Abstract
The University of Florida (UF) Health Personalized Medicine Program launched in 2012 with CYP2C19 genotyping for clopidogrel response at UF Health Shands Hospital. We have since expanded CYP2C19 genotyping to UF Health Jacksonville and established the infrastructure at UF Health to support clinical implementation for five additional gene–drug pairs: TPMT–thiopurines, IFNL3 (IL28B)–PEG IFN-α-based regimens, CYP2D6–opioids, CYP2D6/CYP2C19–antidepressants and CYP2C19–proton pump inhibitors. We are contributing to the evidence based on outcomes with genotype-guided therapy through pragmatic studies of our clinical implementations. In addition, we have developed a broad array of educational programs for providers, trainees and students that incorporate personal genotype evaluation to enhance participant learning.
Keywords: : clopidogrel, education, genotype, implementation, opioids, PEG-interferon, pharmacogenetics, proton pump inhibitors, selective serotonin uptake inhibitors, thiopurines
The University of Florida (UF) Health Personalized Medicine Program (PMP) was created in the spring of 2011 with a focus on pharmacogenetics. We described our program in an Institutional Profile in 2013 and provide an update in the current review [1]. The PMP is housed within the UF Clinical and Translational Sciences Institute and led by a multidisciplinary team including pharmacists, physicians, informaticians and others. As detailed previously, the guiding principles of our program include the need for informatics support within the electronic health record (EHR) for the application of genotype results to prescribing decisions, with back-up support provided by expert clinical pharmacists. In addition, we continue to believe that the most efficient manner of implementation is through a broad, pre-emptive genotyping chip, such that information can be generated at one time and used across the patient's lifespan. However, at present, the most clinically realistic approach, mainly from a reimbursement standpoint, is to genotype reactively in response to a drug order. Nonetheless, our ultimate goal is to overcome reimbursement and other barriers that hinder adoption of pre-emptive testing. In order to support sustainability of pharmacogenetic implementation, we also recognize the importance of generating data on metrics and clinical outcomes with pharmacogenetic implementation, and educating providers on appropriate interpretation and application of genotype results.
CYP2C19-clopidogrel implementation
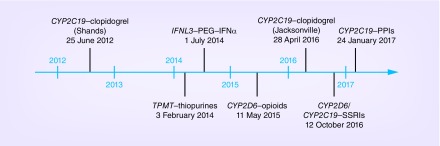
Our initial implementation was launched in June 2012 with CYP2C19 testing to predict clopidogrel response in patients undergoing percutaneous coronary intervention (PCI) at UF Health Shands Hospital (Figure 1). The rationale, process and infrastructure for this implementation have been described [1,2]. In line with our guiding principles, we initially genotyped for CYP2C19 as part of a larger pharmacogenetic panel [3]. The test was incorporated into the routine clinical care for patients undergoing left heart catheterization so that genotype results would be available in the event that the patient proceeded to PCI. Genotype results were entered into the EHR, and additional genotypes were stored in a data repository for potential future clinical use. However, because there is no reimbursement model for pre-emptive pharmacogenetics testing, this approach was not sustainable when we began clinically billing for testing in 2013.
Figure 1. . Timeline of launch of pharmacogenetic implementations at University of Florida Health.
While institutional and grant support was used to establish program infrastructure, CYP2C19 genotyping is now part of routine clinical care. The test order resides on the post-PCI order set, with testing performed in the UF Health Pathology Laboratory and results available within the EHR in 2–3 days. Clinical decision support provides a ‘Best Practice Advisory’ (BPA) when an order for clopidogrel is entered in the EHR for a patient with a CYP2C19 nonfunctional allele, notifying the physician of potentially reduced clopidogrel effectiveness and recommending alternative therapy [2]. In the event that genotype results are returned after patient discharge, a clinical pharmacist communicates with the physician to discuss alternative therapy in those with a nonfunctional allele.
In spring 2016, we expanded testing to UF Health Jacksonville. Consistent with early implementation at UF Health Shands, genotyping is done for patients undergoing left heart catheterization, and testing is paid for with research funding. Testing is done using a rapid genotyping platform (Spartan RX, Spartan Biosciences Inc., ON, USA), with results available in approximately 1 h and entered into the EHR. The rapid availability of test results allows physicians to immediately prescribe genotype-informed antiplatelet therapy.
Additional implementations
We have since implemented the following gene-drug pairs: TPMT–thiopurines, IFNL3 (IL28B)–PEG–IFN-α-based regimens, CYP2D6–opioids, CYP2D6/CYP2C19–antidepressants and CYP2C19–proton pump inhibitors (PPIs). Launch dates are shown in Figure 1. In each case, implementation followed physician requests for genotyping to inform prescribing decisions. Genotype tests for all but IFNL3 were established by the UF Health Pathology Laboratories, and all genotype results are entered into the laboratory section of the EHR.
With the exception of CYP2C19 testing for PPIs, CPIC guidelines are available for each implementation and genotype-guided recommendations provided to UF Health physicians are consistent with these guidelines [4–7]. In the absence of CPIC guidelines for CYP2C19–PPIs, we carefully reviewed the evidence and felt it to be sufficient to support genotype-guided PPI dosing recommendations. In particular, the efficacy of PPIs, which are metabolized by CYP2C19, is highly dependent on plasma concentration [8]. Compared with CYP2C19 poor and intermediate metabolizers, rapid and ultrarapid metabolizers have lower PPI plasma concentrations, lesser gastric acid suppression and an increased risk for treatment failure with usual PPI dosing [9,10]. A recommendation for a dose increase is provided for rapid and ultrarapid metabolizers. Clinicians are advised that usual PPI doses are expected to be effective for normal metabolizers and that lower doses may be sufficient for symptom management in PMs.
