Abstract
The environmental toxin β-N-methylamino-L-alanine (BMAA) has been causatively linked to neurodegenerative disease pathology. In a rat model, neonatal BMAA exposure resulted in selective uptake in the hippocampal formation and caused learning and memory impairments in adult animals. Moreover, high dose neonatal BMAA exposure resulted in formation of protein inclusions in the CA1 region of the adult hippocampus. However the mechanism underlying BMAA induced neuropathology remains elusive. Imaging mass spectrometry is a powerful method for spatial interrogation of biochemical distribution in biological tissue with high chemical specificity. The aim of this study was to therefore characterize the lipid microenvironment of BMAA-induced hippocampal lesions in adult rats using matrix-assisted laser desorption/ionization (MALDI) and time-of-flight SIMS (ToF-SIMS imaging). Multimodal imaging was carried out by ToF-SIMS scans of the hippocampal formation followed by whole tissue scans using MALDI imaging. Multivariate analysis was performed on the SIMS data in order to delineate the spatial biochemistry surrounding the lesions. The data show lesion-specific localization of phosphatidylcholine fragments, suggesting neuroinflammatory glial cell activation. Complementary MALDI imaging data showed increased levels of phosphoethanolamines colocalizing with the proteopathic lesions pointing to macroautophagic mechanisms associated with neurotoxin-induced protein accumulation. Multimodal IMS by means of ToF-SIMS and MALDI mass spectrometry proved to be a powerful technique for neurotoxicological research.
Keywords: Imaging Mass Spectrometry (IMS), beta-N-methylamino-L-alanine (BMAA), ALS/PD-Complex, matrix assisted laser desorption/ionization (MALDI), Time of Flight secondary ion mass spectrometry (ToF-SIMS)
1. Introduction
The environmental toxin β-N-methylamino-L-alanine (BMAA) was shown to be neurotoxic several decades ago and has received attention as a possible risk factor for neurodegenerative disease. Recently it has been revealed that most cyanobacteria are able to produce the non-protein amino acid BMAA.1 Exposure to BMAA has been suggested to be involved in the etiology of Amyotrophic lateral sclerosis/Parkinsonism-dementia complex (ALS/PDC) on the island of Guam2 and in ALS and Alzheimer’s disease in North America.3
BMAA exposure to neonatal rats (PND; postnatal days 9–10) induced long-term cognitive impairments and changes in neuronal protein expression at adult age.4 Developmental exposure to a high dose (460 mg/kg) also induced severe lesions in the adult hippocampus accompanied by neuronal degeneration, cell loss, calcium deposits and astrogliosis.4 In order to understand the mechanisms of BMAA-induced neurobehavioral alterations, more studies on the biomolecular effects of BMAA in the hippocampus are needed, since this brain area is essential for learning and memory. For probing spatio-temporal dynamics of ongoing biochemical processes, advanced analytical techniques are needed. Imaging mass spectrometry (IMS) is a powerful technique for probing the molecular architecture of biological tissue in an anatomical context.5,6 The technology allows the direct and comprehensive identification and spatial profiling of biomolecular species in situ, while maintaining high molecular specificity.7
Time of flight secondary mass spectrometry (ToF-SIMS) based imaging MS allows analyses at submicron scale (<500 nm), which is unprecedented despite recent advances in other IMS techniques. In contrast, matrix assisted laser desorption ionization (MALDI) IMS has limitations with respect to spatial resolution (typically >10 um) but provides more intact biomolecular information. Complementary application of both IMS techniques is therefore a promising approach. IMS has been demonstrated to be a valuable approach in neuroscience research8 and represents a powerful technology for spatially profiling lipids, neuropeptides and proteins in complex biological matrices such as the nervous system9. The aim of this study was to investigate neurotoxin-induced neurochemical changes in the hippocampal formation using multimodal (i.e. ToF-SIMS + MALDI) IMS.
