Abstract
Chemotherapy treatments are considered essential tools to defeat cancer progression and dissemination to improve patients’ quality of life and survival. Although most malignancies initially respond to chemotherapeutic treatments, after an unpredictable period, tumor cells develop mechanisms of resistance to the treatment. Different cell compartments are involved in the mechanism of chemoresistance, and multiple mechanisms can be activated by single cells at different times of the cancer progression. Alteration of drug metabolism, derangement of intracellular pathways’ signaling, cross-talk between different membrane receptors, and modification of apoptotic signaling and interference with cell replication are all mechanisms that the cell uses to overcome the effect of pharmacological compounds.
In this review, we describe different adaptation, mostly at the level of the proteome, which cancer cells use to develop resistance to cancer treatment.
Keywords: Proteomics, drug metabolism, intracellular signaling
During the 1950s, with the beginning of “modern era” of cancer chemotherapy, Goodman and colleagues1 realized that chemical compounds might have an initial drastic effect on tumor growth, but often because of novel molecular adaptation within the malignancy, cancer cells acquire the capability of overcoming the momentary toxic effect of chemotherapeutic agents. In the mid-60s, Frei and colleagues proposed a different therapeutic approach to overcome the development of chemo-resistance.2 They hypothesized that combinations of drugs, each with a distinctive mechanism of action, should overcome the development of mechanism of resistance described with single-agent regimens. Indeed, by combing 4 different agents, they were able to produce long-term remissions in children with acute lymphoblastic leukemia. Since than, scientists and clinicians have experienced rewards and frustrations in developing novel therapeutic strategies to improve patients’ quality of life and survival.2
Although important advancements in the field have been reached, thanks to the development of more effective agents and to a better understanding of the underlying biology, the multifactorial nature of this phenomenon still represents a central inquiry for the development of more successful chemo-therapeutic strategies. Recent studies have demonstrated that, although 90% of breast cancer, and 50% of its metastatic lesions, initially respond to chemotherapeutic treatment, after an unpredictable period, tumor cells develop diverse mechanisms of resistance to the treatment.3
Because of tumors’ heterogeneity (site of origin, stage, and other molecular characteristics) and different adaptation of tumor cells to chemotherapeutic agents, evaluation of successful regimens and improvement of the response rate to chemotherapeutic agents are complicated and sometimes deceptive. Indeed, it is currently common practice to focus on response rate rather than success rate of chemotherapy. Reduction or stabilization of tumor mass, decline of symptoms, and decrease of serum level of specific biomarkers, rather than 5-year overall survival, represent the main goals in treating and monitoring patients’ response to adjuvant and neoadjuvant regimens.
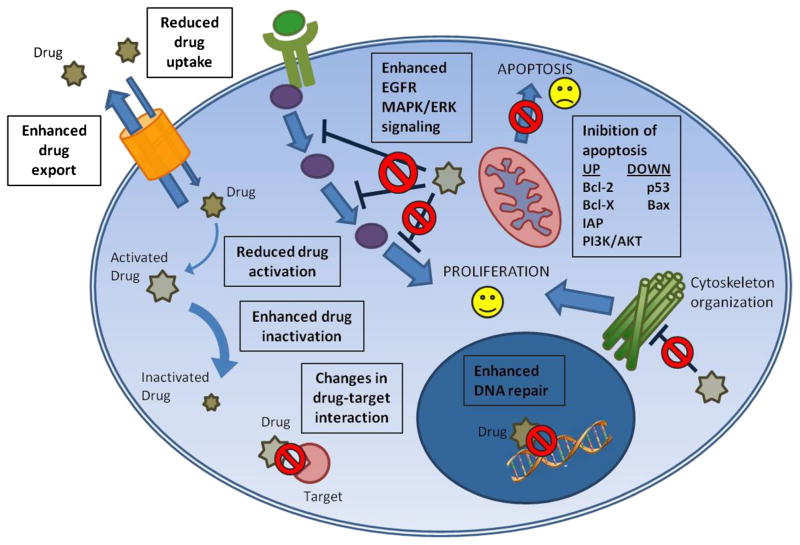
Anatomic characteristic of the tumor, pharmaceutical properties of different compounds, and the interaction between host and drugs are all elements that play a central role in the success of the therapeutic strategies. The neoplastic cells need to constantly readapt to different stimulations to preserve cells survival and tumor homeostasis in unfavorable conditions. To overcome the toxic effects of curative compounds, cancer cells have to continuously develop the capability to implement and strengthen normal physiological functions or to mature de novo mechanisms of resistance against single selected compounds or multiple agents, often apparently unrelated. Nearly any type of chemoresistances is a multifactorial process involving induction of drug-detoxifying mechanism, quantitative and qualitative modification of drug targets, arrest of cell cycle, regulation of DNA replication or reparation mechanisms, and modulation of apoptosis4 (Fig. 1). These modifications are acquired in response to a selection pressure by the drug treatment (acquired resistance) or expressed by cells already resistant and that will never respond to the drug treatment (intrinsic resistance).
FIGURE 1.
Schematic representation of the main cellular mechanisms of chemioresistance. Efficacy of anticancer drugs is driven by multiple factors that go from alteration of drug processing to the regulation of the main pathway involved in cell survival and proliferation. During drug processing, different conditions favor alteration in drug uptake and export. Genetic variants of enzymes involved in drug processing reduce activation or enhance inactivation. Genetic mutations affect drug target genes causing modifications in drug-target interaction. Cancer cells respond to stress conditions by activating feedback pathway that keep them alive, such as enhanced proliferation signal (by epidermal growth factor receptor and mitogen-activated protein kinase/ extracellular single-regulated kinase pathway), deranged DNA repair mechanisms, and inhibition of apoptosis. All of these are mechanisms that counteract drugs’ action, making the cell resistant to them.
