 1. In MaxQuant search reports, the PIF was determined based on the peak list in the isolation window from the closest full scan of the tandem mass spectrum. It was previously reported that setting the PIF filter at 75% could efficiently improve overall quantification.17
4.13. Due to the fact that TMT reporter ions fall in the range from 126 to 131 m/z, it is acceptable as long as the MS/MS scan range covers the TMT reporter ion range. Therefore, it is a good strategy to fix the low end of the scan range and make it smaller than m/z 126 so the scan range can cover all TMT reporter ions. We chose m/z 115 in our experiment. Other setting should be acceptable as well provided that they are less than 126 Da.
4.14. In TMT10plex experiment, four pairs of reporter ions are very similar in mass, which are TMT-127N at 127.124760 Da and TMT-127C at 127.131079 Da; TMT-128N at 128.128114 Da and TMT-128C at 128.134433 Da; TMT-129N at 129.131468 Da and TMT-129C at 129.137787 Da; and TMT-130N at 130.134822 Da and TMT-130C at 130.141141 Da. For these four pairs of reporter ions, the mass difference is only the difference between a 13C atom and a 15N atom, which is 0.006319 Da (∼50 ppm). Therefore, it is very important for the instrument to resolve the close reporter ion peaks to achieve good quantification. A minimum resolution of at least 30,000 in the MS2 scan has been reported6 to be required for adequate separation of these similar reporter ions.Although higher resolution provides better quantification, this takes more transient time when using an orbitrap. More transient time for a single MS/MS acquisition reduces the total number of peptides that can be analyzed for a given LC gradient length. To balance resolution and transient time, a resolution of 35,000 is a relatively optimized parameter, which could adequately separate the similar reporter ion peaks while maintaining a reasonable duty cycle. The similar reporter ions also affect data processing. In the database search setup, the reporter ion mass tolerance has to be set to <0.006319 Da. In our experiment, we set the reporter mass tolerance to be 0.003 Da.
4.15. The proteins only identified by site are these proteins identified only by a modification site. In general, these protein identifications or lower confidence assignments and should be eliminated from quantification.
4.16. TMT reporter ion intensities quantify the relative intensities of a given protein across the different samples analyzed in a single multiplexed experiment. In our study, we used relative quantifications based on the assumption that every plasma sample should have the same overall protein amounts, which means the total protein intensities for every TMT label channel should be the same in theory. Although the protein intensities were similar among different channels as a result of our pilot LC-MS/MS runs to check reporter ion yield across samples, they were adjusted to exactly the same level. An alternative strategy this is not shown here is to base relative protein amounts across samples on the same plasma volume. For this approach, if equal volumes of all samples are processed identically and recoveries prior to mixing the TMT tagged samples are expected to be constant, the pilot mixing experiments and this normalization should not be used. Instead, equal volumes of each tagged sample should be mixed and only the internal normalization to the common reference should be used (see 4.17)
4.17. In simple to compare the plasma proteins across different experimental sets, the protein intensities need to be further normalized according to the reference channel that is common to all experiments as it is a single pooled plasma sample that has been tagged in a single modification experiment with the 126 reporter ion. This normalization was performed by dividing every individual protein intensity by the corresponding protein intensity in the TMT reference channel (TMT-126 channel in our study) from the same experiment. By performing scaling and normalization, the protein intensities from different TMT label channels were able to be directly compared across the entire study.
Figure 1.
Results of the first pilot ratio check for a study involving six TMT10plex experimental sets. The total intensity of each reporter ion channel is shown after 2 μL of each differentially tagged sample were combined and analyzed in a single LC-MS/MS run. Reporter ions are color-coded from lowest (yellow=126 Da) to highest mass from left to right respectively for each experimental set.
Figure 2.
Results of the second pilot ratio check for a study involving six TMT10plex experimental sets. The total intensity of each reporter ion channel is shown after 1 to 5 μL of each differentially tagged sample were combined and analyzed in a single LC- MS/MS run. The volumes combined were adjusted based upon the data in Figure 1. Reporter ions are color-coded from lowest (yellow=126 Da) to highest mass from left to right respectively for each experimental set.
Acknowledgments
This work was supported by NIH Grants RO1HD076279, RO1CA131582, and WW Smith Charitable Trust Grants H1205 and H1305 (D.W. Speicher), PA Department of Health Commonwealth Universal Research Enhancement (CURE) Program Grant (B. Ky) as well as CA10815 (NCI core grant to the Wistar Institute).