2. Experimental
Chemicals and Reagents
All chemicals including β-N-Methylamino-L-alanine (L-BMAA) hydrochloride (≥97%) were of pro-analysis grade and purchased from Sigma Aldrich (St. Louis, USA). TissueTek optimial cutting tool (OCT) was purchased from Sakura Finetek (AJ Alphen aan den Rijn, The Netherlands). Water was obtained from a Milli-Q (Millipore, Bedford, MA, USA) purification system.
Animal Experiments
Pregnant outbred Wistar rats were obtained from Scanbur BK AB (Sollentuna, Sweden).10,11 Male pups were given one daily sec injection (20 μL/g) of BMAA 460 mg/kg (corresponding to 600 mg/kg BMAA HCl, n=7) freshly dissolved in Hanks’ balanced salt solution, or vehicle (n = 7), for 2 days on PND 9–10 (Figure 1A). The short- and long-term behavioral effects of BMAA in these animals have been published separately.11,12 All animal experiments were approved by the Uppsala Ethical Committee on Animal Experiments and followed the guidelines of Swedish legislation on animal experimentation (Animal Welfare Act SFS1998:56) and European Union legislation (Convention ETS123 and Directive 86/609/EEC).
Figure 1. Experimental layout.
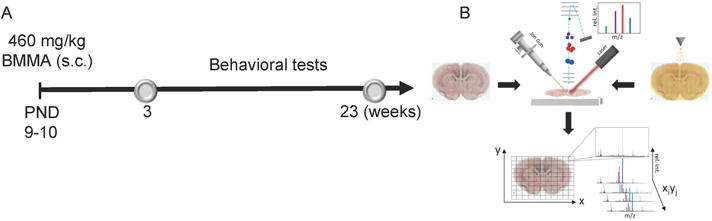
(A) The study design comprised injection of male pups at PND 9-10 followed by behavioral evaluation on weeks 3 and 23. Adult animals were then sacrificed followed by tissue dissection and sectioning. (B) The brain sections were first analyzed by means of ToF-SIMS followed by matrix application and MALDI imaging MS. Systematic analysis of the whole sections in a predefined raster allows image reconstruction for all individual ion signals.
Sample Preparation
The animals were killed by decapitation at 23 weeks of age, the brains were quickly frozen in dry ice and stored at −80°C. Coronal cryosections were obtained from the right hemisphere of all animals at the level of hippocampus (−3.36 mm relative to the bregma13). The brain sections (12 μm) were thaw mounted on conductive glass slides (indium tin oxide, ITO, Bruker Daltonics, Bremen, Germany), dried under vacuum for 10 min and stored at −20°C until further use.
ToF-SIMS Analysis
Prior IMS, samples were thawed under vacuum for 15 min. SIMS imaging was performed with an ION-TOF V ToF-SIMS instrument (IONTOF GmbH, Münster, Germany) equipped with a Bi3+ cluster ion primary ion source. High resolution images from the inclusions were collected in burst-alignment mode14 with a pulsed primary ion current of 0.1 pA at 25 keV and an ion dose of 4.8 × 1011 ions/cm2 with a mass resolution of M/ΔM = 350 full width at half maximum at m/z 500. Here, 50 scans were acquired from a predefined quadratic scan area with 238 μm in edge length. Images comprising 512 × 512 pixels were acquired resulting in a spatial resolution of 464 nm. All spectra were acquired and processed with the Surface Lab software (v. 6.3 ION-TOF). All spectra were calibrated internally and signals for [C]+, [CH]+, [CH2]+, [CH3]+, [C5H15PNO4]+ and [C27H45]+ were used as calibration points. Mass interval lists were created by peak search of all individual samples according to the following search parameters: S/N>3, width 0.8 Da. A mass interval list was created that contained the list of m/z values to be included in the multivariate analysis. The image data were reconstructed, exported into the *.bif6 format and loaded into PLS-Toolbox (v. 7.02, Eigenvector Research Inc., Wenatchee, WA) running under MatLab (v.12, The MathWorks Inc., Natick, MA). Here data were subjected to maximum autocorrelation factor analysis (MAF).
