Abstract
Amyotrophic lateral sclerosis (ALS) is a devastating neurodegenerative disorder characterized by loss of upper and lower motor neurons. Different mechanisms contribute to the disease initiation and progression, including mitochondrial dysfunction which has been proposed to be a central determinant in ALS pathogenesis. Indeed, while mitochondrial defects have been mainly described in ALS-linked SOD1 mutants, it is now well established that mitochondria become also dysfunctional in other ALS conditions. In such context, the mitochondrial quality control system allows to restore normal functioning of mitochondria and to prevent cell death, by both eliminating and replacing damaged mitochondrial components or by degrading the entire organelle through mitophagy. Recent evidence shows that ALS-related genes interfere with the mitochondrial quality control system. This review highlights how ineffective mitochondrial quality control may render motor neurons defenseless towards the accumulating mitochondrial damage in ALS.
Keywords: C9orf72, FUS, SOD1, Optineurin, Parkin, PGC-1α, PINK1, TDP-43 proteinopathies, TBK1, VCP
Introduction
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disorder characterized by a progressive degeneration of motor neurons in the spinal cord, brain stem and cerebral cortex. An important histopathological hallmark of ALS is the presence of cytosolic protein aggregates in motor neurons and surrounding glia. Patients develop muscle weakness, progressive paralysis and spasticity. Cognitive defects are rare whereas 20% of ALS cases also show frontotemporal dementia. Over time, complications appear with eating difficulties (dysphagia) and respiratory distress (orthopnea and dyspnea). The survival time from diagnosis is 2–5 years. Among treatments, riluzole prolongs life by only a few months but without ameliorating motor functions (Miller et al., 2012) and the antioxidant edavarone has recently been approved by the Federal Drug Administration for ALS.
Most ALS cases (90%) are sporadic with no family history and the remaining 10% are inherited forms, indicating that both environmental and genetic factors are involved in the disease etiology. Because mutations in the copper/zinc superoxide dismutase 1 (SOD1) gene were discovered first in 1993 (Rosen et al., 1993), significant advances in ALS pathogenesis come from animal models expressing mutant forms of SOD1. In the last decade, an important breakthrough was achieved with the identification of new ALS-linked genes. Among these, mutations in two DNA/RNA binding proteins: TAR DNA-binding protein of 43 kDa (TDP-43) and fused in sarcoma (FUS) have put RNA metabolism as a key mechanism in ALS (Sreedharan et al., 2008; Kwiatkowski et al., 2009; Vance et al., 2009). Non-mutated TDP-43 has also been detected in aggregates of the majority of ALS patients, suggesting that deregulation and mislocalization of wild-type TDP-43 mediate both sporadic and familial ALS (Arai et al., 2006). More recently, an abnormal expansion of hexanucleotide (GGGGCC) repeat in a non-coding region of the chromosome 9 open reading frame 72 gene (C9orf72) was identified as the most common cause of familial ALS (40-50%) (DeJesus-Hernandez et al., 2011; Renton et al., 2011). Despite the lack of an ATG start codon, the hexanucleotide repeat expansion is translated into dipeptide repeat proteins (Ash et al., 2013). As a possible mechanism, sequestration of RNA-binding proteins by expanded repeat RNA may synergically contribute to the toxicity along with the accumulation of dipeptide repeat proteins (Rohrer et al., 2015). Beyond RNA processing, components of protein/organelle quality control have emerged as important causal factors of ALS, including p97/valosin-containing protein (VCP), ubiquilin, optineurin and p62/SQSTM1. As a consequence, many cellular processes are affected including abnormal protein aggregations, excitotoxicity, mitochondrial abnormalities, oxidative damage and inflammation. One important challenge in the future will be to determine which among these mechanisms are crucial and common in ALS pathogenesis. This may definitely help to identify relevant therapeutic targets. In this review, we discuss the latest evidence showing that compromised mitochondrial quality control is a common determinant in ALS.
Mitochondrial Quality Control Pathways
The major function of mitochondria is the production of adenosine triphosphate (ATP) through the oxidative phosphorylation system (OXPHOS). During cell respiration, electrons are transferred to oxygen molecules and produce superoxide anions. Because they are highly toxic, superoxide anions are usually neutralized by antioxidant enzymes. However in pathological conditions, mitochondrial dysfunction leads to ATP depletion, superoxide anion overload and release of proapoptotic molecules such as cytochrome c. Cells have nevertheless adapted a mitochondrial quality control (MQC) system to overcome mitochondrial defects. MQC is particularly crucial for neurons which are long living cells and thereby can accumulate damage in mitochondria.
Mitochondrial protein quality control
Quality control of mitochondrial proteins restores the native conformation and function of damaged mitochondrial proteins, or eliminates them if they reach a point of no return. This system therefore assures protein homeostasis in mitochondria, via the ubiquitin-proteasome (UPS) system and mitochondrial chaperones and proteases.
The UPS selectively eliminates proteins of the outer mitochondrial membrane (OMM) by tagging them with lysine 48-linked polyubiquitin chain (K48 chain). Proteins with K48 chains are extracted by the segregase p97/VCP, then delivered to the proteasome for degradation (Tanaka et al., 2010). Strikingly, some studies have suggested that damaged proteins localized at the inner mitochondrial membrane (IMM) or in the mitochondrial matrix can be retrotranslocated to the OMM where they will be ubiquitinated and eliminated by the UPS (Margineantu et al., 2007; Azzu and Brand, 2010).
Heat-shock proteins (Hsp), especially Hsp60 and Hsp70 are known for their role in importing newly synthesized mitochondrial proteins into mitochondrial matrix and conferring their native conformation (Ostermann et al., 1989; Voos et al., 1996; Liu et al., 2001). When exposed to stress, Hsp also stabilize damaged mitochondrial proteins, preventing them from aggregating (Bender et al., 2011). If proteins are highly damaged and become irreparable, then Hsp70 maintains them in a soluble state until they are proteolyzed by mitochondrial proteases (Wagner et al., 1994).
Misfolded mitochondrial proteins which are unable to recover their native form are redirected to the mitochondrial proteolytic system. Proteases Lon and AAA+/FtsH (Filament-forming temperature sensitive) degrade aberrant proteins in the mitochondrial matrix and IMM, respectively. Lon has both chaperone and proteolytic activities and is considered as the main mitochondrial protease, since Lon deletion in yeast results in an increased number of mitochondrial protein aggregates (Bender et al., 2011). Lon mainly targets mitochondrial enzymes with Fe-S clusters which become easily unstable after covalent oxidative modification (Bota and Davies, 2002; Bender et al., 2011). In contrast, AAA+ proteases do not focus on specific proteins but degrade all sorts of misfolded proteins in the IMM (Leonhard et al., 1999), as well as non-assembled proteins such as subunits of respiratory complexes (Arlt et al., 1998).
Mitochondrial biogenesis
Mitochondrial biogenesis, or mitochondriogenesis, is the process by which new mitochondrial components are synthesized to replenish damaged mitochondria. This mechanism also allows the genesis of proteins involved in OXPHOS and in other crucial mitochondrial functions. Activating mitochondriogenesis within cells is beneficial during mitochondrial dysfunction since it preserves mitochondrial energy metabolism and integrity and therefore prevents cell death and pathology occurrence.
Mitochondriogenesis is regulated by the master transcriptional coactivator peroxisome proliferator-activated receptor gamma (PPAR-γ) coactivator-1α (PGC-1α). PGC-1α coordinates the expression of mitochondrial components between the nuclear and mitochondrial genomes, through its binding to nuclear receptors (PPAR-γ, estrogen-related receptor α) or transcription factors such as nuclear respiratory factors (NRFs) (Puigserver and Spiegelman, 2003). Moreover, PGC-1α increases the expression levels of NRFs (Wu et al., 1999). Then, NRFs regulate the expression of respiratory complex subunits (Wu et al., 1999), as well as proteins implicated in mitochondrial import (Blesa et al., 2007) and heme biosynthesis (Braidotti et al., 1993). NRFs also modulate the expression of mitochondrial transcription factor A (TFAM) which is responsible for mitochondrial DNA transcription and replication (Virbasius and Scarpulla, 1994). PGC-1β and PGC-1 related coactivator, both members of the PGC-1α co-activator family, also contribute to mitochondrial biogenesis (Lin et al., 2003; Gleyzer et al., 2005), but their role has not been fully elucidated yet.
Apart from PGC-1α, silent mating type information regulation two-1 (SIRT1) and adenosine monophosphate-activated protein kinase (AMPK) have been shown to contribute to mitochondrial biogenesis. SIRT1 is a NAD-dependent protein deacetylase which regulates the expression of genes involved in mitochondrial respiration through PGC-1α (Lagouge et al., 2006). Indeed, SIRT1 directly interacts with PGC-1α and activates it (Nemoto et al., 2005; Rodgers et al., 2005). Furthermore, the AMPK kinase is an energy cell sensor which interrupts ATP consumption and activates ATP-producing pathways during energy-demanding periods. In this case, AMPK induces mitochondrial biogenesis and activates PGC-1α through direct phosphorylation (Jäger et al., 2007) or indirectly by stimulating SIRT1 (Cantó et al., 2009). Consequently, the AMPK/SIRT1/PGC-1α axis plays a key role in maintaining the mitochondrial and cellular energy metabolism in stress conditions.
