Abstract
Classification of the human gut microbiome into distinct types, or “enterotypes,” provides an attractive framework for understanding microbial variation in health and disease. However, as discussed here, several different methods of collapsing enterotype variation into a few discrete clusters suggest that enterotype distribution is continuous and can vary widely within an individual.
Introduction
Interindividual variation in the human gut microbiome is large when considering relative shifts in both dominant and rare taxa (Costello et al., 2009; Huttenhower et al., 2012; Yatsunenko et al., 2012) and, with deeper sequencing, has been related to diverse human diseases (Clemente et al., 2012). Understanding the nature of microbial variation in healthy adults, how this variation becomes altered in human disease, and whether we can use microbial features to predict specific conditions are among the key challenges in the field at present.
It has been suggested that human gut microbiomes fall into three distinct types or “enterotypes” (Arumugam et al., 2011). Although in the original finding these clusters were reported as “densely populated areas in a multidimensional space of community composition” that are “not as sharply delimited as, for example, human blood groups,” the popular press and secondary literature have tended to focus on the idea of discrete types. The utility of discrete clustering in microbiome analyses remains a topic of debate. Here we describe several conceptual qualifications that should be considered when using an enterotype framework to analyze data. As suggested previously by Jeffery et al. (2012), we find that most human gut microbiome data collected to date support continuous gradients of dominant taxa rather than discrete enterotypes. Our analysis also indicates that an individual’s enterotype can be highly variable and that putative discrete clusters are less effective as disease biomarkers than a predictive model constructed from the raw taxon-relative abundances. Because the concept of enterotypes has important implications for how to conduct microbiome-related disease research, and because similar analyses continue to be performed in more recently published studies, we believe it is important to describe alternative interpretations of the enterotype concept and the assumptions that underlie these different interpretations so that investigators can choose the model that best fits their study system.
Why Should We Care Whether There Are Discrete Clusters?
To nonspecialists, the argument over enterotypes might seem somewhat esoteric: why does it matter if variation tends to be continuous or discrete? This argument is important because our model of how microbial diversity is structured has a large impact on framing research questions, and informing the approaches we should take in order to understand the considerable variability in the human microbiome.
One of the most surprising—and at times baffling—findings from culture-independent observations of taxonomic microbiome variation has been the extraordinary within- and between-individual diversity in the human gut. It is now well established that a single human gut microbiome can harbor hundreds of unique species. Furthermore, individuals share little of their microbial communities (Costello et al., 2009; Huttenhower et al., 2012) and a single person has persistent and distinctive strains of bacteria (Faith et al., 2013; Schloissnig et al., 2013). If human microbiomes could indeed be divided into three separate groups, we could collapse this highly multidimensional human microbiome variation into just a few easily understood categories.
The existence of discrete enterotypes would have broad implications in the study of microbiome-related human disease. For example, if patients could be grouped according to enterotype (as with blood type), we could more readily pursue personalized microbiome-based diagnostics and therapeutics. This could greatly simplify the tasks of inferring biomarkers for disease, predicting the effects of perturbation on the microbiome, and mapping the complex network of interactions between microbial taxa. On the other hand, if human microbiomes fall along multidimensional gradients, the task of discovering biomarkers for disease requires more sophisticated statistical methodologies, and substantially larger sample sizes to support hypothesis testing.
There are also cases where properties of specific genes or microbes in the microbiome are related to health outcomes, as in classic single-pathogen paradigms. In such cases, focusing on overall community-based categories or patterns may be less helpful, because these broad categories could mask important underlying variation in individual strains that drives phenotypes or clinical outcomes.
How Well Do Discrete Enterotypes Link to Human Disease?
One potential advantage of enterotype analysis is that enterotypes may relate to human disease. However, collapsing global microbiome variation into dominant clusters need not necessarily identify disease associations better than a more directly data- or hypothesis-driven approach. For example, if there is a bacterium whose increased abundance is associated with a given disease and with a putative enterotype cluster, then relying on the cluster membership for diagnosis and biomarker discovery may mask potentially important disease-related variation within each putative cluster (Figure 1). In a cluster-based approach, a person in the disease-related cluster would be classified as being high risk, while people in the other clusters would be classified as low risk, regardless of the individual’s position along the spectrum of intracluster variation. In contrast, an approach directly modeling association of the disease with specific bacteria can support more sensitive and specific diagnostic tools. Although disease status might be statistically associated with discrete enterotype membership in some cases, this need not imply that discrete clusters are the best biomarkers for a given disease relative to other descriptions of the data (for example, abundances of particular taxa or locations of samples in multivariate spaces defined by different distance metrics and dimensionality reduction criteria). It may still be useful to discretize a biomarker to make it actionable for clinical purposes, but we believe that it makes more sense to discover the biomarker in a supervised way, linking it directly to disease risk, rather than relying on unsupervised clusters found in population structure.