Because the response to specific therapeutic regimens induces unpredictable and dynamic cell derangement to overcome the toxic effect of treatment, it is fundamental in oncologic research to discover reliable and early secreted predictive and prognostic biomarkers able to signal novel adaption of the tumor.5 In the past decades, the development of more sophisticated molecular techniques has played a pivotal role in the investigation and understanding of molecular derangement responsible for this phenomenon. In this review, we report the main mechanisms involved in the development of resistance focusing on the role of different proteins and cell compartments in the ontogenesis of chemoresistance.
DRUG METABOLISM
Chemical Drug Modification
Metabolic enzymes involved in drug and xenobiotic detoxification influence individual drug response and resistance. This process is divided in 3 phases. The first one is mediated by the cytochrome P450 (CYP450), a superfamily of enzymes that catalyze the oxidation of organic substances. The second one is the formation of conjugates between these substances and glutathione, glucuronic acid, or sulfate due to glutathione S-transferase (GST), UDP-glucuronosyl transferase, and sulfatase, respectively. These enzymes are expressed in all tissues but prevalently in the liver and in enterocytes that are the first physical barrier that the xenobiotics have to go through to be absorbed and enter into the organism. The third phase of this process of detoxification consists in the exportation of metabolized substances through transmembrane pumps such as P glycoprotein and multidrug resistance (MDR) protein (MRP) family members.
Different regulation or genetic variations occurring on these enzymes and transporters can affect their action on drug processing, resulting in abnormalities in medicament absorption, distribution, metabolism, and excretion. CYP450 class of genes is encoded for more than 50 proteins that differ in their structure and expression. Cytochrome P450 functional differences are considered to be major determinants in organotrophism, responsible for organ-specific susceptibility to a given toxicant,6 and interindividual variability.7 Cytochrome P 3A4 is the most abundantly expressed CYP450 in human liver and small intestine and is known to metabolize more than 120 different drugs. A large number of human genetic polymorphisms have been described to occur within the CYP450 superfamily, and they influence protein functionality and consequentially susceptibility to drugs and eventually load to intrinsic resistance.
Frequently, coordinated deregulation of drug-activating enzymes of phase I and up-regulation of drug-conjugating enzymes of phase II are observed in drug-resistant tumor cells.8
Platinum derived drugs such as cispatin, oxaliplatin, and carboplatin are alkylating agents that bind covalently to DNA after conversion in an active form. Glutathione S-transferase conjugates these substances with glutathione,9 and the interaction causes inactivation of the drugs by formation of adducts that is subsequently eliminated from cells through ABC (ATP binding cassette) pumps.10 Enhanced conjugating activity of GST may determine resistance to alkylating agents. Sensitivity to cis-platin is also associated with an enhanced presence of glutathione in colon cancer cells and with the expression of the enzymes responsible for its synthesis in different tumors.11
Efflux Pumps
Several proteins, members of the ATP binding cassette protein family (ABC protein), have been identified throughout the years as central players for the development of drug resistance mediated by alteration of detoxification mechanisms based on efflux pumps. This superfamily of proteins has the capability to actively promote, through the hydrolysis of ATP, the translocation of a wide group of substrates (drugs, steroids, metabolites) across cellular membranes.12 This group of protein typically creates unidirectional flow of substrates from the cytosol to the extracellular matrix or to intracellular compartments. ABC proteins are associated with several genetic diseases such as cystic fibrosis (CFTR), Stargardt disease (ABCR), and Dubin-Johnson syndrome (MRP-2).13
It has been amply demonstrated that ABC proteins are involved in the development of MDR, a phenomenon in which the tumor acquires cross-resistance to an array of different compounds that are structurally and functionally unrelated.14 ABC proteins’ mechanism of action is given by increased cell detoxification, decreased uptake, and increased efflux and modification of the drugs. All these modifications are responsible for an overall reduction of delivery to the target molecules.
Four proteins have been identified as central player in the development of MDR, 3 of which are ABC protein: classic MDR (or PgP), non–P glycoprotein MDR (MRP), and breast cancer resistance protein (BRCP). The fourth protein involved in the process is the lung resistance–related protein (LRP), although LRP does not belong to the ABC superfamily of protein, because of its involvement in the MDR process. The nature of LRP and its function are described at the end of this section.
Multidrug resistance is an ABC membrane protein bound to a P glycoprotein.15 It acts as a detoxifying agent for the cell by directly pumping, through ATP hydrolysis, toxins and xenobiotics out of the cell. Etoposide, doxorubicin, and taxanes are all substrates involved in the MDR-mediated efflux.16 According to recent evidences, the overexpression of the MDR protein has also an indirect impact on passive drug diffusion.17 Indeed, this protein acts indirectly on electrical membrane potential, ion transport regulation, and signal transduction, creating a significant alternation of compartmental biophysical parameters such as pH equilibrium.