MALDI imaging
Following the SIMS experiments, MALDI matrix (alpha-cyano-4-hydroxycinnamic acid (CHCA), 7 g/l, 50% acetonitrile, 0.2% trifluoroacetic acid) was applied with a commercial pneumatic nebulizer (Image Prep II, Bruker Daltonics, Bremen, Germany). The FlexImaging software (v.3.0, Bruker) was used for imaging sequence generation, data acquisition and evaluation of single ion images. MALDI experiments were performed in negative-reflector mode on an ultraflextreme TOF/TOF instrument (Bruker) at a lateral resolution of 50 um. A mass range from 500–2000 Da was analyzed with shots/pixel: 1000 at 1 kHz repetition rate and a laser beam size of 20 μm (“small”). The spectra were calibrated externally with data acquired from calibration solution (PeptideMix1, Bruker) spotted adjacent to the sections (Figure 1B).
3. Results and Discussion
Neonatal BMAA exposure on PND 9–10 resulted in selective neuropathological changes in the hippocampal region of adult rats six months of age. The most prominent changes comprised formation of severe lesions in the adult hippocampus (Figure 2A,B) of BMAA-treated animals. In the present study, we therefore employed multimodal IMS for probing the native neurochemical distribution in the hippocampus upon neonatal exposure to BMAA.
Figure 2. MALDI IMS in negative-ion mode reveals unknown lipid masses that colocalize with BMAA-induced lesions.
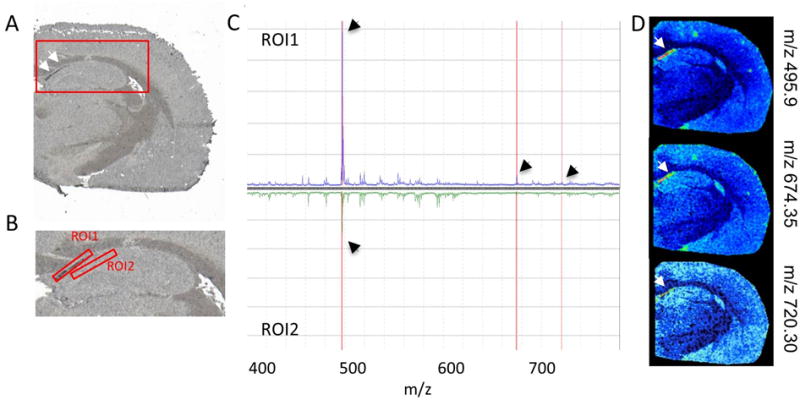
(A) Lesions were observed (arrows) in the hippocampus (red square) of adult rats following neonatal BMAA exposure. (B) ROI assignment and differential analysis of average spectra (hippocampus region magnified). (C) The average spectra of the inclusion ROI (1) was compared to the adjacent control region (2) as well as an adjacent region within the CA1 part of the hippocampus. (D) Single ion images of masses that displayed characteristic changes in between the two ROI’s were verified localization of distinct m/z to the lesions (arrows).
In situ analysis using MALDI IMS on whole tissue sections did not only uncover animals with neuronal lesions but also pinpointed CA1 as the most severely affected region in the hippocampus (Figure 2). For all BMAA treated animals, displaying lesions in the CA1, region of interest (ROI) analysis of these inclusions was performed. The lesions were assigned as distinct ROI for generation of the respective average spectra (Figure 2B,C). Comparative analysis of all detected peaks in the average spectra (t-test, p<0.05) of the lesion region (ROI1) to surrounding CA1 regions (ROI2) allowed identification of individual peaks that were specific to the lesion ROI. The resulting m/z-peaks were further validated by generating single ion images (Figure 2D). For three m/z values, (m/z 495.93, m/z 674.35, m/z 720.35) distinct localization to the inclusions was verified. Accurate mass matching suggests that the two later mass peaks can be assigned as phosphoethanolamines (PEs) with m/z 674.3 (PE 31:1) and m/z 720.3 (PE 35:6).