Mitochondrial dynamics: fission versus fusion
Mitochondria form a highly dynamic network. Fission and fusion events control the shape, size and number of mitochondria and allow the exchange of proteins and lipids. The fission-fusion balance is necessary to regulate mitochondrial distribution during cell division and differentiation, but also to maintain mitochondrial integrity and cell survival during stress periods (Twig et al., 2008). Indeed, whereas fission facilitates segregation and elimination of defective mitochondria, fusion repairs the damaged mitochondrial DNA and mixes the content of both functional and defective mitochondria.
Mitochondrial fission is mainly regulated by the cytoplasmic protein, dynamin-related protein 1 (DRP1) (Smirnova et al., 2001). While still debated, DRP1 binding on mitochondria may require receptors on the OMM such as mitochondrial fission factor (Mff), Fission 1 (Fis1) and mitochondrial elongation factors (MIEF) (Palmer et al., 2011; Zhao et al., 2011a). Then, DRP1 oligomerizes and forms a constrictive ring around the OMM (Smirnova et al., 2001). GTP hydrolysis by DRP1 triggers the mitochondrion fragmentation. Interestingly, mitochondria-associated membranes (MAMs) with endoplasmic reticulum (ER) facilitate DRP1 recruitment to the OMM and define the position of mitochondrial division sites (Friedman et al., 2011).
Mitochondrial fusion is an important mechanism of the MQC system, mostly because it permits the exchange of mitochondrial DNA and other constituents such as respiratory complexes between adjacent mitochondria to conserve their integrity (Legros et al., 2004). In addition, fusing intact and dysfunctional mitochondria together allows the “dilution” of local damaged components. Fusion between IMM and OMM occurs through distinct yet complementary mechanisms, via optic atrophy 1 (OPA1) and mitofusins (Mfns), respectively.
Long isoforms of OPA1 (L-OPA1) are subjected to proteolytic cleavage by intramitochondrial proteases, generating short isoforms called S-OPA1 (Song et al., 2007). It is proposed that L-OPA1 is anchored to the IMM whereas S-OPA1 is enriched in the intermembrane space (Satoh et al., 2003; Cipolat et al., 2006; Ishihara et al., 2006). L-OPA1 and S-OPA1 interact together to regulate IMM fusion (Song et al., 2007; Zick et al., 2009), although it was initially thought that only L-OPA1 is required for fusion (Ishihara et al., 2006; Anand et al., 2014). However, in stress conditions, reduced mitochondrial membrane potential (ΔΨm) promotes massive cleavage of L-OPA1 to S-OPA1, therefore altering the fusion process (Head et al., 2009). Of note, the presence of OPA1 on only one of the two adjacent mitochondria is sufficient to allow their fusion (Song et al., 2009).
OMM fusion is regulated by mitofusin 1 (Mfn1) and mitofusin 2 (Mfn2). Mfns are characterized by very similar sequences (77% of similarity, 60% of identity) with a slight difference in the number of amino acids (741 amino acids for Mfn1 versus 757 amino acids for Mfn2) (Santel and Fuller, 2001). Mfn1 yet seems to be more indispensable for mitochondrial fusion than Mfn2 (Cipolat et al., 2004; Ishihara et al., 2004). At the structural level, Mfns essentially possess a GTPase domain, as well as two «coiled-coil» domains with heptad repeats (Rojo et al., 2002) which allow Mfns to interact with each other and form homo/heterodimers (Chen et al., 2003). Following GTP hydrolysis, Mfns change their conformation and force the outer membranes of adjacent mitochondria to fuse together (Chen et al., 2003). Interestingly, the absence of Mfns in vitro slows down the fusion of both outer and inner mitochondrial membranes, whereas OPA1 is dispensable for OMM fusion (Song et al., 2009). This indicates that Mfns are also required for IMM fusion. Apart from their fusogenic role, Mfns exert other mitochondrial functions. For instance, they promote mitochondrial transport by interacting with the mitochondrial membrane GTPase Miro and the kinesin adaptor Milton (Misko et al., 2010). Strikingly, Mfn2 is not exclusively mitochondrial; it is also present on ER surface where it interacts with mitochondrial Mfn2 to tether the two organelles. Accordingly, Mfn2 deficiency increases the distance between ER and mitochondria and alters the efficacy of calcium import from ER to mitochondria (De Brito and Scorrano, 2008).
Mitochondrial-derived vesicles
A recently discovered mechanism involving structures called mitochondrial-derived vesicles (MDV) has been shown to eliminate damaged mitochondrial components (for review, see Roberts et al., 2016). MDVs are small vesicles of 70 to 150 nm which contain damaged proteins and lipids originating from different compartments of mitochondria (Neuspiel et al., 2008). The generation of MDVs is considered as an early response of mitochondria to oxidative stress, since they appear in the first minutes after the stress-inducing agent has been applied (Soubannier et al., 2012a). Moreover, this process does not require neither a dissipation of the ΔΨm nor the intervention of fission factor DRP1 (Neuspiel et al., 2008; Soubannier et al., 2012a; McLelland et al., 2014). Indeed, it has been shown that MDVs appear before the stress-induced mitochondrial fragmentation. In vitro experiments have established that the nature of the cargo in MDVs depends on the mitochondrial target of the stress. For instance, the application of antimycin A, an inhibitor of respiratory chain complex III, increases the formation of MDVs which are particularly enriched in oxidized complex III subunits (Soubannier et al., 2012b). Following their split from mitochondria, MDVs are sent to be degraded in lysosomes (Soubannier et al., 2012a; McLelland et al., 2014). They can also be transported to peroxisomes (Neuspiel et al., 2008), although the purpose of this mechanism is still unclear. The formation of MDVs would therefore constitute a first line of defense against total mitochondrial dysfunction. However in extreme conditions, and to insure its survival, cells are forced to entirely eliminate defective mitochondria through a process involving lysosomes but also, unlike MDVs, the autophagy machinery, so called mitophagy (Soubannier et al., 2012a; McLelland et al., 2014).
Mitophagy
Mitophagy is a selective macroautophagy process during which irreparable mitochondria are degraded. It therefore prevents the accumulation of defective mitochondria and limits the release of reactive oxygen species and proapoptotic factors. During mitophagy, mitochondria are engulfed by a structure called phagophore, forming an autophagosome which then fuses with a lysosome before its content is degraded. As the other types of autophagy, mitophagy occurs in the perinuclear space of cells where lysosomes are particularly enriched. It is mainly orchestrated by the PINK1/Parkin pathway (Martinez-Vicente, 2017; Rüb et al, 2017; Whitworth and Pallanck, 2017). These genes have been largely studied in the context of Parkinson's disease. Indeed, mutations in PINK1 (PARK6) and parkin (PARK2) genes contribute to familial forms of this disease (Kitada et al., 1998; Valente et al., 2004).
PTEN-induced kinase 1 (PINK1) is a mitochondrial serine/threonine kinase which in physiological conditions is cleaved by different intramitochondrial proteases (Gakh et al., 2002; Deas et al., 2011). However, following an abnormal dissipation of the ΔΨm, PINK1 mitochondrial import and proteolysis are blocked. PINK1 therefore accumulates on the OMM and recruits its partner Parkin, a cytoplasmic E3 ubiquitin ligase (Matsuda et al., 2010; Narendra et al., 2010). The existence of a genetic epistasis between PINK1 and Parkin has been essentially highlighted in Drosophila (Ziviani et al., 2010). PINK1 phosphorylates Parkin, stimulating its ubiquitin ligase activity and its translocation to impaired mitochondria (Shiba-Fukushima et al., 2014). Lately, additional Parkin-recruiting mechanisms have been brought to light. Indeed, it has been demonstrated that PINK1 is able to phosphorylate ubiquitin on serine 65 (S65), which then binds to Parkin, activates it and induces its translocation to mitochondria (Kane et al., 2014; Okatsu et al., 2015). This explains why Parkin is found polyubiquitinated during mitophagy (Geisler et al., 2010). Conversely, Parkin recruitment to mitochondria is inhibited when a non-phosphorylable form of ubiquitin (ubiquitin S65A) is expressed. Moreover, Kazlauskaite et al. (2015) and Koyano et al. (2014) have shown that, in addition of Parkin phosphorylation and activation by PINK1, its interaction with phospho-ubiquitins helps it gain a maximal level of activity. Ubiquitin is therefore considered as both an activator and a substrat of Parkin. Finally, proteins located on the OMM, such as Mfn2, can be phosphorylated by PINK1 and therefore serve as receptors to recruit Parkin to deficient mitochondria (Ziviani et al., 2010; Chen and Dorn, 2013). After its activation, Parkin ubiquitinates several proteins on the OMM mainly via K63 and K48 ubiquitin chains: K63 chains allow the recognition of damaged mitochondria by phagophores, whereas proteins tagged with K48 chains are degraded by the proteasome (Geisler et al., 2010; Chan et al., 2011).