It is well known that cancer cells have the capacity of growing in hypoxic microenvironment. In addition, because of enhanced metabolic rate, these cells often have increased glycolysis and increased production of lactic acid, which results in intracellular accumulation of protons.18 Hyperproduction of protons in the cellular environment causes activation of the proton excludes (V-ATP ase, Na+/H+, and carbonic anhydrase) and consequent alteration of intracellular–extracellular pH gradient.19 Malignant cells, especially in metastatic setting, present an inverted gradient with acidosis in the extracellular compartment and alkalosis in the intracellular one. This new biophysical setting deeply affects cell capability to uptake and process pharmacological compounds. Indeed, low pH in the extracellular domain causes protonation and neutralization of basic chemo-therapeutic agents preventing them from entering into the targeted cells and explicating their cytotoxic function. In addition, perturbation of the intracellular pH leads to the formation of cytoplasmic acidic vesicles. Drugs’ sequestration within these organelles causes protonation of the chemicals agents and elimination of the drug through vesicle degranulation.20
Non–P glycoprotein MDR (MRP), also known as multi-specific anion-transporter, profoundly differs in its functions and substrate specificity from the MDR.21 Indeed, whereas MDR mostly expels neutral or basic hydrophobic compounds, MRP-1 presents high affinity for conjugated organic anions. Several evidences suggest that MRP-1 requires the presence of reduced glutathione (GSH) to promote expulsion of toxic agents from the cytosol.22 The interaction between MRP-1 and GSH in the development of drug resistance is currently not completely unveiled; however, several authors have shown that GSH concentration in cancer cells stimulates the uptake of vincristine in MRP-enriched membrane vesicles.23,24 Vincristine, doxorubicin, and antifolates represent some of the substrates that are actively transported by MRP-mediated efflux.16 The third protein involved in MDR is the BCRP, a half transponder of the ABC family presenting a single-nucleotide–binding domain. In breast, colon, and gastric cell lines, overexpression of BRCP solely has been found associated with drug resistance to mitoxantrone, topotecan, and doxorubicin.25 Another protein known to mediate chemoresistance is LRP, which belongs to the major vault protein family.26 Vault proteins are the main component of the complex ribonucleoprotein particles. Although its role in chemoresistance is still under investigation, it is well known that vault proteins are involved in controlling DNA exposure to cytotoxic agent. Recent studies have suggested that, rather than acting as an active transporter, this protein is responsible for the uptake of pharmacological compounds present in the cytosol and sequestration of the drugs into exocytosis vesicles. Etoposide, doxorubicin, vincristine, paclitaxel, and cisplatin/carboplatin are all targets of LRP.27,28
INTRACELLULAR DRUG ACTIVITY
RTKs and Cell Signaling
In the last decades, the development of more sophisticated biomolecular investigations has led to a deeper and more comprehensive understanding of protein signaling networks that regulate cellular function. Several studies have highlighted that many of these cellular protein “circuits” are deregulated in cancer cells and that overexpression and activation (phosphorylation) of key proteins within the neoplastic lesion actually drive cell adaptation and survival throughout the different phases of cancer progression and development of resistance to chemotherapeutic agents.29 For these reasons, in the past 3 decades, new chemotherapeutic strategies aiming at targeting specific molecules involved in signal transduction from the cell surface to the nucleus have been developed.30–32
During cancer progression and response to treatment, cancer cells are characterized by the development of intense interactions between different members of the RTKs superfamily. This interaction between multiple members of the family guarantees and intensifies the transmission of the signal from the cell surface to common downstream key effectors. For the presence of a complex and interconnected protein network, cancer cells easily overcome blockage at single receptor level by rerouting survival and proliferation signals through different members of the RTK family and their downstream effectors.33
First, RTKs that were targeted by monoclonal antibody were members the EGFR family.34 In human cells, there are 4 types of EGFR receptors: EGFR (ErbB-1), HER2/neu (ErbB-2), HER3 (ErbB-3), and HER4 (ErbB-4).35 Receptors’ activation occurs thorough homodimerization and/or heterodimerization that cause activation of specific signaling cascades leading to the activation of key prosurvival pathways such as PI3K/Akt, Ras/ Raf/MEK/Erk, and STAT.36,37
RTKs heterodimerizations occur between members of the same subfamily of receptors or between different subgroups. For example, heterodimerization of ErbB-2, ErbB-3, and IGF-1R generates enhanced cross-activation of 2 independent signaling pathways. This cross-activation is a mechanism that cancer cells use to overcome the inhibition produced by monoclonal antibodies and small molecules targeting RTKs.38 Indeed, heterodimerization of ErbB-2, ErbB-3, and IGF-1R or compensative overexpression of IGF-1R has been reported as an elective mechanism of resistance to EGFR inhibitors in several types of cancer cell lines.39 Moreover, several in vitro studies have reported that the administration of a selected EGFR inhibitor causes almost exclusively deactivation of the Ras/Raf/MEK/Erk, leaving the PI3K/Akt still able to compensate. In addition, rapidly compensatory mechanisms provided by the simultaneous activation of the EGFR and IGF-1R stimulate countervailing activation of the Ras/Raf/MEK/ Erk and PI3K/Akt pathways through activation of mediator proteins such as Shc, Src, or IRS-1. In in vitro models, combination therapies in which EGFR and IGF-1R are both inhibited have shown promising results in inducing down-regulation of PI3K/ Akt and Ras/Raf/MEK/Erk.40
Activation of the EGFR/c-erbB2/MAPK signaling pathway has been recently demonstrated to be involved not only in direct activation of cell growth, but also in the development of resistance against hormonal therapy in breast cancer. Indeed, MAPK activation leads to phosphorylation of key serine residues of the estrogen receptor in breast cancer cells. ERK1/2-mediated estrogen receptor phosphorylation promotes activation of the receptor and ligand-independent stimulation of transcriptional activity. It has been suggested that such activation could play a role in developing resistance against drugs that antagonize the estrogen receptor itself.41,42
Other mechanisms of resistance that cancer cells develop to escape treatment with RTKs inhibitors are given by amplification and overexpression of compensatory RTK receptors, such as MET,43 acquisition of ex novo secondary mutation on the receptors or of downstream effectors (i.e., EGFR, PI3K, K-Ras),44 loss of tumor suppressor such as PTEN,45,46 and derangement of downstream effectors.
One of the most important pathways involved in the development of chemoresistance is the PI3K/Akt. Activation of the PI3K/Akt pathway occurs through growth factor receptor signaling, interleukin and other proteins involved in cell survival. Activation of the PI3K leads to recruitment of the receptor at the cellular membrane and consequent production of phosphoinositol-3 phosphates.47 This second messenger is responsible for translocation of Akt to the cell membrane and consequent activation through phosphorylation by PDK. At the same time, PI3K, by involving of Grb2/SOS, IRS-1 and Shc, causes activation of Ras and consequently the Raf/MEK/Erk pathway.48
Once Akt is activated, it modulates cell survival and apoptosis through different pathways: BAD, ASK-1, cyclin-dependent kinase inhibitors p21 and p27,49 GSK3, FKHR members, and mTOR. For the central role that PI3K/Akt pathway plays in cell survival, it also has a tremendous impact on tumor response to chemotherapeutic agents. Several studies have demonstrated that the cytotoxic effect of chemotherapy often correlates with the capability of the agent to inhibit Akt. Indeed, pharmacological compounds able to decrease Akt activity are usually associated with increased cell death in numerous cell lines. The exact mechanism of action by which Akt induces chemoresistance is still under investigation. Experimental evidence has demonstrated that overexpression or hyperactivation of Akt plays a central role in the response to chemotherapeutic agents.