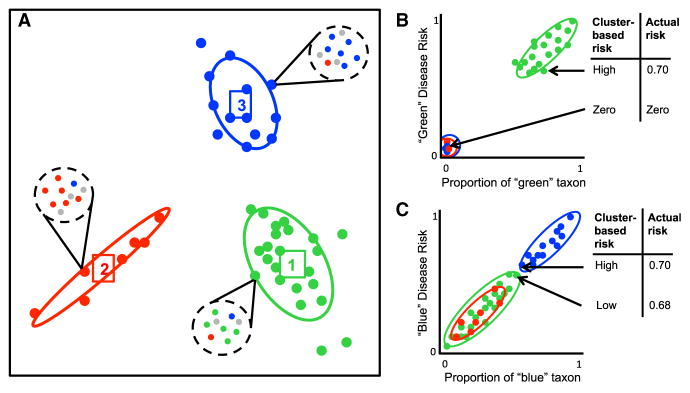
Figure 1. Clustering Continuous Data May Mask Within-Cluster Variation.
(A) Hypothetical clustering of complex bacterial communities. Each cluster is colored by its dominant strain (green, red, or blue). In this example, most taxa are present in every cluster, with the exception of the green taxon, which only appears in cluster one. It is assumed that the blue and green taxa are respectively associated with “blue disease” and “green disease.”
(B and C) Disease risk for samples plotted on a continuous axis showing proportion of a given representative taxon. (B) When disease risk is correlated with taxa found in only one cluster, associations between disease risk and enterotype will be strong, but clustering may still mask meaningful variation within the disease cluster. (C) When disease risk is correlated with taxa found in more than one cluster, clustering may cause even more extreme masking of important disease risk variation within clusters.
We expect that a predictive model using taxon-relative abundances would be more effective than a model using cluster labels from unsupervised enterotype clustering. To test this hypothesis, we compared the performance of a machine-learning-based classifier (Knights et al., 2011), trained on the full relative abundance measurements for all taxa, to the performance of a classifier that used comembership information from unsupervised clusters, on two different health-related human gut microbiome classification tasks. The cluster-based classifier estimated the probability that a sample belonged to a particular category using the fraction of coclustered samples that also belonged to the category of interest. The classification tasks were classifying lean versus obese adults (Turnbaugh et al., 2009) and classifying healthy subjects versus patients with Crohn’s disease (Morgan et al., 2012). In each case, the taxon-based classifier outperformed substantially the cluster-based classifier (t test comparing areas under the receiver operating characteristic curves, p = 2.2 × 10−12, p < 2.2 × 10−16, respectively; see Supplemental Information available online), with an increased predictive strength of 7.1% and 35.0%, respectively (increased area under the receiver operating characteristic curve; Figures S1 and S2).
Several more subtle aspects of analysis should be considered when exploring potential enterotype-disease associations. First, diseases associated with the gastrointestinal tract can involve substantial shifts in microbiome taxonomic profile, as in the case of inflammatory bowel disease (Morgan et al., 2012). Subjects with such diseases might therefore cluster separately from healthy individuals, but the disease cluster would represent less a naturally occurring discrete enterotype than simply an altered host state. Another consideration when using the discrete enterotype paradigm for disease associations is how well it can incorporate temporal dynamics. As described below, some healthy individuals’ enterotypes are highly variable over time. This lack of stability itself could be a biomarker or precondition for certain diseases and is much harder to characterize in a discrete framework. Relying on discrete cluster membership only allows us to determine whether a subject’s microbiome crossed an established threshold toward another profile but does not permit characterization of the extent or trajectory of change. It would also mask within-subject variability when the microbiome profile remained within a putative cluster, although this variability might also be important.
Why Might We Think There Are Clusters when There Are Not?
The current balance of evidence indicates that human gut microbial communities vary continuously along a complex multidimensional distribution. Furthermore, the existence of discrete clusters in the human microbiomes is a strong claim requiring substantial evidence.