The PINK1/Parkin pathway manages mitophagy through several mechanisms. Firstly, it induces the fragmentation of dysfunctional mitochondria, therefore segregating damaged mitochondria: daughter mitochondria with a weak ΔΨm are eliminated, while those with a high ΔΨm undergo fusion (Twig et al., 2008). Mitochondrial fragmentation during mitophagy is mainly triggered by Parkin-mediated ubiquitination and degradation of Mfns (Gegg et al., 2010; Ziviani et al., 2010). Secondly, PINK1 and Parkin immobilize defective mitochondria by degrading Miro which links mitochondria to microtubule motors (Kane and Youle, 2011). Thus, blocking movement of impaired mitochondria facilitates their engulfment by autophagic phagophores. Thirdly, the PINK1/Parkin pathway induces the formation and maturation of phagophores near dysfunctional mitochondria by recruiting autophagy initiators such as Unc51-Like Kinase l (ULK1) (Lazarou et al., 2015) and Beclin1 (Michiorri et al., 2010). Generated phagophores can then recognize Parkin-induced ubiquitinated mitochondria via cytoplasmic adaptor proteins, of which the most studied are Optineurin (Wong and Holzbaur, 2014; Heo et al., 2015) and p62/SQSTM1 (Geisler et al., 2010). Adaptors are structurally able to bind ubiquitinated OMM proteins via their ubiquitin-binding domain and the autophagosome-associated protein LC3 (microtubule-associated protein 1A/1B-Light Chain 3) via their LC3-interacting region domain. The binding of autophagosomes to adaptors depends on their phosphorylation by the kinase TANK-binding kinase 1 (TBK1), which is co-recruited along with her partner Optineurin (Matsumoto et al., 2015; Moore and Holzbaur, 2016). Once mitochondria-containing autophagosomes are formed and fused with lysosomes, their content is digested via lysosomal hydrolases. Interestingly, PINK1 and Parkin have been shown to induce the formation of lysosome-degraded MDVs before mitophagy activation (McLelland et al., 2014), indicating that the PINK1/Parkin pathway regulates mitochondrial integrity at different levels.
Finally, mitophagy can also occur through a PINK1/Parkin-independent manner. Novel autophagy receptors located in mitochondria and capable of direct interaction with LC3 have been identified, including Bcl-2 and adenovirus E1B 19 kDa-interacting protein 3 (BNIP3) and Nix/BNIP3L (BNIP3-Like) (Novak et al., 2010; Rikka et al., 2011), or FUNDC1 (FUN14 domain-containing protein 1) (Liu et al., 2012). Apart from mitophagy receptors, cardiolipin redistribution in mitochondria turned out to be a potential mitophagy signal. When mitochondria become dysfunctional, cardiolipin is retrotranslocated from the IMM to the OMM where it recruits phagophores by associating with LC3 (Chu et al., 2013).
Mitochondrial Quality Control Defects in ALS
MQC defects in ALS have been previously reported in mutant SOD1 models (for reviews, see Shi et al., 2010; Palomo and Manfreddi, 2015; Edens et al., 2016). Indeed, mutant SOD1 has an increased tendency to localize in mitochondria and leads to the accumulation of defective mitochondria. In this review, we mostly focus on new ALS genes with a particular interest in TDP-43 which is recognized as a major cause of the disease. MQC defects are summarized in Additional Table 1 (60KB, pdf) .
Mitochondrial quality control processes that are altered by ALS causing genes
Mitochondrial protein quality control in ALS
So far, the state of mitochondrial protein quality control in ALS has not been fully investigated. It has been demonstrated on mice spinal cord and in vitro neuronal models that ALS mutant SOD1 alters the expression of Hsp70 (Fukada et al., 2004; Yamashita et al., 2007) and downregulates the mitochondrial protease Lon (Fukada et al., 2004). On the other hand, TDP-43 and FUS abnormally interact with the mitochondrial chaperone Hsp60 in different ALS models (Freibaum et al., 2010; Deng et al., 2015), exacerbating mitochondrial dysfunction (Deng et al., 2015).
Mitochondrial biogenesis in ALS
It is becoming increasingly evident that mitochondrial biogenesis is impaired in ALS. Current studies are aiming to find possible therapeutic targets to restore normal mitochondriogenesis and slow down the disease progression, and PGC-1α seems to be a suitable candidate. Indeed, PGC-1α and its downstream effector NRF1 are downregulated in spinal cord and muscles tissues of patients and mutant SOD1 mice (Thau et al., 2012; Russell et al., 2013). Interestingly, it has been shown that single-nucleotide polymorphisms reported in the brain-specific promoter region of PGC-1α modify age of onset and survival of ALS patients and mutant SOD1 mice (Eschbach et al., 2013). So far, strategies aiming to increase PGC-1α expression in ALS models were very fruitful. PGC-1α overexpression protected against mitochondrial fragmentation and neuronal death in rat motor neurons expressing mutant SOD1 (Song et al., 2013). It also slowed the disease progression in mutant SOD1 mice by preserving their motor activity and attenuating motor neuron degeneration, even though the effect on survival is debated (Liang et al., 2011; Zhao et al., 2011b; Golko-Perez et al., 2017). PGC-1α upregulation in mutant SOD1 mouse muscles also delays muscle atrophy but without affecting survival (Da Cruz et al., 2012). Furthermore, treating mutant SOD1 mice with PPAR-γ agonist pioglitazone ameliorated their survival and motor performance and reduced motor neuron loss (Kiaei et al., 2005; Schutz, 2005).
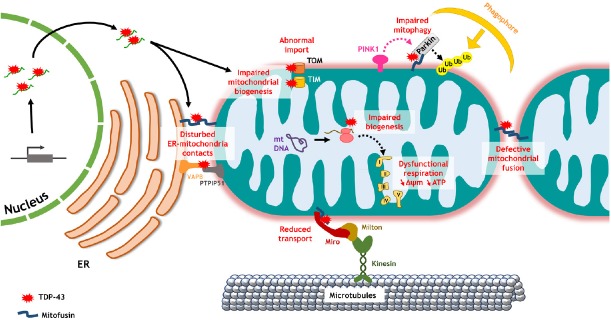
The toxic effect of TDP-43 on mitochondrial biogenesis in ALS has been lately brought to light. AMPK activity was found drastically diminished in spinal cords and brains of transgenic mice carrying the A315T mutation (Perera et al., 2014). Transcriptome analyses performed on A315T mutant TDP-43 mice revealed a dysregulation of RNA regulating OXPHOS or other mitochondrial functions (Stribl et al., 2014). More recently, Wang et al. (2016) demonstrated a direct role of TDP-43 in mitochondrial biogenesis impairment using different cell models, including rat primary neurons and patient fibroblasts (Figure 1). In physiological conditions, wild-type TDP-43 is poorly localized at mitochondria. However, when it is overexpressed or carries ALS mutations, TDP-43 abundantly accumulates on the IMM. In this case, it binds to mitochondria-transcribed mRNA encoding for the complex I subunits ND3 and ND6 and blocks their protein translation (Wang et al., 2016). Therefore, TDP-43 specifically reduces complex I assembly and impairs ATP production (Wang et al., 2016). The same group also showed that TDP-43 abnormally interacts with translocases of the OMM, Tom20 and Tom70, or of the IMM, Tim77, altering the import of nuclear-encoded mitochondrial proteins (Wang et al., 2016) (Figure 1). We and others have demonstrated that TDP-43 also deregulates the expression of nuclear-encoded mitochondrial proteins such as Mfns in rat and fly brain neurons (Sephton et al., 2011; Khalil et al., 2017). TDP-43 therefore perturbs the expression of mitochondrial proteins encoded by both nuclear and mitochondrial genomes. Thus increasing mitochondrial biogenesis is expected to counteract TDP-43-induced toxicity. Accordingly, PPAR-γ agonist pioglitazone rescues locomotor defects in Drosophila expressing TDP-43 in motor neurons (Joardar et al., 2015).
Figure 1.
Mitochondrial quality control defects mediated by TDP-43 dysregulation.
TDP-43 impairs biogenesis of mitochondrial proteins by altering nuclear-encoded or mitochondria-transcribed mRNA, or by preventing protein import to mitochondrial matrix through TOM/TIM translocases. In particular, TDP-43 perturbs the expression and assembly of complex I subunits leading to mitochondrial dysfunction, as well as mitofusin which causes excessive fragmentation of mitochondria. Mitofusin downregulation may also affect mitochondrial transport and mitophagy. TDP-43 disturbs ER-mitochondria contacts as well by blocking the interaction between VAPB and PTPIP51, and probably between both organelle's mitofusins. ΔΨm: mitochondrial membrane potential; ER: endoplasmic reticulum; mt: mitochondrial; PINK1: PTEN-induced kinase 1; PTPIP51: protein tyrosine phosphatase interacting protein 51; TDP-43: TAR DNA-binding protein of 43 kDa; TIM: translocase of the inner membrane; TOM: translocase of the outer membrane; Ub: ubiquitin; VAPB: vesicle-associated membrane protein-associated protein B.
On the other hand, decreased levels of PGC-1α have been reported in mice expressing mutant FUS in brain stem and spinal cord (Bayer et al., 2017), and application of PPAR-γ agonist rosiglitazone rescues dendrite loss and spatial memory of mutant FUS rats (Huang et al., 2012). While the effect of C9orf72 mutation on mitochondrial biogenesis is still clouded, recent studies showed that mutant C9orf72 enhances PGC-1α expression in patient fibroblasts (Onesto et al., 2016) and interacts with the mitochondrial import translocase Tim50 in Neuro2A cells (Blokhuis et al., 2016).
Mitochondrial dynamics in ALS
Studies on ALS patients and animal models have agreed that mitochondrial dynamics balance is altered in this pathology, leaning towards excessive fragmentation of mitochondria. Smaller mitochondria have been described in different models expressing ALS mutant SOD1 (Raimondi et al., 2006; Magrané et al., 2009, 2012; Vande Velde et al., 2011; Liu et al., 2013; Song et al., 2013; Finelli et al., 2015) and linked to a misexpression of mitochondrial dynamics genes. Indeed, studies on mouse spinal cord and skeletal muscles have shown that mutant SOD1 downregulates Mfns (Liu et al., 2013; Russell et al., 2013) as well as OPA1, while levels of phosphorylated DRP1 and Fis1 were particularly elevated (Liu et al., 2013). Interestingly, DRP1 inactivation recovers normal morphology of mitochondria and prevents death of mutant SOD1-expressing neurons (Song et al., 2013).