Cytoskeleton Organization
The cytoskeleton is a cellular dynamic structure capable of maintaining cell plasticity, and it is responsible for constant changes within the cell setting that lead to cell division, intracellular trafficking, and cell migration. Microtubules are dynamic microfilaments constituted by heterodimerization of α-/β-tubulin. They create protein complex with microtubule-binding proteins.50 The dynamic equilibrium in the microtubules is maintained by constant polymerization and depolymerization of their constituents. Because of all the different functions in which this cellular compartment is involved, microtubules have been considered for a long time ideal targets for antitubulin chemotherapeutic compounds: taxanes and vinca alkaloids. Antitubulin agents are known to have the ability to interfere with cell division by blocking the transition between metaphase and anaphase during mitosis, and as a consequence, they induce apoptosis. Antitubulin agents act either as microtubules stabilizers or as destabilizers.51 The first group of drugs (vinca alkaloids) interferes with poly-merization by binding 1 or 2 specific domains of the tubulin and preventing the formation of the microfilaments and the mitotic spindle. The second group (taxanes) increases polymerization by binding specific sites on the b-tubulin contained within the surface of the microfilaments suppressing the dynamic function of the microtubules.
Antimicrotubule resistance develops through several mechanisms: MDR, alteration of the microtubules components, and deficient apoptotic signaling (p53, bcl-2, blc-xl mediated).52 Alterations of the microtubules’ component can either be qualitative or quantitative. Examples of qualitative modification include alteration of the α-/β-tubulin proteins or derangement of associated to microtubule proteins (MAP2 and MAP 4, STOP, survivin, and caveolin 1), alteration of the cellular localization, and posttranslational modifications.53 Quantitative modifications of the microtubules include increased level of 1 specific tubulin isotope (i.e., bIII-tubulin) or mutation of the tubulin gene.
Apoptosis and DNA Repair
Chemotherapeutic agents act via different pathways all aiming at the same goal: to induce selective tumor cell death by inducing different cellular damages. Treatment efficacy depends not only on the direct cell impairment, but also on the cells’ ability to respond to these damages by inducing the apoptotic machinery. Therefore, drugs’ effect is associated with expression of specific death genes, coding, for example, for Bak, Bax, SMAC/DIABLO, PTEN, and p53, and down-regulation of survival counterparts, such as IAP proteins, Bcl-2 family members, proteins involved in PI3K/AKT signaling, and p53.54 The susceptibility of tumor cells to drug-induced apoptosis depends on the balance between proapoptotic and survival (antiapoptotic) signals. Blockade or down-regulation of former pathway or up-regulation of the latter one is a mechanism of MDR.
Resistance to doxorubicin, a DNA intercalating drug used in the treatment of a wide range of tumors, was observed, for example, in endometrial cancer, and it is due to up-regulation of the antiapoptotic PI3K/AKT signaling pathway.54 Indeed, it was previously demonstrated that overexpression of PTEN in Ishikawa cells significantly enhances doxorubicin chemosensitivity due to PTEN ability to impair AKT activation.55 This evidence confirms that one of the mechanisms of resistance to doxorubicin resides in cancer cells’ capacity to escape apoptosis.
A key factor in the induction of apoptosis in response to radiotherapy and chemotherapy is the tumor suppressor protein p53. Approximately 50% of the tumors express mutation on the p53 gene.56 Normal p53 is activated in response to different cell injuries; its activation results in cell cycle arrest and apoptosis. Activation of p53 is mediated by different cellular mechanisms such as DNA damaging agents, altered ribonucleotide pools, changes in redox potential, disruption of mitotic spindle, and so on. Thus, it is evident that most cytostatic drugs have to be p53 activators. When p53 is mutated, cells are less prone to initiate apoptosis and more resistant to DNA insult, such as that induced by chemotherapeutic agents. Indeed presence of abnormally functioning p53 is a common finding in MDR tumor cells. A large study done at the National Cancer Institute revealed a positive correlation between p53 status and cell sensitivity to cytotoxic drugs. P53 mutant cells are more often resistant to unrelated chemotherapeutic agents than p53 wild-type cells.57
In vivo tumor cells are exposed to stress conditions, such as inadequate vascularization in large solid tumors, glucose and other nutrient deprivation, hypoxia, and low pH, which may contribute to the selection of subclones with decreased apoptotic potential and thereby lead to resistance to antitumor drugs. These stress conditions induce drug resistance through the activation of cell defense mechanisms that usually protect the cell from injury and promote its survival in case of nutrient deprivation. For instance, glucose-regulated stress response is associated with the development of cellular resistance to topoisomerase II (topoII) poisons, such as etoposide and doxorubicin. Topoisomerases are nuclear enzymes that regulate DNA supercoiling catalyzing the formation of transient single- (topoI) or double-stranded (topoII) DNA breaks, central for DNA transcription and replication. For these reasons, the formation of DNA-topoisomerase-drug complexes initiates the production of lethal DNA strand breaks and induces accumulation of those stable breaks in S-phase triggering checkpoint mechanism that leads to the activation of the intrinsic apoptotic pathway.58,59 Under stress conditions, like glucose starvation, the proteasome accumulates in the nucleus. This agglomeration induces degradation of nuclear proteins, including topoII, making the substrate unavailable for drug interaction. This mechanism leads the cell to etoposide resistance. Indeed, in vitro studies showed that proteasome inhibitors (i.e., lactacystin) or the presence of a mutant proteasome effectively confers etoposide sensitivity by reducing its translocation into the nucleus.60,61
Stress conditions induce drug resistance not only by adaptive changes, but also through genetic alterations that can occur during cancer cell proliferation. For instance, DNA mutations can modify drug molecular targets. For example, resistance to irinotecan or etoposide, 2 drugs that act on topoI and topoII, respectively, can be caused by mutations of the topoisomerase active site. This active site is fundamental for DNA cleavage and for the interaction between topoisomerase and pharmacological compounds.62 Many cytostatic drugs cause direct or indirect activation of the DNA. Resistance toward these agents may be mediated by the enhanced capability of tumor cells to repair DNA damages. Cells exposed to genotoxic agents must repair DNA injury to survive, but if the damage is very severe and the cells cannot reverse the insult, they activate apoptosis. Nucleotide excision repair (NER) is one of the major mechanisms involved in the repair of DNA adducts, such as those resulting from the use of alkylating agents or those that irreversibly bind to DNA, such as cisplatin.63 Twelve proteins are involved in NER, and up-regulation of some of them can increase the cell DNA repair activity and drug resistance.