A major challenge in clustering high-dimensional data is accurately determining whether discrete clusters are actually present. Appropriate statistical tests based on established thresholds for cluster quality evaluation should be used to determine whether cluster structure exists (Koren et al., 2013). A thorough quantitative investigation of established clustering methods and tests for microbiome data, for example from the gene expression literature, would be a useful resource for the community.
We find that continuous gradients of several dominant genera are strongly associated with interindividual variation in a number of published human gut microbiome data sets. Dominant genera including Bacteroides and Ruminococcus tend to vary continuously between and within putative enterotypes. There is evidence that some dominant taxa, most notably Prevotella, are absent from a fraction of the human population, leading to a discrete effect; however, we have found that even Prevotella varies substantially within putative enterotypes. These taxa increase toward one extreme margin of a putative enterotype and decrease toward the other, therefore implying that the putative discrete clusters may be masking potentially important variation.
We also find that certain visualizations such as scatter diagrams or “starburst” plots can cause the eye to perceive discrete clusters to be stronger than they are, as demonstrated by comparison in Figure 2. We recommend that such plots be accompanied by unmodified unsupervised ordination plots. Supervised ordination plots can be used to find the projection that most clearly shows clustering assessed using other approaches. However, this approach may show such clusters visually even if they are not statistically significant, so it should be used with caution when the presence of cluster structure is under consideration. We find that supervised ordination plots are likely to show false cluster structure when the number of features in the data is much higher than the number of samples (Figures 2A and 2D), as is often the case in microbiome analyses.
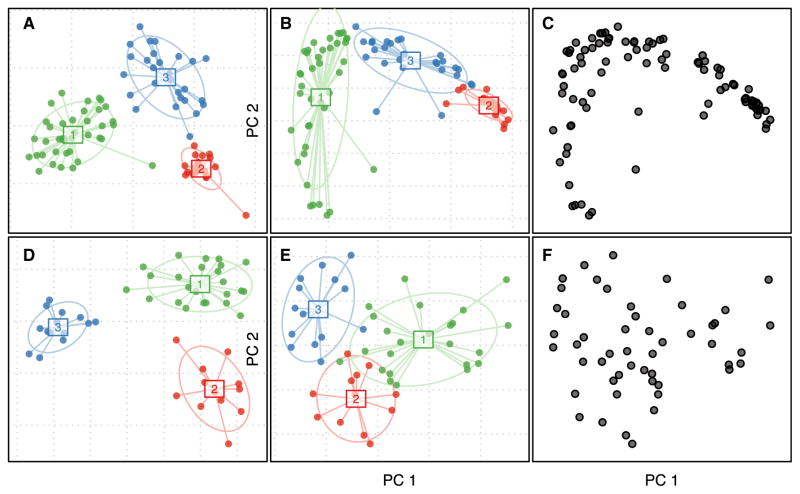
Figure 2. Common Visualizations Can Support Different Conclusions.
(A) Between-class analysis of soil samples with varying pH (Lauber et al., 2009). The ordination method used is supervised, meaning that the plot is intended to make the clusters look as separate as possible, while assuming that the clusters are valid based on simulated data.
(B) Unsupervised principal coordinates plot of soil samples in (A), colored by membership in putative, but nonsignificant, clusters.
(C) Exactly the same plot as (B) but without colors or annotation, revealing a lack of clear discrete structure in the data.
(D–F) The same types of plots as (A)–(C) but using simulated data with three dominant taxa and with no discrete cluster structure (Supplemental Information).
Skewed or biased sampling frames can also confound discrete cluster analysis. We know that certain host and environmental factors have large effects on the gut microbiome profile, and choice of sampling frame across these factors is strongly linked to the resulting conclusions about the nature and extent of microbiome variation. For example, discrete cluster analysis in a hypothetical study involving adults with and without recent antibiotic usage would probably produce one or more clusters linked to the treatment group, but this would not indicate that discrete clusters were present in the normal variation of the healthy untreated gut microbiome.