Studies realized on in vitro and in vivo TDP-43 models have shown a tendency of mitochondria towards fragmentation (Xu et al., 2010, 2011; Wang et al., 2013, 2016; Magrané et al., 2014; Finelli et al., 2015; Khalil et al., 2017). Few groups sought to decipher the mechanisms behind the abnormal TDP-43-induced mitochondrial fission. For that purpose, we have measured expression levels of genes regulating mitochondrial dynamics. We have shown that expression of pro-fission factor DRP1 remains unchanged in Drosophila brains expressing TDP-43 (Khalil et al., 2017), while Xu et al. (2010) reported an increase in DRP1 phosphorylation in transgenic mice. On the other hand, we reported that TDP-43 does not modify the expression of fusogenic factor OPA1 in Drosophila neurons, while another group discovered in mutant TDP-43 mice that OPA1 expression increases after birth then decreases with time (Stribl et al., 2014). The use of either a mutant or a wild-type form of TDP-43 might justify this difference. More importantly, we and others agree that TDP-43 decreases the expression of the fusogenic factor Mfn in patient muscles (Russell et al., 2013), transgenic mice (Xu et al., 2010) and Drosophila neurons (Khalil et al., 2017) (Figure 1). TDP-43 downregulates mfn expression by binding directly to its mRNA (Khalil et al., 2017). Interestingly, mfn overexpression restores mitochondrial length, movement and function in TDP-43-expressing rat motor neurons (Wang et al., 2013) and ameliorates neuromuscular junction dysfunction and locomotor defects in TDP-43-expressing flies (Khalil et al., 2017). Hence, we propose that TDP-43 induces excessive mitochondrial division by holding up the fusion process between mitochondria.
Mitochondrial fragmentation has been reported in motor neurons expressing wild-type or mutant FUS as well as in fly models (Tradewell et al., 2012; Deng et al., 2015). Patient fibroblasts expressing ALS mutant C9orf72 also show shortened mitochondria (Onesto et al., 2016). Mutations in CHCHD10 which is enriched at mitochondrial cristae junctions have been linked to ALS (Bannwarth et al., 2014). Again, skin fibroblasts from patients with a CHCHD10 mutation exhibit reduced mitochondrial length (Bannwarth et al., 2014). Thus, unbalanced mitochondrial dynamics seems to be a common feature in ALS.
Mitophagy in ALS
Mitophagy can be considered as the most affected MQC mechanism in ALS. Indeed, mutations in genes regulating the mitophagy process such as VCP, optineurin and TBK1 are directly linked to ALS (for review, see Majcher, 2015). ALS mutant VCP is unable to migrate to damaged mitochondria and segregate ubiquitinylated proteins, therefore causing abnormal mitochondrial accumulation in mouse embryonic fibroblasts and fly muscles (Kim et al., 2013; Kimura et al., 2013). Moreover, Moore and Holzbaur showed in HeLa cells that mutations in optineurin and TBK1 interfere with LC3 recruitment to depolarized mitochondria, leading to reduced mitophagic rate (Moore and Holzbaur, 2016).
In other forms of ALS, mitophagy is also likely altered since accumulation of autophagic vacuoles colocalized with mitochondria have been reported in mutant SOD1 mouse motor neurons (Xie et al., 2015) and in patient fibroblasts expressing mutant C9orf72 (Onesto et al., 2016). Of interest, physical interaction between C9orf72 with the autophagy initiator ULK1 strongly suggests a role of C9orf72 in the autophagy process (Sullivan et al., 2016). However, how it regulates autophagy remains unclear.
Several studies on patient tissues and murine models indicate that TDP-43 leads to abnormal aggregation of mitochondria, mainly in the perinuclear somatic space of motor neurons (Shan et al., 2010; Xu et al., 2010, 2011; Janssens et al., 2013; Wang et al., 2013; Magrané et al., 2014). Mitochondrial clusters have also been reported in transgenic rat cortical neurons expressing mutant FUS (Huang et al., 2012). Such perinuclear clusters of mitochondria are reminiscent of deficient mitochondria undergoing mitophagy. Accordingly, PINK1 and Parkin expression levels are increased in HEK293 cells overexpressing FUS, and downregulating either protein partially rescues the abnormal phenotype of Drosophila expressing FUS (Chen et al., 2016). However, contradictory data concerning the effect of TDP-43 on Parkin expression have been collected on A315T mutant TDP-43 mice (Hebron et al., 2013; Stribl et al., 2014). On the other hand, clusters of mitochondria in TDP-43 or FUS animal models may not be ubiquitinated (Shan et al., 2010; Cannon et al., 2012; Huang et al., 2012). In addition, whereas Hong et al. (2012) showed that TDP-43 increases the localization of autophagic marker LC3 on mitochondria in NSC-34 cells, recent evidence reported that LC3 is not even activated in the presence of TDP-43 (Zhang et al., 2010; Janssens et al., 2013; Onesto et al., 2016).
Another hypothesis would be that TDP-43 disrupts the anterograde transport of mitochondria, which could justify their accumulation in the neuronal soma as well as their absence in neuromuscular junctions (Shan et al., 2010). Since kinesin-associated proteins have been detected in TDP-43-positive cytoplasmic aggregates in mouse motor neurons (Shan et al., 2010), their sequestration might indicate a probable deregulation of mitochondrial axonal transport (Figure 1). It has also been reported that TDP-43 and FUS can prevent the interaction between vesicle-associated membrane protein-associated protein B (VAPB) and protein tyrosine phosphatase interacting protein 51 (PTPIP51) in NSC-34 cells and mouse spinal cords, decreasing the ER-mitochondria contacts (Stoica et al., 2014, 2016) (Figure 1). This might interrupt the calcium flux between both organelles and cause mitochondria to stagnate in neuronal regions enriched in calcium. It is also worth mentioning that mutations in VAPB have been described in ALS, with mutant VAPB blocking the anterograde transport of mitochondria in rat cortical axons (Mórotz et al., 2012). Therefore, further works are required to definitely demonstrate that mitophagy is perturbed in TDP-43 and FUS models and possibly involved in ALS pathogenesis. If it is proven that mitophagy is the main altered MQC in ALS, then pharmacological agents should be more than ever designed to restore this process.
Conclusion
In the last decade, major insights into the molecular basis of ALS have arisen from the identification of new ALS-linked genes. In view of the huge number of genes, one major challenge is to unravel common pathways in ALS. This is particularly critical to develop therapeutic targets with a large spectrum. MQC is emerging as a key common pathway directly or secondarily impaired by ALS causing genes, but also in other neurodegenerative diseases such as Parkinson's disease. Indeed, these pathologies share common mitochondrial alterations while affecting distinct neuronal populations. This includes excessive mitochondrial fission and perturbed mitophagy, as well as reduced complex I expression and activity which have been reported in familial and sporadic forms of Parkinson's disease (for review, see Hu and Wang, 2016) and lately in ALS by Wang et al. (2016). In such a condition, neurons may quickly reach a saturation state of defective mitochondria. Nevertheless, this raises an important question: how alterations of MQC result in selective motor neuronal damage in ALS while dopaminergic neurons are more vulnerable in Parkinson's disease? Additional investigations are needed in the upcoming years to answer this challenging question and deepen our understanding of ALS pathogenesis.
Additional file: Additional Table 1 (60KB, pdf) Mitochondrial quality control processes that are altered by ALS causing genes
Footnotes
Conflicts of interest: None declared.
Contributor agreement: A statement of “Publishing Agreement” has been signed by an authorized author on behalf of all authors prior to publication.
Plagiarism check: This paper has been checked twice with duplication-checking software iThenticate.
Peer review: A double-blind and stringent peer review process has been performed to ensure the integrity, quality and significance of this paper.