Fluorodeoxyuridine monophosphate is the active metabolite of 5-fluorouracil (5-FU) that inhibits purine and thymidylate biosynthesis by binding and blocking thymidylate synthase. This results in an imbalance of the intracellular nucleotide pool, which causes profound alteration in DNA synthesis and affects the repair processes.64 Mismatches, generated by the abundance of dUTP, are corrected by uracil-DNA-glycosidase as part of NER. Resistance to thymidylate synthase inhibitors is due to increase in dUTPases that limit the accumulation of dUTP and the consequent damage. Alterations of several DNA repair mechanisms are involved in the development of resistance. In vitro studies have shown that cancer cells acquire resistance to 5-FU when point mutations are developed during cell replication. Modification of the mismatch repair (MMR) system is one of the mechanisms used by tumor cells to overcome 5-FU mechanism of action. Several studies have reported increased 5-FU resistance in cells with deficiencies in the MMR system that is another mechanism of DNA repair. It corrects the errors generated by mismatched nucleotides or wrongly matched nucleotides occurring during replication.65 Defect in this mechanism increases the occurrence of genetic mutations and gives the cells a higher tolerance to chemical modification of DNA. This accounts for the enhanced resistance to genotoxic anticancer agents. Moreover, the loss of some elements of the MMR system also favors cell ability to elude apoptosis when DNA is highly damaged DNA.66
CONCLUSIONS
Chemoresistance to anticancer agents is a consequence of multiple factors that include a wide variety of circumstances, such as individual variability and sensitivity to drug, tissue lineage, environmental localization of the tumor, its aggressiveness, and intracellular molecular alteration. We discussed different mechanisms of resistance related to drug processing and mechanisms of action. Indeed, resistance can arise at different levels, from the 3 different phases of drug processing, to drug intracellular activity, and more widely to the regulation of the main pathway involved in cell survival and proliferation (Table 1).
TABLE 1.
Mechanism of Chemoresistance and its Targets
| Mechanisms | Resistance to | |
|---|---|---|
| Drug metabolism | ||
| Cytochrome P450 | Oxidation of therapeutic agents is compromised because of genetic polymorphisms inducing alteration in protein functionality and susceptibility to drugs | Ifosfamide, vinblastine, etoposide, and doxorubicin |
| GST | Enhanced conjugating activity | Alkylating agents |
| Cell membrane and intracellular mediated resistance | ||
| MDR | Increased efflux of chemotherapeutic agent from the cytosol to the extracellular matrix or intracellular organelles | Anthracyclines, taxanes, Vinca alkaloids, mitoxantrone |
| Receptor tyrosine kinase | Heterodimerizations and receptor cross-activation; amplification or overexpression of compensatory RTKs; ex novo secondary mutation; derangement of tumor suppressors | Monoclonal antibody, small tyrosine kinase inhibitors, tamoxifen |
| Cytoskeleton | Excessive stabilization or destabilization of microtubule dynamic structure | Taxanes and vinca alkaloids |
| Cell division and programmed death | ||
| Defective apoptosis | Mutation on tumor suppressors, hyperactivation of prosurvival pathways | Doxorubicin, alkylating agents |
| Alteration of DNA replication or repair | Interference with DNA “interacting” enzymes such as topoisomerase | Etoposide, irinotecan, doxorubicin, gemcitabine |
Different cell compartments and organelles are involved in the development of resistance to pharmacological compounds.
The investigation of the mechanisms participating in the regulation of cell viability and the evaluation of the connections between different signaling pathways is relevant and is the major aim of proteomic studies in cancer research. Characterization of mechanisms and alterations occurring in survival pathways that lead the escape from apoptosis in response to drugs is useful for identifying new therapeutic targets and sets of biomarkers that could guide clinicians in the selection of treatment. Furthermore, there are a series of new promising proteomic techniques that will allow investigators to study the real-time variation of protein expression and activation according to different pathological or physiological conditions and cell response to a given stimulus, such as drugs.
In addition, because proteins play a central role in drug development because they are most often the drug targets for the new classes of molecularly targeted inhibitors, it is imperative to contemporarily monitor numerous biochemical pathways and to develop bimolecular techniques capable of creating accurate portrait of ongoing cellular signaling at a network scale. To achieve a more inclusive understanding of molecular adaptation to novel stimuli and to integrate molecular profiling into clinical practice, in the last 2 decades many innovative biomolecular technologies have been developed. One such method, which was invented in our laboratories, is the RPMA (reverse-phase protein microarray). The RPMA is a multiplex, high-throughput, highly sensitive technique capable of evaluating concurrently hundreds of different analytes not only in their native forms but also in their posttranslational modification.