References
- 1.Zhou Li, Rachel MA, C K, H GB, H RL, P C. Systematic Comparison of Label-Free, Metabolic Labeling, and Isobaric Chemical Labeling for Quantitative Proteomics on LTQ Orbitrap Velos. Journal of Proteome Research. 2012;11(3):1582–1590. doi: 10.1021/pr200748h. [DOI] [PubMed] [Google Scholar]
- 2.Ong SE, B B, K I, K DB, S H, P A, M M. Stable Isotope Labeling by Amino Acids in Cell Culture, SILAC, as a Simple and Accurate Approach to Expression Proteomics. Molecular & Cellular Proteomics. 2002;1(5):376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 3.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Multiplexed Protein Quantitation in Saccharomyces cerevisiae Using Amine-reactive Isobaric Tagging Reagents. Molecular & Cellular Proteomics. 2004;3(12):1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 4.Thompson A, Schäfer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Hamon C. Tandem Mass Tags: A Novel Quantification Strategy for Comparative Analysis of Complex Protein Mixtures by MS/MS. Analytical Chemistry. 2003;75(8):1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 5.Dayon L, Hainard A, Licker V, Turck N, Kuhn K, Hochstrasser DF, Burkhard PR, Sanchez JC. Relative Quantification of Proteins in Human Cerebrospinal Fluids by MS/MS Using 6-Plex Isobaric Tags. Analytical Chemistry. 2008;80(8):2921–2931. doi: 10.1021/ac702422x. [DOI] [PubMed] [Google Scholar]
- 6.McAlister GC, Huttlin EL, Haas W, Ting L, Jedrychowski MP, Rogers JC, Kuhn K, Pike I, Grothe RA, Blethrow JD, Gygi SP. Increasing the Multiplexing Capacity of TMTs Using Reporter Ion Isotopologues with Isobaric Masses. Analytical Chemistry. 2012;84(17):7469–7478. doi: 10.1021/ac301572t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Werner T, Becher I, Sweetman G, Doce C, Savitski MM, Bantscheff M. High-Resolution Enabled TMT 8-plexing. Analytical Chemistry. 2012;84(16):7188–7194. doi: 10.1021/ac301553x. [DOI] [PubMed] [Google Scholar]
- 8.Viner R, Bomgarden R, Blank M, Rogers J. Increasing the Multiplexing of Protein Quantitation from 6- to 10-Plex with Reporter Ion Isotopologues. PN_AMAS_W617_RViner_R1. 2013 [Google Scholar]
- 9.Everley RA, Kunz RC, McAllister FE, Gygi SP. Increasing Throughput in Targeted Proteomics Assays: 54-Plex Quantitation in a Single Mass Spectrometry Run. Analytical Chemistry. 2013;85(11):5340–5346. doi: 10.1021/ac400845e. [DOI] [PubMed] [Google Scholar]
- 10.Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M, Baselga J, Norton L. Use of Chemotherapy plus a Monoclonal Antibody against HER2 for Metastatic Breast Cancer That Overexpresses HER2. New England Journal of Medicine. 2001;344(11):783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 11.Telli ML, Hunt SA, Carlson RW, Guardino AE. Trastuzumab-Related Cardiotoxicity: Calling Into Question the Concept of Reversibility. Journal of Clinical Oncology. 2007;25(23):3525–3533. doi: 10.1200/JCO.2007.11.0106. [DOI] [PubMed] [Google Scholar]
- 12.Villen J, Gygi SP. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat Protocols. 2008;3(10):1630–1638. doi: 10.1038/nprot.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao Z, Tang HY, Wang H, Liu Q, Speicher DW. Systematic Comparison of Fractionation Methods for In-depth Analysis of Plasma Proteomes. Journal of Proteome Research. 2012;11(6):3090–3100. doi: 10.1021/pr201068b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.(a) Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. Accurate Proteome-wide Label-free Quantification by Delayed Normalization and Maximal Peptide Ratio Extraction, Termed MaxLFQ. Mol Cell Proteomics. 2014;13(9):2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotech. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 15.Michalski A, Cox J, Mann M. More than 100,000 Detectable Peptide Species Elute in Single Shotgun Proteomics Runs but the Majority is Inaccessible to Data-Dependent LC-MS/MS. Journal of Proteome Research. 2011;10(4):1785–1793. doi: 10.1021/pr101060v. [DOI] [PubMed] [Google Scholar]
- 16.Ting L, Rad R, Gygi SP, Haas W. MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat Meth. 2011;8(11):937–940. doi: 10.1038/nmeth.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wenger CD, Lee MV, Hebert AS, McAlister GC, Phanstiel DH, Westphall MS, Coon JJ. Gas-phase purification enables accurate, multiplexed proteome quantification with isobaric tagging. Nat Meth. 2011;8(11):933–935. doi: 10.1038/nmeth.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]