References
- 1.Anand R, Wai T, Baker MJ, Kladt N, Schauss AC, Rugarli E, Langer T. The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J Cell Biol. 2014;204:919–929. doi: 10.1083/jcb.201308006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H, Mann D, Tsuchiya K, Yoshida M, Hashizume Y, Oda T. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- 3.Arlt H, Steglich G, Perryman R, Guiard B, Neupert W, Langer T. The formation of respiratory chain complexes in mitochondria is under the proteolytic control of the m-AAA protease. EMBO J. 1998;17:4837–4847. doi: 10.1093/emboj/17.16.4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, DeJesus-Hernandez M, Van Blitterswijk MM, Jansen-West K, Paul JW, Rademakers R, Boylan KB, Dickson DW, Petrucelli L. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azzu V, Brand MD. Degradation of an intramitochondrial protein by the cytosolic proteasome. J Cell Sci. 2010;123:578–585. doi: 10.1242/jcs.060004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baldwin KR, Godena VK, Hewitt VL, Whitworth AJ. Axonal transport defects are a common phenotype in Drosophila models of ALS. Hum Mol Genet. 2016;25:2378–2392. doi: 10.1093/hmg/ddw105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bannwarth S, Ait-El-Mkadem S, Chaussenot A, Genin EC, Lacas-Gervais S, Fragaki K, Berg-Alonso L, Kageyama Y, Serre V, Moore DG, Verschueren A, Rouzier C, Le Ber I, Augé G, Cochaud C, Lespinasse F, N’Guyen K, de Septenville A, Brice A, Yu-Wai-Man P, et al. A mitochondrial origin for frontotemporal dementia and amyotrophic lateral sclerosis through CHCHD10 involvement. Brain. 2014;137:2329–2345. doi: 10.1093/brain/awu138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bayer H, Lang K, Buck E, Higelin J, Barteczko L, Pasquarelli N, Sprissler J, Lucas T, Holzmann K, Demestre M, Lindenberg KS, Danzer KM, Boeckers T, Ludolph AC, Dupuis L, Weydt P, Witting A. ALS-causing mutations differentially affect PGC-1α expression and function in the brain vs. peripheral tissues. Neurobiol Dis. 2017;97:36–45. doi: 10.1016/j.nbd.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Bender T, Lewrenz I, Franken S, Baitzel C, Voos W. Mitochondrial enzymes are protected from stress-induced aggregation by mitochondrial chaperones and the Pim1/LON protease. Mol Biol Cell. 2011;22:541–554. doi: 10.1091/mbc.E10-08-0718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blesa JR, Prieto-Ruiz JA, Hernández JM, Hernández-Yago J. NRF-2 transcription factor is required for human TOMM20 gene expression. Gene. 2007;391:198–208. doi: 10.1016/j.gene.2006.12.024. [DOI] [PubMed] [Google Scholar]
- 11.Blokhuis AM, Koppers M, Groen EJN, van Den Heuvel DMA, Dini Modigliani S, Anink JJ, Fumoto K, van Diggelen F, Snelting A, Sodaar P, Verheijen BM, Demmers JAA, Veldink JH, Aronica E, Bozzoni I, Den Hertog J, van Den Berg LH, Pasterkamp RJ. Comparative interactomics analysis of different ALS-associated proteins identifies converging molecular pathways. Acta Neuropathol. 2016;132:1–22. doi: 10.1007/s00401-016-1575-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bota DA, Davies KJ. Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat Cell Biol. 2002;4:674–680. doi: 10.1038/ncb836. [DOI] [PubMed] [Google Scholar]
- 13.Braidotti G, Borthwick IA, May BK. Identification of regulatory sequences in the gene for 5-aminolevulinate synthase from rat. J Biol Chem. 1993;268:1109–1117. [PubMed] [Google Scholar]
- 14.Cannon A, Yang B, Knight J, Farnham IM, Zhang Y, Wuertzer CA, D’Alton S, Lin WL, Castanedes-Casey M, Rousseau L, Scott B, Jurasic M, Howard J, Yu X, Bailey R, Sarkisian MR, Dickson DW, Petrucelli L, Lewis J. Neuronal sensitivity to TDP-43 overexpression is dependent on timing of induction. Acta Neuropathol. 2012;123:807–823. doi: 10.1007/s00401-012-0979-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cantó C, Gerhart-hines Z, Feige JN, Lagouge M, Milne JC, Elliott PJ, Puigserver P, Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan NC, Salazar AM, Pham AH, Sweredoski MJ, Kolawa NJ, Graham RLJ, Hess S, Chan DC. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum Mol Genet. 2011;20:1726–1737. doi: 10.1093/hmg/ddr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y, Dorn GW., 2nd PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 2013;340:471–475. doi: 10.1126/science.1231031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, Deng J, Wang P, Yang M, Chen X, Zhu L, Liu J, Lu B, Shen Y, Fushimi K, Xu Q, Wu JY. PINK1 and Parkin are genetic modifiers for FUS-induced neurodegeneration. Hum Mol Genet. 2016;25:5059–5068. doi: 10.1093/hmg/ddw310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cipolat S, Martins de Brito O, Dal Zilio B, Scorrano L. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc Natl Acad Sci U S A. 2004;101:15927–15932. doi: 10.1073/pnas.0407043101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cipolat S, Rudka T, Hartmann D, Costa V, Serneels L, Craessaerts K, Metzger K, Frezza C, Annaert W, D’Adamio L, Derks C, Dejaegere T, Pellegrini L, D’Hooge R, Scorrano L, De Strooper B. Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling. Cell. 2006;126:163–175. doi: 10.1016/j.cell.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 22.Chu CT, Ji J, Dagda RK, Jiang JF, Tyurina YY, Kapralov AA, Tyurin VA, Yanamala N, Shrivastava IH, Mohammadyani D, Qiang Wang KZ, Zhu J, Klein-Seetharaman J, Balasubramanian K, Amoscato AA, Borisenko G, Huang Z, Gusdon AM, Cheikhi A, Steer EK, et al. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat Cell Biol. 2013;15:1197–2005. doi: 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Da Cruz S, Parone PA, Lopes VS, Lillo C, McAlonis-Downes M, Lee SK, Vetto AP, Petrosyan S, Marsala M, Murphy AN, Williams DS, Spiegelman BM, Cleveland DW. Elevated PGC-1α activity sustains mitochondrial biogenesis and muscle function without extending survival in a mouse model of inherited ALS. Cell Metab. 2012;15:778–786. doi: 10.1016/j.cmet.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 25.Deas E, Plun-Favreau H, Gandhi S, Desmond H, Kjaer S, Loh SHY, Renton AEM, Harvey RJ, Whitworth AJ, Martins LM, Abramov AY, Wood NW. PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum Mol Genet. 2011;20:867–879. doi: 10.1093/hmg/ddq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GY, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deng J, Yang M, Chen Y, Chen X, Liu J, Sun S, Cheng H, Li Y, Bigio EH, Mesulam M, Xu Q, Du S, Fushimi K, Zhu L, Wu JY. FUS interacts with HSP60 to promote mitochondrial damage. PLoS Genet. 2015;11:e1005357. doi: 10.1371/journal.pgen.1005357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Vos KJ, Chapman AL, Tennant ME, Manser C, Tudor EL, Lau KF, Brownlees J, Ackerley S, Shaw PJ, McLoughlin DM, Shaw CE, Leigh PN, Miller CC, Grierson AJ. Familial amyotrophic lateral sclerosis-linked SOD1 mutants perturb fast axonal transport to reduce axonal mitochondria content. Hum Mol Genet. 2007;16:2720–2728. doi: 10.1093/hmg/ddm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edens BM, Miller N, Ma Y. Impaired autophagy and defective mitochondrial function: converging paths on the road to motor neuron degeneration. Front Cell Neurosci. 2016;10:1–16. doi: 10.3389/fncel.2016.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eschbach J, Schwalenstöcker B, Soyal SM, Bayer H, Wiesner D, Akimoto C, Nilsson AC, Birve A, Meyer T, Dupuis L, Danzer KM, Andersen PM, Witting A, Ludolph AC, Patsch W, Weydt P. PGC-1α is a male-specific disease modifier of human and experimental amyotrophic lateral sclerosis. Hum Mol Genet. 2013;22:3477–3484. doi: 10.1093/hmg/ddt202. [DOI] [PubMed] [Google Scholar]
- 31.Finelli MJ, Liu KX, Wu Y, Oliver PL, Davies KE. Oxr1 improves pathogenic cellular features of ALS-associated FUS and TDP-43 mutations. Hum Mol Genet. 2015;24:3529–3544. doi: 10.1093/hmg/ddv104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Freibaum BD, Chitta RK, High AA, Taylor JP. Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J Proteome Res. 2010;9:1104–1120. doi: 10.1021/pr901076y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fukada K, Zhang F, Vien A, Cashman NR, Zhu H. Mitochondrial proteomic analysis of a cell line model of familial amyotrophic lateral sclerosis. Mol Cell Proteomics. 2004;12:1211–1223. doi: 10.1074/mcp.M400094-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gakh O, Cavadini P, Isaya G. Mitochondrial processing peptidases. Biochim Biophys Acta - Mol Cell Res. 2002;1592:63–77. doi: 10.1016/s0167-4889(02)00265-3. [DOI] [PubMed] [Google Scholar]
- 36.Gegg ME, Cooper JM, Chau KY, Rojo M, Schapira AH, Taanman JW. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum Mol Genet. 2010;19:4861–4870. doi: 10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geisler S, Holmström KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 38.Gleyzer N, Vercauteren K, Scarpulla RC. Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators. Mol Cell Biol. 2005;25:1354–1366. doi: 10.1128/MCB.25.4.1354-1366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Golko-Perez S, Amit T, Bar-Am O, Youdim MBH, Weinreb O. A novel iron chelator-radical scavenger ameliorates motor dysfunction and improves life span and mitochondrial biogenesis in SOD1G93A ALS mice. Neurotox Res. 2017;31:230–244. doi: 10.1007/s12640-016-9677-6. [DOI] [PubMed] [Google Scholar]