We hypothesized that discrete clustering within nested sampling frames in a study involving multiple host factors with nested effect sizes would illustrate the sensitivity of putative discrete clusters to the choice of sampling frame. Using previously published data (Yatsunenko et al., 2012), we performed discrete clustering of gut enterotypes within nested sampling frames from individuals of wide-ranging age in three different countries. We used these host factors to choose nested subsets of the full data set in order of effect size: age, westernized versus nonwesternized diet, and nationality of the host (Yatsunenko et al., 2012). Cluster analysis of all samples produced two clusters strongly linked to age; after removing subjects under the age of two, cluster analysis produced clusters associated with westernized/nonwesternized status of the host diet; after removing subjects from the USA, cluster analysis produced two clusters associated with the two remaining nonwesternized nations (Figure 3C). The cluster quality metrics for the first two sampling frames approached or exceeded the suggested threshold of 0.5 for claiming discrete clusters (average silhouette width = 0.52, 0.49, respectively; final cluster average silhouette width = 0.2). In all cases, the optimal number of clusters chosen via silhouette width was two. This analysis demonstrates that cluster comembership between samples can be driven by sampling frame and selection bias, rather than by inherent natural discrete variation. Furthermore, if one performs cluster analysis in a study comprised of only populations that differ strongly in host and/or environmental factors, then apparent clustering could simply be an artifact of lack of sampling between the extremes. This analysis also demonstrates that putative clusters will not generalize among studies involving different ranges of host factors. We would like to note, however, that the original enterotype clusters claimed were not linked to the country of origin of the subjects (Arumugam et al., 2011), making it less likely that choice of sampling frame influenced the results.
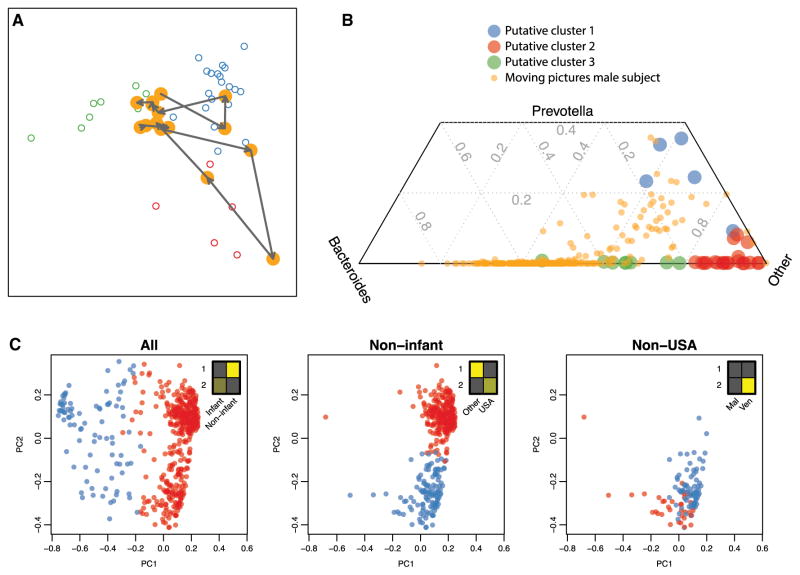
Figure 3. Enterotypes Can Be Unstable, Continuous, and Driven by Sampling Frame.
(A and B) Genus-level enterotype time series superimposed on putative clusters derived from 33 subjects (Arumugam et al., 2011). (A) Two selected trajectories of consecutive daily samples are shown for a single male subject (Caporaso et al., 2011). Meta-HIT samples are colored by putative enterotype cluster. The two selected trajectories show the subject’s microbiome profile “walking” from one putative enterotype to another over the course of several days. (B) Ternary plot of composition of Bacteroides, Prevotella, and other genera daily for a year for a single subject (Caporaso et al., 2011) and for published cross-sectional samples (Arumugam et al., 2011). These analyses demonstrate the temporal fluidity of enterotypes and provide clear proof by counterexample that enterotypes are not discrete states that separate individuals.
(C) Clustering performed on nested sampling frames from individuals of wide-ranging age in three different countries (Yatsunenko et al., 2012). Methods were described previously (Arumugam et al., 2011). Insets show relative sizes of sample subsets (columns) in putative clusters 1 and 2 (rows). Clustering of all individuals, those over age 2, and those over age 2 and not living in the USA identify clusters driven by age, USA versus non-USA citizenship, and Malawi versus Venezuela citizenship, respectively (chi-square test, p = 2.5 × 10−66, 2.5 × 10−63, 8.6 × 10−4, respectively), demonstrating that cluster comembership between samples is driven by sampling frame.
Is an Individual’s Enterotype Stable over Time?