The capability, through multiple tissue biopsies, to detect molecular derangements driving drug resistance allows identifying patients who develop resistance to a treatment before clinical manifestation of the phenomenon. Moreover, direct analysis of cellular derangement and identification of the mechanisms that are developing as a response to cellular damage will allow the medical community to select appropriate single- or multiple-agent treatment. In the future, profiling of individual tumors to predict drug responsiveness and interindividual variability in drug response will underpin the selection of the most appropriate chemotherapeutic regimens. Selective manipulation of signal transduction pathways involved in the mechanisms of resistance in combination with present chemotherapeutic drugs may lead to an increased potency in the clinic and the improvement of the efficiency of existing therapy, resulting in a better prognosis and overall survival.
References
- 1.Goodman LS, Wintrobe MM, Dameshek W, et al. Nitrogen mustard therapy; use of methyl-bis (beta-chloroethyl) amine hydrochloride and tris (beta-chloroethyl) amine hydrochloride for Hodgkin’s disease, lymphosarcoma, leukemia and certain allied and miscellaneous disorders. J Am Med Assoc. 1946;132:126–132. doi: 10.1001/jama.1946.02870380008004. [DOI] [PubMed] [Google Scholar]
- 2.Frei E, 3rd, Karon M, Levin RH, et al. The effectiveness of combinations of antileukemic agents in inducing and maintaining remission in children with acute leukemia. Blood. 1965;26(5):642–656. [PubMed] [Google Scholar]
- 3.Gonzalez-Angulo AM, Morales-Vasquez F, Hortobagyi GN. Overview of resistance to systemic therapy in patients with breast cancer. Adv Exp Med Biol. 2007;608:1–22. doi: 10.1007/978-0-387-74039-3_1. [DOI] [PubMed] [Google Scholar]
- 4.Chapal N, Molina L, Molina F, et al. Pharmacoproteomic approach to the study of drug mode of action, toxicity, and resistance: applications in diabetes and cancer. Fundam Clin Pharmacol. 2004;18:413–422. doi: 10.1111/j.1472-8206.2004.00258.x. [DOI] [PubMed] [Google Scholar]
- 5.Oldenhuis CN, Oosting SF, Gietema JA, et al. Prognostic versus predictive value of biomarkers in oncology. Eur J Cancer. 2008;44:946–953. doi: 10.1016/j.ejca.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 6.Roos PH, Bolt HM. Cytochrome P450 interactions in human cancers: new aspects considering CYP1B1. Expert Opin Drug Metab Toxicol. 2005;1:187–202. doi: 10.1517/17425255.1.2.187. [DOI] [PubMed] [Google Scholar]
- 7.Luqmani YA. Mechanisms of drug resistance in cancer chemotherapy. Med Princ Pract. 2005;14(suppl 1):35–48. doi: 10.1159/000086183. [DOI] [PubMed] [Google Scholar]
- 8.Thorgeirsson SS, Huber BE, Sorrell S, et al. Expression of the multidrug-resistant gene in hepatocarcinogenesis and regenerating rat liver. Science. 1987;236:1120–1122. doi: 10.1126/science.3576227. [DOI] [PubMed] [Google Scholar]
- 9.Teicher BA, Holden SA, Kelley MJ, et al. Characterization of a human squamous carcinoma cell line resistant to cis-diamminedichloroplatinum(II) Cancer Res. 1987;47:388–393. [PubMed] [Google Scholar]
- 10.Meijer C, Mulder NH, Timmer-Bosscha H, et al. Relationship of cellular glutathione to the cytotoxicity and resistance of seven platinum compounds. Cancer Res. 1992;52:6885–6889. [PubMed] [Google Scholar]
- 11.Siddik ZH. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22:7265–7279. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- 12.Kos V, Ford RC. The ATP-binding cassette family: a structural perspective. Cell Mol Life Sci. 2009;66:3111–3126. doi: 10.1007/s00018-009-0064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dean M, Hamon Y, Chimini G. The human ATP-binding cassette (ABC) transporter superfamily. J Lipid Res. 2001;42:1007–1017. [PubMed] [Google Scholar]
- 14.Ernst R, Kueppers P, Stindt J, et al. Multidrug efflux pumps: substrate selection in ATP-binding cassette multidrug efflux pumps—first come, first served? FEBS J. 2010;277:540–549. doi: 10.1111/j.1742-4658.2009.07485.x. [DOI] [PubMed] [Google Scholar]
- 15.Roepe PD. What is the precise role of human MDR 1 protein in chemotherapeutic drug resistance? Curr Pharm Des. 2000;6:241–260. doi: 10.2174/1381612003401163. [DOI] [PubMed] [Google Scholar]
- 16.Sève P, Dumontet C. Chemoresistance in non-small cell lung cancer. Curr Med Chem Anticancer Agents. 2005;5:73–88. doi: 10.2174/1568011053352604. [DOI] [PubMed] [Google Scholar]
- 17.Roepe PD, Wei LY, Hoffman MM, et al. Altered drug translocation mediated by the MDR protein: direct, indirect, or both? J Bioenerg Biomembr. 1996;28:541–555. doi: 10.1007/BF02110444. [DOI] [PubMed] [Google Scholar]
- 18.Young CD, Anderson SM. Sugar and fat—that’s where it’s at: metabolic changes in tumors. Breast Cancer Res. 2008;10:202. doi: 10.1186/bcr1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Izumi H, Torigoe T, Ishiguchi H, et al. Cellular pH regulators: potentially promising molecular targets for cancer chemotherapy. Cancer Treat Rev. 2003;29:541–549. doi: 10.1016/s0305-7372(03)00106-3. [DOI] [PubMed] [Google Scholar]