- 40.Head B, Griparic L, Amiri M, Gandre-Babbe S, Van Der Bliek AM. Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. J Cell Biol. 2009;187:959–966. doi: 10.1083/jcb.200906083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hebron ML, Lonskaya I, Sharpe K, Weerasinghe PPK, Algarzae NK, Shekoyan AR, Moussa CE. Parkin ubiquitinates tar-DNA binding protein-43 (TDP-43) and promotes its cytosolic accumulation via interaction with histone deacetylase 6 (HDAC6) J Biol Chem. 2013;288:4103–4115. doi: 10.1074/jbc.M112.419945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heo JM, Ordureau A, Paulo JA, Rinehart J, Harper JW. The PINK1-PARKIN mitochondrial ubiquitylation pathway drives a program of OPTN/NDP52 recruitment and TBK1 activation to promote mitophagy. Mol Cell. 2015;60:7–20. doi: 10.1016/j.molcel.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hong K, Li Y, Duan W, Guo Y, Jiang H, Li W, Li C. Full-length TDP-43 and its C-terminal fragments activate mitophagy in NSC34 cell line. Neurosci Lett. 2012;530:144–149. doi: 10.1016/j.neulet.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 44.Hu Q, Wang G. Mitochondrial dysfunction in Parkinson's disease. Transl Neurodegener. 2016;5:14. doi: 10.1186/s40035-016-0060-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang C, Tong J, Bi F, Wu Q, Huang B, Zhou H, Xia XG. Entorhinal cortical neurons are the primary targets of FUS mislocalization and ubiquitin aggregation in FUS transgenic rats. Hum Mol Genet. 2012;21:4602–4614. doi: 10.1093/hmg/dds299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ishihara N, Eura Y, Mihara K. Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity. J Cell Sci. 2004;117:6535–6546. doi: 10.1242/jcs.01565. [DOI] [PubMed] [Google Scholar]
- 47.Ishihara N, Fujita Y, Oka T, Mihara K. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J. 2006;25:2966–2977. doi: 10.1038/sj.emboj.7601184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jäger S, Handschin C, St-Pierre J, Spiegelman BM. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc Natl AcadSci U S A. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Janssens J, Wils H, Kleinberger G, Joris G, Cuijt I, Ceuterick-De Groote C, Van Broeckhoven C, Kumar-Singh S. Overexpression of ALS-associated p. M337V human TDP-43 in mice worsens disease features compared to wild-type human TDP-43 mice. Mol Neurobiol. 2013;48:22–35. doi: 10.1007/s12035-013-8427-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Joardar A, Menzl J, Podolsky TC, Manzo E, Estes PS, Ashford S, Zarnescu DC. PPAR gamma activation is neuroprotective in a Drosophila model of ALS based on TDP-43. Hum Mol Genet. 2015;24:1741–1754. doi: 10.1093/hmg/ddu587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kane LA, Youle RJ. PINK1 and Parkin flag miro to direct mitochondrial traffic. Cell. 2011;147:721–723. doi: 10.1016/j.cell.2011.10.028. [DOI] [PubMed] [Google Scholar]
- 52.Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, Banerjee S, Youle RJ. PINK1 phosphorylates ubiquitin to activate parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205:143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kazlauskaite A, Martínez-torres RJ, Wilkie S, Kumar A, Peltier J, Johnson C, Zhang J, Hope AG, Peggie M, Trost M, Mf D, Aalten V, Alessi DR, Prescott AR, Knebel A, Walden H, Muqit MM. Binding to serine 65-phosphorylated ubiquitinprimes Parkin for optimal PINK1-dependent phosphorylation and activation. EMBO Rep. 2015;16:939–954. doi: 10.15252/embr.201540352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khalil B, Cabirol-Pol MJ, Miguel L, Whitworth AJ, Lecourtois M, Liévens JC. Enhancing Mitofusin/Marf ameliorates neuromuscular dysfunction in Drosophila models of TDP-43 proteinopathies. Neurobiol Aging. 2017;54:71–83. doi: 10.1016/j.neurobiolaging.2017.02.016. [DOI] [PubMed] [Google Scholar]
- 55.Kiaei M, Kipiani K, Chen J, Calingasan NY, Beal MF. Peroxisome proliferator-activated receptor-gamma agonist extends survival in transgenic mouse model of amyotrophic lateral sclerosis. Exp Neurol. 2005;191:331–336. doi: 10.1016/j.expneurol.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 56.Kim NC, Tresse E, Kolaitis RM, Molliex A, Ruth E, Alami NH, Wang B, Joshi A, Smith RB, Ritson GP, Winborn BJ, Moore J, Lee J, Yao T, Pallanck L, Kundu M, Taylor JP. VCP is essential for mitochondrial quality control by PINK1/Parkin and this function is impaired by VCP mutations. Neuron. 2013;78:65–80. doi: 10.1016/j.neuron.2013.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kimura Y, Fukushi J, Hori S, Matsuda N, Okatsu K, Kakiyama Y, Kawawaki J, Kakizuka A, Tanaka K. Different dynamic movements of wild-type and pathogenic VCPs and their cofactors to damaged mitochondria in a Parkin-mediated mitochondrial quality control system. Genes Cells. 2013;18:1131–1143. doi: 10.1111/gtc.12103. [DOI] [PubMed] [Google Scholar]
- 58.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokoshi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 59.Kong J, Xu Z. Massive mitochondrial degeneration in motor neurons triggers the onset of amyotrophic lateral sclerosis in mice expressing a mutant SOD1. J Neurosci. 1998;18:3241–3250. doi: 10.1523/JNEUROSCI.18-09-03241.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, Kimura Y, Tsuchiya H, Yoshihara H, Hirokawa T, Endo T, Fon EA, Trempe J, Saeki Y, Tanaka K, Matsuda N. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 61.Kwiatkowski TJ, Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, Valdmanis P, Rouleau GA, Hosler BA, Cortelli P, de Jong PJ, Yoshinaga Y, Haines JL, Pericak-Vance MA, Yan J, Ticozzi N, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Sciences. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 62.Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1α. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 63.Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, Sideris DP, Fogel AI, Youle RJ. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Legros F, Malka F, Frachon P, Lombès A, Rojo M. Organization and dynamics of human mitochondrial DNA. J Cell Sci. 2004;117:2653–2662. doi: 10.1242/jcs.01134. [DOI] [PubMed] [Google Scholar]
- 65.Leonhard K, Stiegler A, Neupert W, Langer T. Chaperone-like activity of the AAA domain of the yeast Yme1 AAA protease. Nature. 1999;398:3478–351. doi: 10.1038/18704. [DOI] [PubMed] [Google Scholar]
- 66.Li Y, Ray P, Rao EJ, Shi C, Guo W, Chen X, Woodruff EA, 3rd, Fushimi K, Wu JY. A Drosophila model for TDP-43 proteinopathy. Proc Natl Acad Sci U S A. 2010;107:3169–3174. doi: 10.1073/pnas.0913602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liang H, Ward WF, Jang YC, Bhattacharya A, Bokov AF, Li Y, Jernigan A, Richardson A, van Remmen H. PGC-1α protects neurons and alters disease progression in an amyotrophic lateral sclerosis mouse model. Muscle Nerve. 2011;44:947–956. doi: 10.1002/mus.22217. [DOI] [PubMed] [Google Scholar]
- 68.Lin J, Tarr PT, Yang R, Rhee J, Puigserver P, Newgard CB, Spiegelman BM. PGC-1α in the regulation of hepatic glucose and energy metabolism. J Biol Chem. 2003;278:30843–30848. doi: 10.1074/jbc.M303643200. [DOI] [PubMed] [Google Scholar]
- 69.Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, Ma Q, Zhu C, Wang R, Qi W, Huang L, Xue P, Li B, Wang X, Jin H, Wang J, Yang F, Liu P, Zhu Y, Sui S, et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14:177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 70.Liu Q, Krzewska J, Liberek K, Craig EA. Mitochondrial Hsp70 Ssc1: role in protein folding. J Biol Chem. 2001;276:6112–6118. doi: 10.1074/jbc.M009519200. [DOI] [PubMed] [Google Scholar]
- 71.Liu W, Yamashita T, Tian F, Morimoto N, Ikeda Y, Deguchi K, Abe K. Mitochondrial fusion and fission proteins expression dynamically change in a murine model of amyotrophic lateral sclerosis. Curr Neurovasc Res. 2013;10:222–230. doi: 10.2174/15672026113109990060. [DOI] [PubMed] [Google Scholar]
- 72.Magrané J, Hervias I, Henning MS, Damiano M, Kawamata H, Manfredi G. Mutant SOD1 in neuronal mitochondria causes toxicity and mitochondrial dynamics abnormalities. Hum Mol Genet. 2009;18:4552–4564. doi: 10.1093/hmg/ddp421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Magrané J, Sahawneh MA, Przedborski S, Estévez ÁG, Manfredi G. Mitochondrial dynamics and bioenergetic dysfunction is associated with synaptic alterations in mutant SOD1 motor neurons. J Neurosci. 2012;32:229–242. doi: 10.1523/JNEUROSCI.1233-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Magrané J, Cortez C, Gan WB, Manfredi G. Abnormal mitochondrial transport and morphology are common pathological denominators in SOD1 and TDP43 ALS mouse models. Hum Mol Genet. 2014;23:1413–1424. doi: 10.1093/hmg/ddt528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Majcher V, Goode A, James V, Lay R. Autophagy receptor defects and ALS-FTLD. Mol Cell Neurosci. 2015;66:43–52. doi: 10.1016/j.mcn.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 76.Margineantu DH, Emerson CB, Diaz D, Hockenbery DM. Hsp90 inhibition decreases mitochondrial protein turnover. PLoS One. 2007;2:e1066. doi: 10.1371/journal.pone.0001066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martinez-vicente M. Neuronal mitophagy in neurodegenerative diseases. Front Mol Neurosci. 2017;10:1–13. doi: 10.3389/fnmol.2017.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, Sou YS, Saiki S, Kawajiri S, Sato F, Kimura M, Komatsu M, Hattori N, Tanaka K. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Matsumoto G, Shimogori T, Hattori N, Nukina N. TBK1 controls autophagosomal engulfment of polyubiquitinated mitochondria through p62/SQSTM1 phosphorylation. Hum Mol Genet. 2015;24:4429–4442. doi: 10.1093/hmg/ddv179. [DOI] [PubMed] [Google Scholar]