One implicit assumption in the discrete enterotype claim is that an individual’s enterotype is relatively fixed over time. If an individual were to switch enterotypes regularly, then that individual’s microbiome would have to transition between clusters, leading to intermediate states and a blurring of putative cluster boundaries. Essentially every study that has addressed the question has shown that serial samples from the same individual tend to be relatively similar compared to differences among individuals. However, because of substantial variation observed within individuals in studies with a small number of time points for each individual (Costello et al., 2009; Huttenhower et al., 2012), we hypothesized that some individuals cross the putative enterotype boundaries on a regular basis. To test this hypothesis, we projected a dense time series of 1 year’s worth of daily gut microbiome samples from a single individual (Caporaso et al., 2011) onto the published putative enterotype clusters (Arumugam et al., 2011). We performed ordination and putative cluster identification according to the exact methods described in the original finding and then overlaid two courses of consecutive days in which the microbiome of the time series individual traverses from one putative cluster to another (Figure 3A). We also compared the mixtures of Bacteroides, Prevotella, and other genera in the time series individual to those in the putative clusters (Figure 3B). Bacteroides and Prevotella were the dominant genera, representing the two most robust clusters in the previously claimed discrete enterotypes. The comparison indicates that the microbiome of the single time series individual occupies, at times, nearly every region of the space of mixtures observed in the multisubject, single time point data. Although this analysis requires validation in a large cohort, it demonstrates that for some healthy subjects, enterotype can vary widely and continuously over time.
Conclusions
In light of our findings, we believe that previous analyses produced overconfidence in the claim of discrete enterotypes and that continuous variation is the simpler and therefore better-supported conclusion. Furthermore, we have demonstrated that discrete clustering methodologies can be sensitive to sampling frame bias and selection bias. We also evaluated the utility of unsupervised putative discrete clusters for building predictive biomarkers but found strong evidence that they are outperformed by predictors that model complex and multidimensional taxon distributions. Consequently, although discrete clusters may be significantly correlated with a disease state, they may not be appropriate for predicting that disease state due to masking of important within-cluster variation in critical taxa. Finally, in a meta-analysis including both dense single-individual time series data and cross-sectional multiple-individual data, we demonstrated that a healthy adult human’s microbiome can traverse much of the total variation space of healthy human gut microbiomes throughout the course of a year, providing evidence that enterotypes are fluid and continuous.
In particular, it is critical to note the following confounding factors in considering the existence of discrete community types:
Confounding environmental variables where only the extremes of the range are sampled.
Null models that provide apparent support for clustering because of poor model fit rather than because clustering is an appropriate statistical description of the data.
Stability over time, which could arise because people resemble themselves over time in general rather than because there are specific barriers to switching cluster types.
Association of clusters with clinical variables, which may mask more precise underlying relationships but still yield a statistically significant result.
We conclude that although the enterotype hypothesis is a conceptually appealing one, and microbial community variation will certainly be important for diagnosing and predicting many microbiome-associated diseases, the appropriate statistical description of the microbiome is still an emerging area of inquiry. Also, the evidence against discrete community types, including those with barriers preventing switching among them, is accumulating rapidly.
Supplementary Material
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes one table and two figures and can be found with this article online at http://dx.doi.org/10.1016/j.chom.2014.09.013.
References
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, et al. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J, Knights D, Gajer P, Ravel J, Fierer N, et al. Genome Biol. 2011;12:R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente JC, Ursell LK, Parfrey LW, Knight R. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, et al. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, Creasy HH, Earl AM, Fitzgerald MG, Fulton RS, et al. Nature. 2012;486:207–214. [Google Scholar]
- Jeffery IB, Claesson MJ, O’Toole PW, Shanahan F. Nat Rev Microbiol. 2012;10:591–592. doi: 10.1038/nrmicro2859. [DOI] [PubMed] [Google Scholar]
- Knights D, Parfrey LW, Zaneveld J, Lozupone C, Knight R. Cell Host Microbe. 2011;10:292–296. doi: 10.1016/j.chom.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren O, Knights D, Gonzalez A, Waldron L, Segata N, Knight R, Huttenhower C, Ley RE. PLoS Comput Biol. 2013;9:e1002863. doi: 10.1371/journal.pcbi.1002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber CL, Hamady M, Knight R, Fierer N. Appl Environ Microbiol. 2009;75:5111–5120. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, et al. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloissnig S, Arumugam M, Sunagawa S, Mitreva M, Tap J, Zhu A, Waller A, Mende DR, Kultima JR, Martin J, et al. Nature. 2013;493:45–50. doi: 10.1038/nature11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.