- 20.Fais S. Proton pump inhibitor–induced tumour cell death by inhibition of a detoxification mechanism. J Intern Med. 2010;267:515–525. doi: 10.1111/j.1365-2796.2010.02225.x. [DOI] [PubMed] [Google Scholar]
- 21.Hipfner DR, Deeley RG, Cole SP. Structural, mechanistic and clinical aspects of MRP1. Biochim Biophys Acta. 1999;1461:359–376. doi: 10.1016/s0005-2736(99)00168-6. [DOI] [PubMed] [Google Scholar]
- 22.Loe DW, Almquist KC, Deeley RG, et al. (MRP)-mediated transport of leukotriene C4 and chemotherapeutic agents in membrane vesicles. Demonstration of glutathione-dependent vincristine transport. J Biol Chem. 1996;271:9675–9682. doi: 10.1074/jbc.271.16.9675. [DOI] [PubMed] [Google Scholar]
- 23.Akan I, Akan S, Akca H, et al. Multidrug resistance–associated protein 1 (MRP1) mediated vincristine resistance: effects of N-acetylcysteine and buthionine sulfoximine. Cancer Cell Int. 2005;5:22. doi: 10.1186/1475-2867-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Jong MC, Slootstra JW, Scheffer GL, et al. Peptide transport by the multidrug resistance protein MRP1. Cancer Res. 2001;61:2552–2557. [PubMed] [Google Scholar]
- 25.Mao Q, Unadkat JD. Role of the breast cancer resistance protein (ABCG2) in drug transport. AAPS J. 2005;7:E118–E133. doi: 10.1208/aapsj070112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rimsza LM, Campbell K, Dalton WS, et al. The major vault protein (MVP), a new multidrug resistance associated protein, is frequently expressed in multiple myeloma. Leuk Lymphoma. 1999;34:315–324. doi: 10.3109/10428199909050956. [DOI] [PubMed] [Google Scholar]
- 27.Dalton WS, Scheper RJ. Lung resistance–related protein: determining its role in multidrug resistance. J Natl Cancer Inst. 1999;91:1604–1605. doi: 10.1093/jnci/91.19.1604. [DOI] [PubMed] [Google Scholar]
- 28.Zurita AJ, Diestra JE, Condom E, et al. Lung resistance–related protein as a predictor of clinical outcome in advanced testicular germ-cell tumours. Br J Cancer. 2003;88:879–886. doi: 10.1038/sj.bjc.6600803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cruzalegui F. Protein kinases: from targets to anti-cancer drugs. Expert Opin Ther Targets. 2003;7:215–234. [Google Scholar]
- 30.Traxler P. Tyrosine kinases as targets in cancer therapy—successes and failures. Expert Opin Ther Targets. 2003;7:215–234. doi: 10.1517/14728222.7.2.215. [DOI] [PubMed] [Google Scholar]
- 31.Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scaltriti M, Baselga J. The epidermal growth factor receptor pathway: a model for targeted therapy. Endocr Relat Cancer. 2005;12(suppl 1):S99–S111. doi: 10.1158/1078-0432.CCR-05-1554. [DOI] [PubMed] [Google Scholar]
- 33.Bennasroune A, Gardin A, Aunis D, et al. Tyrosine kinase receptors as attractive targets of cancer therapy. Crit Rev Oncol Hematol. 2004;50:23–38. doi: 10.1016/j.critrevonc.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 34.Drebin JA, Link VC, Weinberg RA, et al. Inhibition of tumor growth by a monoclonal antibody reactive with an oncogene-encoded tumor antigen. Proc Natl Acad Sci U S A. 1986;83:9129–9133. doi: 10.1073/pnas.83.23.9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kruser TJ, Wheeler DL. Mechanisms of resistance to HER family targeting antibodies. Exp Cell Res. 2010;316:1083–1100. doi: 10.1016/j.yexcr.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 36.Scaltriti M, Baselga J. Epidermal growth factor receptor/HER2/insulin-like growth factor receptor signalling and oestrogen receptor activity in clinical breast cancer. Clin Cancer Res. 2006;12:5268–5272. doi: 10.1158/1078-0432.CCR-05-1554. [DOI] [PubMed] [Google Scholar]
- 37.Quesnelle KM, Boehm AL, Grandis JR. STAT-mediated EGFR signaling in cancer. J Cell Biochem. 2007;102:311–319. doi: 10.1002/jcb.21475. [DOI] [PubMed] [Google Scholar]
- 38.Erjala K, Sundvall M, Junttila TT, et al. Signaling via ErbB2 and ErbB3 associates with resistance and epidermal growth factor receptor (EGFR) amplification with sensitivity to EGFR inhibitor gefitinib in head and neck squamous cell carcinoma cells. Clin Cancer Res. 2006;12:4103–4111. doi: 10.1158/1078-0432.CCR-05-2404. [DOI] [PubMed] [Google Scholar]
- 39.Gee JM, Robertson JF, Gutteridge E, et al. receptor/HER2/insulin-like growth factor receptor signalling and oestrogen receptor activity in clinical breast cancer. Endocr Relat Cancer. 2005;12(suppl 1):S99–S111. doi: 10.1677/erc.1.01005. [DOI] [PubMed] [Google Scholar]
- 40.Guix M, Faber AC, Wang SE, et al. Acquired resistance to EGFR tyro-sine kinase inhibitors in cancer cells is mediated by loss of IGF-binding proteins. J Clin Invest. 2008;118:2609–2619. doi: 10.1172/JCI34588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Britton DJ, Hutcheson IR, Knowlden JM, et al. Bidirectional cross talk between ERalpha and EGFR signalling pathways regulates tamoxifen-resistant growth [published online ahead of print October 27, 2005] Breast Cancer Res Treat. 2006;96:131–146. doi: 10.1007/s10549-005-9070-2. [DOI] [PubMed] [Google Scholar]