High levels of PEs have been previously linked to macroautophagy.15 PEs are conjugated to an autophagosomic protein MAP1LC3-II and serves as membrane anchor.15,16 Macroautophagy is a major pathway for bulk degradation of cellular constituents, including proteins and organelles. More recently, autophagic dysfunction has been implicated with neurodegeneration through disturbances of intracellular protein turnover that might ultimately result in neurotoxic protein aggregation.17 Elevated PE levels point therefore to macroautophagic activity in response to neurotoxin-induced abnormal protein accumulations. The exact mechanisms underlying neuronal macroautophagy as well as disturbances in autophagic protein degradation are still not fully understood.17
To further elucidate the neurochemistry of BMAA induced lesions, targeted SIMS imaging was performed in burst-alignment mode followed by multivariate image analysis (Figure 3). The MAF scores reveal distinct localization of chemical species to the lesions. Inspection of the corresponding loadings can be used to identify the specific m/z values (i.e. variables). The results show distinct localization of phophatidylcholine (m/z 184.1) as well as its major fragments choline (m/z 104.1) and trimethylethylimine (m/z 86.1) (Figure 3). An elevated phosphatidylcholine level indicates accumulation of cell membrane and is most likely the result of inflammatory cell activation and recruitment. This process typically involves astrocyte activation and recruitment as a consequence of tissue damage and inflammation.18 Phosphatidylcholine lipids can therefore serve as a marker for neuroglial cell activation in neurotoxin-induced nerve cell degeneration.
Figure 3. ToF SIMS single ion images of hippocampal lesions.
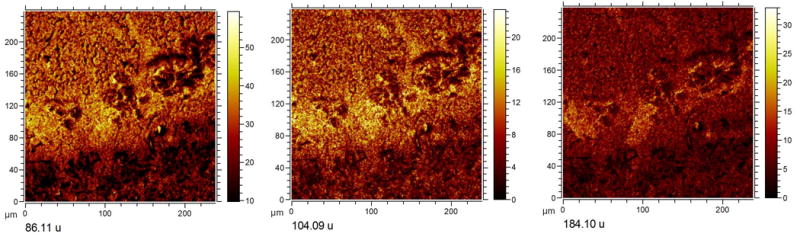
Manual inspection of variables (m/z) revealed by the loadings. The MAF data suggest elevated levels of phosphatidylcholine (PC, m/z 184.1) and its fragments (choline 104.09 and trimethylethylimine, at m/z 86.11). PC and choline fragments show localization to the lesions, suggesting elevated phospholipid levels to be associated with neurodegenerative mechanisms underlying BMAA neurotoxicity.
Taken together, these data suggest elevated phospholipid levels to be associated with neurodegenerative mechanisms underlying BMAA neurotoxicity. These comprise both neuro glial cell activation as well as neuronal macroautophagy.
5. Conclusions
Multimodal imaging in conjunction with multivariate statistical analysis tools were used for molecular histology-based dissection of anatomical features. We used these approaches to identify regional changes in neuronal lipid profiles in BMAA-induced neurodegeneration. This analytical strategy is a powerful approach for studying neuronal lipid chemistry and metabolism in situ. It offers a promising alternate to classical histological techniques because it provides more complex biochemical information while maintaining spatial information.
Acknowledgments
The European Research Council (AGE, ERC advanced grant), the Wallenberg Foundation (AGE, Wallenberg Fellow), the Swedish Research Council VR (AGE, JH), the USA National Institutes of Health (AGE, R01 EB002016-20A1) and FORMAS (EB, OK) are acknowledged for financial support.