- 80.McLelland G, Soubannier V, Chen CX, Mcbride HM, Fon EA. Parkin and PINK 1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO J. 2014;33:282–295. doi: 10.1002/embj.201385902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Michiorri S, Gelmetti V, Giarda E, Lombardi F, Romano F, Marongiu R, Nerini-Molteni S, Sale P, Vago R, Arena G, Torosantucci L, Cassina L, Russo MA, Dallapiccola B, Valente EM, Casari G. The Parkinson-associated protein PINK1 interacts with Beclin1 and promotes autophagy. Cell Death Differ. 2010;17:962–974. doi: 10.1038/cdd.2009.200. [DOI] [PubMed] [Google Scholar]
- 82.Miller RG, Mitchell JD, Moore DH. Riluzole for amyotrophic lateral sclerosis (ALS)/motor neuron disease (MND) Cochrane Database Syst Rev. 2012;3:CD001447. doi: 10.1002/14651858.CD001447.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Misko A, Jiang S, Wegorzewska I, Milbrandt J, Baloh RH. Mitofusin 2 is necessary for transport of axonal mitochondria and interacts with the Miro/Milton complex. J Neurosci. 2010;30:4232–4240. doi: 10.1523/JNEUROSCI.6248-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Moore AS, Holzbaur ELF. Dynamic recruitment and activation of ALS-associated TBK1 with its target optineurin are required for efficient mitophagy. Proc Natl Acad Sci U S A. 2016;113:3349–3358. doi: 10.1073/pnas.1523810113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mórotz GM, De Vos KJ, Vagnoni A, Ackerley S, Shaw CE, Miller CC. Amyotrophic lateral sclerosis-associated mutant VAPBP56s perturbs calcium homeostasis to disrupt axonal transport of mitochondria. Hum Mol Genet. 2012;21:1979–1988. doi: 10.1093/hmg/dds011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1α. J Biol Chem. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 88.Neuspiel M, Schauss AC, Braschi E, Zunino R, Rippstein P, Rachubinski RA, Andrade-navarro MA, McBride HM. Cargo-selected transport from the mitochondria to peroxisomes is mediated by vesicular carriers. Curr Biol. 2008;18:102–108. doi: 10.1016/j.cub.2007.12.038. [DOI] [PubMed] [Google Scholar]
- 89.Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, Rogov V, Löhr F, Popovic D, Occhipinti A, Reichert AS, Terzic J, Dötsch V, Ney PA, Dikic I. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Okatsu K, Koyano F, Kimura M, Kosako H, Saeki Y, Tanaka K, Matsuda N. Phosphorylated ubiquitin chain is the genuine Parkin receptor. J Cell Biol. 2015;209:111–128. doi: 10.1083/jcb.201410050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Onesto E, Colombrita C, Gumina V, Borghi MO, Dusi S, Doretti A, Fagiolari G, Invernizzi F, Moggio M, Tiranti V, Silani V, Ratti A. Gene-specific mitochondria dysfunctions in human TARDBP and C9ORF72 fibroblasts. Acta Neuropathol Commun. 2016;4:47. doi: 10.1186/s40478-016-0316-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ostermann J, Horwich AL, Neupert W, Hartl FU. Protein folding in mitochondria requires complex formation with hsp60 and ATP hydrolysis. Nature. 1989;341:125–130. doi: 10.1038/341125a0. [DOI] [PubMed] [Google Scholar]
- 93.Palmer CS, Osellame LD, Laine D, Koutsopoulos OS, Frazier AE, Ryan MT. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep. 2011;12:565–573. doi: 10.1038/embor.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Palomo GM, Manfredi G. Exploring new pathways of neurodegeneration in ALS: The role of mitochondria quality control. Brain Res. 2015;1607:36–46. doi: 10.1016/j.brainres.2014.09.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Perera ND, Sheean RK, Scott JW, Kemp BE, Horne MK, Turner BJ. Mutant TDP-43 deregulates AMPK activation by PP2A in ALS models. PLoS One. 2014;9:e95549. doi: 10.1371/journal.pone.0090449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Puigserver P, Spiegelman BM. Peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α): Transcriptional coactivator and metabolic regulator. Endocr Rev. 2003;24:78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- 97.Raimondi A, Mangolini A, Rizzardini M, Tartari S, Massari S, Bendotti C, Francolini M, Borgese N, Cantoni L, Pietrini G. Cell culture models to investigate the selective vulnerability of motoneuronal mitochondria to familial ALS-linked G93ASOD1. Eur J Neurosci. 2006;24:387–399. doi: 10.1111/j.1460-9568.2006.04922.x. [DOI] [PubMed] [Google Scholar]
- 98.Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, Scholz SW, Duckworth J, Ding J, Harmer DW, Hernandez DG, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rikka S, Quinsay MN, Thomas RL, Kubli DA, Zhang X, Murphy AN, Gustafsson ÅB. Bnip3 impairs mitochondrial bioenergetics and stimulates mitochondrial turnover. Cell Death Differ. 2011;18:721–731. doi: 10.1038/cdd.2010.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Roberts RF, Tang MY, Fon EA, Durcan TM. Defending the mitochondria: The pathways of mitophagy and mitochondrial-derived vesicles. Int J Biochem Cell Biol. 2016;79:427–436. doi: 10.1016/j.biocel.2016.07.020. [DOI] [PubMed] [Google Scholar]
- 101.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1a and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 102.Rohrer JD, Isaacs AM, Mizlienska S, Mead S, Lashley T, Wray S, Sidle K, Fratta P, Orrell RW, Hardy J, Holton J, Revesz T, Rossor MN, Warren JD. C9orf72 expansions in frontotemporal dementia and amyotrophic lateral sclerosis. Lancet Neurol. 2015;14:291–301. doi: 10.1016/S1474-4422(14)70233-9. [DOI] [PubMed] [Google Scholar]
- 103.Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O’Regan JP, Deng HX, Rahmani Z, Krizus A, Mckenna-Yasek D, Cayabyab A, Gaston SM, Berger R, Tanzi RE, Halperin JJ, Herzfeldt B, van den Bergh R, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 104.Rojo M, Legros F, Chateau D, Lombès A. Membrane topology and mitochondrial targeting of mitofusins, ubiquitous mammalian homologs of the transmembrane GTPaseFzo. J Cell Sci. 2002;115:1663–1674. doi: 10.1242/jcs.115.8.1663. [DOI] [PubMed] [Google Scholar]
- 105.Rüb C, Wilkening A, Voos W. Mitochondrial quality control by the Pink1/Parkin system. Cell Tissue Res. 2017;367:111–123. doi: 10.1007/s00441-016-2485-8. [DOI] [PubMed] [Google Scholar]
- 106.Russell AP, Wada S, Vergani L, Hock MB, Lamon S, Léger B, Ushida T, Cartoni R, Wadley GD, Hespel P, Kralli A, Soraru G, Angelini C, Akimoto T. Disruption of skeletal muscle mitochondrial network genes and miRNAs in amyotrophic lateral sclerosis. Neurobiol Dis. 2013;49:107–117. doi: 10.1016/j.nbd.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 107.Santel A, Fuller MT. Control of mitochondrial morphology by a human mitofusin. J Cell Sci. 2001;114:867–874. doi: 10.1242/jcs.114.5.867. [DOI] [PubMed] [Google Scholar]
- 108.Satoh M, Hamamoto T, Seo N, Kagawa Y, Endo H. Differential sublocalization of the dynamin-related protein OPA1 isoforms in mitochondria. Biochem Biophys Res Commun. 2003;300:482–493. doi: 10.1016/s0006-291x(02)02874-7. [DOI] [PubMed] [Google Scholar]
- 109.Schutz B. The oral antidiabetic pioglitazone protects from neurodegeneration and amyotrophic lateral sclerosis-like symptoms in superoxide dismutase-G93A transgenic mice. J Neurosci. 2005;25:7805–7812. doi: 10.1523/JNEUROSCI.2038-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sephton CF, Cenik C, Kucukural A, Dammer EB, Cenik B, Han Y, Dewey CM, Roth FP, Herz J, Peng J, Moore MJ, Yu G. Identification of neuronal RNA targets of TDP-43-containing ribonucleoprotein complexes. J Biol Chem. 2011;286:1204–1215. doi: 10.1074/jbc.M110.190884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shan X, Chiang PM, Price DL, Wong PC. Altered distributions of Gemini of coiled bodies and mitochondria in motor neurons of TDP-43 transgenic mice. Proc Natl Acad Sci. 2010;107:16325–16330. doi: 10.1073/pnas.1003459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shi P, Wei Y, Zhang J, Gal J, Zhu H. Mitochondrial dysfunction is a converging point of multiple pathological pathways in amyotrophic lateral sclerosis. J Alzheimers Dis. 2010;20:311–324. doi: 10.3233/JAD-2010-100366. [DOI] [PubMed] [Google Scholar]