- 42.Kato S, Endoh H, Masuhiro Y, et al. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science. 1995;270:1491–1494. doi: 10.1126/science.270.5241.1491. [DOI] [PubMed] [Google Scholar]
- 43.Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karapetis CS, Khambata-Ford S, Jonker DJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 45.Loupakis F, Pollina L, Stasi I, et al. PTEN expression and KRAS mutations on primary tumors and metastases in the prediction of benefit from cetuximab plus irinotecan for patients with metastatic colorectal cancer. J Clin Oncol. 2009;27:2622–2629. doi: 10.1200/JCO.2008.20.2796. [DOI] [PubMed] [Google Scholar]
- 46.Barton S, Starling N, Swanton C. Predictive molecular markers of response to epidermal growth factor receptor (EGFR) family–targeted therapies. Curr Cancer Drug Targets. 2010;10:799–812. doi: 10.2174/156800910793357925. [DOI] [PubMed] [Google Scholar]
- 47.Testa JR, Tsichlis PN. AKT signaling in normal and malignant cells. Oncogene. 2005;24:7391–7393. doi: 10.1038/sj.onc.1209100. [DOI] [PubMed] [Google Scholar]
- 48.Grandage VL, Gale RE, Linch DC, et al. PI3-kinase/Akt is constitutively active in primary acute myeloid leukaemia cells and regulates survival and chemoresistance via NF-kappaB, MAPkinase and p53 pathways. Leukemia. 2005;19:586–594. doi: 10.1038/sj.leu.2403653. [DOI] [PubMed] [Google Scholar]
- 49.West KA, Castillo SS, Dennis PA. Activation of the PI3K/Akt pathway and chemotherapeutic resistance. Drug Resist Updat. 2002;5:234–248. doi: 10.1016/s1368-7646(02)00120-6. [DOI] [PubMed] [Google Scholar]
- 50.Nogales E. Structural insights into microtubule function. Annu Rev Biochem. 2000;69:277–302. doi: 10.1146/annurev.biochem.69.1.277. [DOI] [PubMed] [Google Scholar]
- 51.Jordan MA. Mechanism of action of antitumor drugs that interact with microtubules and tubulin. Curr Med Chem Anticancer Agents. 2002;2:1–17. doi: 10.2174/1568011023354290. [DOI] [PubMed] [Google Scholar]
- 52.Galmarini CM, Kamath K, Vanier-Viornery A, et al. Drug resistance associated with loss of p53 involves extensive alterations in microtubule composition and dynamics. Br J Cancer. 2003;88:1793–1799. doi: 10.1038/sj.bjc.6600960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dumontet C, Jordan MA. Microtubule-binding agents: a dynamic field of cancer therapeutics. Nat Rev Drug Discov. 2010;9:790–803. doi: 10.1038/nrd3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chaundry P, Asselin E. Resistance to chemotherapy and hormone therapy in endometrial cancer. Endocr Relat Cancer. 2009;16:363–380. doi: 10.1677/ERC-08-0266. [DOI] [PubMed] [Google Scholar]
- 55.Wan X, Li J, Xie X, et al. PTEN augments doxorubicin-induced apoptosis in PTEN-null Ishikawa cells. Int J Gynecol Cancer. 2007;17:808–812. doi: 10.1111/j.1525-1438.2007.00890.x. [DOI] [PubMed] [Google Scholar]
- 56.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 57.O’Connor PM, Jackman J, Bae I, et al. Characterization of the p53 tumor suppressor pathway in cell lines of the National Cancer Institute anti-cancer drug screen and correlations with the growth-inhibitory potency of 123 anticancer agents. Cancer Res. 1997;57:4285–4300. [PubMed] [Google Scholar]
- 58.Rubin EH, Li TK, Duann P, et al. Cellular resistance to topoisomerase poisons. Cancer Treat Res. 1996;87:243–260. doi: 10.1007/978-1-4613-1267-3_10. [DOI] [PubMed] [Google Scholar]
- 59.Liu LF, Desai SD, Li TK, et al. Mechanism of action of camptothecin. Ann N Y Acad Sci. 2000;922:1–10. doi: 10.1111/j.1749-6632.2000.tb07020.x. [DOI] [PubMed] [Google Scholar]
- 60.Ogiso Y, Tomida A, Lei S, et al. Proteasome inhibition circumvents solid tumor resistance to topoisomerase II–directed drugs. Cancer Res. 2000;60:2429–2434. [PubMed] [Google Scholar]
- 61.Ogiso Y, Tomida A, Tsuruo T. Nuclear localization of proteasomes participates in stress-inducible resistance of solid tumor cells to topoisomerase II–directed drugs. Cancer Res. 2002;62:5008–5012. [PubMed] [Google Scholar]
- 62.Li XG, Haluska P, Jr, Hsiang YH, et al. Involvement of amino acids 361 to 364 of human topoisomerase I in camptothecin resistance and enzyme catalysis. Biochem Pharmacol. 1997;53:1019–1027. doi: 10.1016/s0006-2952(96)00899-4. [DOI] [PubMed] [Google Scholar]
- 63.Furuta T, Ueda T, Aune G, et al. Transcription-coupled nucleotide excision repair as a determinant of cisplatin sensitivity of human cells. Cancer Res. 2002;62:4899–4902. [PubMed] [Google Scholar]
- 64.Houghton JA, Tillman DM, Harwood FG. Ratio of 2′-deoxyadenosine–5′-triphosphate/thymidine-5′-triphosphate influences the commitment of human colon carcinoma cells to thymineless death. Clin Cancer Res. 1995;1:723–730. [PubMed] [Google Scholar]
- 65.Meyers M, Wagner MW, Hwang HS, et al. Role of the hMLH1 DNA mismatch repair protein in fluoropyrimidine-mediated cell death and cell cycle responses. Cancer Res. 2001;61:5193–5201. [PubMed] [Google Scholar]
- 66.Marin JJ, Romero MR, Martinez-Becerra P, et al. Overview of the molecular bases of resistance to chemotherapy in liver and gastrointestinal tumours. Curr Mol Med. 2009;9:1108–1129. doi: 10.2174/156652409789839125. [DOI] [PubMed] [Google Scholar]