Footnotes
The authors declare no conflict of interest.
References
- 1.Banack SA, Johnson HE, Cheng R, Cox PA. Mar Drugs. 2007;5:180–96. doi: 10.3390/md504180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banack SA, Cox PA. Neurology. 2003;61:387–9. doi: 10.1212/01.wnl.0000078320.18564.9f. [DOI] [PubMed] [Google Scholar]; Spencer PS, Nunn PB, Hugon J, Ludolph AC, Ross SM, Roy DN, Robertson RC. Science. 1987;237:517–22. doi: 10.1126/science.3603037. [DOI] [PubMed] [Google Scholar]
- 3.Pablo J, Banack SA, Cox PA, Johnson TE, Papapetropoulos S, Bradley WG, Buck A, Mash DC. Acta neurologica Scandinavica. 2009;120:216–25. doi: 10.1111/j.1600-0404.2008.01150.x. [DOI] [PubMed] [Google Scholar]
- 4.Karlsson O, Berg AL, Lindstrom AK, Hanrieder J, Arnerup G, Roman E, Bergquist J, Lindquist NG, Brittebo EB, Andersson M. Toxicol. Sci. 2012;130:391–404. doi: 10.1093/toxsci/kfs241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanrieder J, Phan NTN, Kurczy ME, Ewing AG. ACS chemical neuroscience. 2013;4:666–679. doi: 10.1021/cn400053c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cornett DS, Reyzer ML, Chaurand P, Caprioli RM. Nature Methods. 2007;4:828–833. doi: 10.1038/nmeth1094. [DOI] [PubMed] [Google Scholar]
- 7.Benabdellah F, Seyer A, Quinton L, Touboul D, Brunelle A, Laprevote O. Anal Bioanal Chem. 2009 doi: 10.1007/s00216-009-3031-2. [DOI] [PubMed] [Google Scholar]
- 8.Hanrieder J, Ekegren T, Andersson M, Bergquist J. J Neurochem. 2013;124:695–707. doi: 10.1111/jnc.12019. [DOI] [PubMed] [Google Scholar]
- 9.Fletcher JS. Analyst. 2009;134:2204–15. doi: 10.1039/b913575h. [DOI] [PubMed] [Google Scholar]
- 10.Karlsson O, Roman E, Berg AL, Brittebo EB. Behavioural Brain Research. 2011;219:310–20. doi: 10.1016/j.bbr.2011.01.056. [DOI] [PubMed] [Google Scholar]
- 11.Karlsson O, Roman E, Brittebo EB. Toxicol. Sci. 2009;112:185–95. doi: 10.1093/toxsci/kfp196. [DOI] [PubMed] [Google Scholar]
- 12.Karlsson O, Lindquist NG, Brittebo EB, Roman E. Toxicol. Sci. 2009;109:286–95. doi: 10.1093/toxsci/kfp062. [DOI] [PubMed] [Google Scholar]
- 13.Paxinos G, Watson C. The rat brain in stereotaxic coordinates. 6th. Academic press; Oxford: 2007. [Google Scholar]
- 14.Sodhi RNS. Analyst. 2004;129:483–487. doi: 10.1039/b402607c. [DOI] [PubMed] [Google Scholar]
- 15.Jaeger PA, Wyss-Coray T. Molecular neurodegeneration. 2009;4:16. doi: 10.1186/1750-1326-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. Molecular biology of the cell. 2004;15:1101–11. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez-Vicente M, Cuervo AM. Lancet neurology. 2007;6:352–61. doi: 10.1016/S1474-4422(07)70076-5. [DOI] [PubMed] [Google Scholar]
- 18.Raivich G, Bohatschek M, Kloss CU, Werner A, Jones LL, Kreutzberg GW. Brain research reviews. 1999;30:77–105. doi: 10.1016/s0165-0173(99)00007-7. [DOI] [PubMed] [Google Scholar]