- 113.Shiba-Fukushima K, Inoshita T, Hattori N, Imai Y. PINK1-mediated phosphorylation of parkin boosts parkin activity in Drosophila. PLoS Genet. 2014;10:e1004391. doi: 10.1371/journal.pgen.1004391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Smirnova E, Griparic L, Shurland D-L, Bliek van der AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Song Z, Chen H, Fiket M, Alexander C, Chan DC. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J Cell Biol. 2007;178:749–755. doi: 10.1083/jcb.200704110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Song Z, Ghochani M, McCaffery JM, Frey TG, Chan DC. Mitofusins and OPA1 mediate sequential steps in mitochondrial membrane fusion. Mol Biol Cell. 2009;20:3525–3532. doi: 10.1091/mbc.E09-03-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Song W, Song Y, Kincaid B, Bossy B, Bossy-Wetzel E. Mutant SOD1G93A triggers mitochondrial fragmentation in spinal cord motor neurons: Neuroprotection by SIRT3 and PGC-1α. Neurobiol Dis. 2013;51:72–81. doi: 10.1016/j.nbd.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Soubannier V, Mclelland G, Zunino R, Braschi E, Rippstein P, Fon EA, Mcbride HM. A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Curr Biol. 2012a;22:135–141. doi: 10.1016/j.cub.2011.11.057. [DOI] [PubMed] [Google Scholar]
- 119.Soubannier V, Rippstein P, Kaufman BA, Shoubridge EA, Mcbride HM. Reconstitution of mitochondria derived vesicle formation demonstrates selective enrichment of oxidized cargo. PLoS One. 2012b;7:e52830. doi: 10.1371/journal.pone.0052830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, Baralle F, de Belleroche J, Mitchell JD, Leigh PN, Al-Chalabi A, Miller CC, Nicholson G, Shaw CE. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Stoica R, De Vos KJ, Paillusson S, Mueller S, Sancho RM, Lau KF, Vizcay-Barrena G, Lin WL, Xu YF, Lewis J, Dickson DW, Petrucelli L, Mitchell JC, Shaw CE, Miller CCJ. ER-mitochondria associations are regulated by the VAPB-PTPIP51 interaction and are disrupted by ALS/FTD-associated TDP-43. Nat Commun. 2014;5:3996. doi: 10.1038/ncomms4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Stoica R, Paillusson S, Gomez-suaga P, Mitchell JC, Lau DH, Gray EH, Sancho RM, Vizcay-barrena G, Vos De KJ, Shaw CE, Hanger DP, Noble W, Miller CC. ALS/FTD-associated FUS activates GSK-3β to disrupt the VAPB-PTPIP51 interaction and ER-mitochondria associations. EMBO Rep. 2016;17:1–17. doi: 10.15252/embr.201541726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Stribl C, Samara A, Trümbach D, Peis R, Neumann M, Fuchs H, Gailus-Durner V, Hrabĕ de Angelis M, Rathkolb B, Wolf E, Beckers J, Horsch M, Neff F, Kremmer E, Koob S, Reichert AS, Hans W, Rozman J, Klingenspor M, Aichler M, et al. Mitochondrial dysfunction and decrease in body weight of a transgenic knock-in mouse model for TDP-43. J Biol Chem. 2014;289:10769–10784. doi: 10.1074/jbc.M113.515940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sullivan PM, Zhou X, Robins AM, Paushter DH, Kim D, Smolka MB. The ALS/FTLD associated protein C9orf72 associates with SMCR8 and WDR41 to regulate the autophagy-lysosome pathway. Acta Neuropathol Commun. 2016;4:51. doi: 10.1186/s40478-016-0324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, Youle RJ. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Thau N, Knippenberg S, Körner S, Rath KJ, Dengler R, Petri S. Decreased mRNA expression of PGC-1α and PGC-1α-regulated factors in the SOD1G93A ALS mouse model and in human sporadic ALS. J Neuropathol Exp Neurol. 2012;71:1064–1074. doi: 10.1097/NEN.0b013e318275df4b. [DOI] [PubMed] [Google Scholar]
- 127.Tradewell ML, Yu Z, Tibshirani M, Boulanger MC, Durham HD, Richard S. Arginine methylation by prmt1 regulates nuclear-cytoplasmic localization and toxicity of FUS/TLS harbouring ALS-linked mutations. Hum Mol Genet. 2012;21:136–149. doi: 10.1093/hmg/ddr448. [DOI] [PubMed] [Google Scholar]
- 128.Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, Corkey BE, Shirihai OS. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, Albanese A, Nussbaum R, González-Maldonado R, Deller T, Salvi S, Cortelli P, Gilks WP, Latchman DS, Harvey RJ, Dallapiccola B, et al. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 130.Vance C, Rogelj B, Hortobágyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, Ganesalingam J, Williams KL, Tripathi V, Al-Saraj S, Al-Chalabi A, Leigh PN, Blair IP, Nicholson G, de Belleroche J, Gallo JM, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Vande Velde C, McDonald KK, Boukhedimi Y, McAlonis-Downes M, Lobsiger CS, Hadj SB, Zandona A, Julien JP, Shah SB, Cleveland DW. Misfolded SOD1 associated with motor neuron mitochondria alters mitochondrial shape and distribution prior to clinical onset. PLoS One. 2011;6:e22031. doi: 10.1371/journal.pone.0022031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Virbasius J V, Scarpulla RC. Activation of the human mitochondrial transcription factor A gene by nuclear respiratory factors: a potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc Natl Acad Sci U S A. 1994;91:1309–1313. doi: 10.1073/pnas.91.4.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Voos W, von Ahsen O, Müller H, Guiard B, Rassow J, Pfanner N. Differential requirement for the mitochondrial Hsp70-Tim44 complex in unfolding and translocation of preproteins. EMBO J. 1996;15:2668–2677. [PMC free article] [PubMed] [Google Scholar]
- 134.Wagner I, Arlt H, van Dyck L, Langer T, Neupert W. Molecular chaperones cooperate with PIM1 protease in the degradation of misfolded proteins in mitochondria. EMBO J. 1994;13:5135–5145. doi: 10.1002/j.1460-2075.1994.tb06843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang W, Li L, Lin WL, Dickson DW, Petrucelli L, Zhang T, Wang X. The ALS disease-associated mutant TDP-43 impairs mitochondrial dynamics and function in motor neurons. Hum Mol Genet. 2013;22:4706–4719. doi: 10.1093/hmg/ddt319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wang W, Wang L, Lu J, Siedlak SL, Fujioka H, Liang J, Jiang S, Ma X, Jiang Z, da Rocha EL, Sheng M, Choi H, Lerou PH, Li H, Wang X. The inhibition of TDP-43 mitochondrial localization blocks its neuronal toxicity. Nat Med. 2016;22:869–878. doi: 10.1038/nm.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Whitworth AJ, Pallanck LJ. PINK1/Parkin mitophagy and neurodegeneration — what do we really know in vivo? Curr Opin Genet Dev. 2017;44:47–53. doi: 10.1016/j.gde.2017.01.016. [DOI] [PubMed] [Google Scholar]
- 138.Wong YC, Holzbaur EL. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc Natl Acad Sci U S A. 2014;111:4439–4448. doi: 10.1073/pnas.1405752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 140.Xie Y, Zhou B, Lin MY, Wang S, Foust KD, Sheng ZH. endolysosomal deficits augment mitochondria pathology in spinal motor neurons of asymptomatic fALS mice. Neuron. 2015;87:355–370. doi: 10.1016/j.neuron.2015.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Xu YF, Gendron TF, Zhang YJ, Lin WL, D’Alton S, Sheng H, Casey MC, Tong J, Knight J, Yu X, Rademakers R, Boylan K, Hutton M, McGowan E, Dickson DW, Lewis J, Petrucelli L. Wild-type human TDP-43 expression causes TDP-43 phosphorylation, mitochondrial aggregation, motor deficits, and early mortality in transgenic mice. J Neurosci. 2010;30:10851–10859. doi: 10.1523/JNEUROSCI.1630-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Xu YF, Zhang YJ, Lin WL, Cao X, Stetler C, Dickson DW, Lewis J, Petrucelli L. Expression of mutant TDP-43 induces neuronal dysfunction in transgenic mice. Mol Neurodegener. 2011;6:73. doi: 10.1186/1750-1326-6-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yamashita H, Kawamata J, Okawa K, Kanki R, Nakamizo T, Hatayama T, Yamanaka K, Takahashi R, Shimohama S. Heat-shock protein 105 interacts with and suppresses aggregation of mutant Cu/Zn superoxide dismutase: Clues to a possible strategy for treating ALS. J Neurochem. 2007;102:1497–1505. doi: 10.1111/j.1471-4159.2007.04534.x. [DOI] [PubMed] [Google Scholar]
- 144.Zhang YJ, Gendron TF, Xu YF, Ko LW, Yen SH, Petrucelli L. Phosphorylation regulates proteasomal-mediated degradation and solubility of TAR DNA binding protein-43 C-terminal fragments. Mol Neurodegener. 2010;5:1–13. doi: 10.1186/1750-1326-5-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhao J, Liu T, Jin S, Wang X, Qu M, Uhlén P, Tomilin N, Shupliakov O, Lendahl U, Nistér M. Human MIEF1 recruits Drp1 to mitochondrial outer membranes and promotes mitochondrial fusion rather than fission. EMBO J. 2011a;30:2762–2778. doi: 10.1038/emboj.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhao W, Varghese M, Yemul S, Pan Y, Cheng A, Marano P, Hassan S, Vempati P, Chen F, Qian X, Pasinetti GM. Peroxisome proliferator activator receptor gamma coactivator-1alpha (PGC-1α) improves motor performance and survival in a mouse model of amyotrophic lateral sclerosis. Mol Neurodegener. 2011b;6:51. doi: 10.1186/1750-1326-6-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zick M, Duvezin-Caubet S, Schäfer A, Vogel F, Neupert W, Reichert AS. Distinct roles of the two isoforms of the dynamin-like GTPase Mgm1 in mitochondrial fusion. FEBS Lett. 2009;583:2237–2243. doi: 10.1016/j.febslet.2009.05.053. [DOI] [PubMed] [Google Scholar]
- 148.Ziviani E, Tao RN, Whitworth AJ. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc Natl Acad Sci U S A. 2010;107:5018–5023. doi: 10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mitochondrial quality control processes that are altered by ALS causing genes