Best Practice Advisories were built in the UF Health Epic system for TPMT and CYP2D6 genotypes relative to thiopurines and opioids, respectively, and additional BPAs are in process to support the CYP2D6/CYP2C19-selective serotonin reuptake inhibitor implementation. Collectively the approach with BPAs is that they are only triggered when the patient carries a genotype that warrants an alternative approach. For TPMT, the BPA recommends lower thiopurine doses for those with a single nonfunctional allele and either lower doses or consideration of alternative therapy for those homozygous for two nonfunctional alleles. For CYP2D6 genotype, the BPA is in response to a codeine or tramadol order for a patient with a genotype associated with poor or ultrarapid metabolism. The advisory warns about the risk for reduced effectiveness for poor metabolizers or risk for toxicity for ultrarapid metabolizers and suggests that an alternative opioid that does not rely on CYP2D6 metabolism be prescribed. The clinical pharmacist also tracks those with actionable CYP2D6 genotypes relative to other pain medications (e.g., oxycodone, hydrocodone) and may also recommend alternative treatment approaches. For IFNL3 testing, pharmacists are alerted to the test order by the EHR and follow-up with the ordering provider to give guidance on test interpretation and application.
Table 1 shows the number of genotype tests ordered through December 2016 for each implementation described above. PMP pharmacists are also alerted via the EHR to inpatient and outpatient genotypes ordered outside of a clinical implementation (e.g., CYP2C19 testing for clopidogrel response by neurology). For these, PMP pharmacists consult with the ordering clinician to provide recommendations for interpreting and applying the genetic test results in the patient's care plan.
Table 1. . Pharmacogenetic tests ordered at University of Florida Health.
| Test | Drug | Number of tests† | Ordering service |
|---|---|---|---|
| CYP2C19 | Clopidogrel | 1694 | Cardiology, UF Health Shands |
| 634 | Cardiology, UF Health Jacksonville | ||
| |
|
13 |
Neurology, neurosurgery |
| TPMT | Thiopurines | 43 | Hematology/oncology |
| 67 | Gastroenterology | ||
| |
|
42 |
Other (e.g., dermatology, rheumatology, neurology, internal medicine, family medicine, surgery, neurology) |
|
IFNL3 |
PEG–IFN-α-based regimens |
96 |
Gastroenterology |
| CYP2D6 | Opioids | 202 | Family medicine |
| 33 | Internal medicine | ||
| 34 | Chronic Pain Clinic | ||
| |
|
3 |
Adult oncology |
|
CYP2D6 |
SSRIs |
12 |
Psychiatry |
|
CYP2C19 |
SSRIs |
13 |
Psychiatry |
| CYP2C19 | Proton pump inhibitors | No data yet available | Gastroenterology |
†Number of orders through 31 December 2016. All tests are available clinically, and the number of orders reflects both those ordered in association with research protocols and those ordered for routine clinical care. All orders for CYP2C19–clopidogrel, TPMT and IFNL3 tests at UF Health in Gainesville are for routine clinical care.
ALL: Acute lymphoblastic leukemia; GERD: Gastroesphogeal reflux disease; PCI: Percutaneous coronary intervention; SSRI: Selective serotonin reuptake inhibitor.
Building evidence for sustainability of pharmacogenetics implementation
Our view is that evidence of clinical utility from randomized controlled trials is not required prior to clinical pharmacogenetic implementation. Rather, we believe that strong and consistent genotype associations with drug response and the availability of alternative therapy or dosing for patients with genotypes predictive of poor response or increased toxicity to traditional therapy are sufficient to support implementation. However, recognizing that evidence that genotype-guided therapy positively impacts outcomes is important for sustainability of the program, we have taken a pragmatic approach to examining patient outcomes with several of our implementations.
CYP2C19–clopidogrel
After the first 2 years of program launch, we examined the occurrence of major adverse cardiovascular events in patients undergoing PCI and CYP2C19 testing. We found that, among patients with a nonfunctional allele, the risk for the composite outcome of death, myocardial infarction, stroke or stent thrombosis was significantly lower among patients treated with alternative therapy (e.g., prasugrel or ticagrelor) versus clopidogrel [11]. More recently, we collaborated with six other institutions as part of the NIH-funded IGNITE Network Pharmacogenetics Working Group to examine outcomes with CYP2C19 genotype-guided antiplatelet therapy in a larger population. Results were consistent with our earlier observation and reported at the 2016 American Heart Association Scientific Sessions [12].
CYP2D6–opioids
Outcomes with genotype-guided opioid prescribing are being evaluated in two separate studies. In one study, adult patients with chronic pain are recruited from primary care clinics, serving as implementation or control sites (ClinicalTrials.gov identifier: NCT02335307). Patients from implementation sites undergo CYP2D6 genotyping, with results placed in the EHR and genotype-guided recommendations provided by a pharmacist via a consult note. The other study focuses on cancer-related pain and is being conducted in collaboration with H Lee Moffitt Cancer Center and Research Institute in Tampa, Florida. Patients with advanced cancer are randomized to genotype-guided versus traditional pain management (ClinicalTrials.gov identifier: NCT02664350). Genotype results for the genotype-guided arm are placed in the EHR and accompanied by recommendations for consideration of opioids that are not metabolized by CYP2D6 for poor, intermediate and ultrarapid metabolizers. The primary end point for both studies is patient-reported pain-related outcomes, with the hypothesis that genotype-guided pain management will lead to better pain control compared with usual pain management. The studies are expected to be completed in late 2017.
CYP2C19–PPIs
A comparative effectiveness study, in collaboration with Nemours Children's Hospital in Orlando, Florida, is testing the hypothesis that genotype-supported treatment of gastroesophageal reflux disease and dyspepsia will lead to better symptom control compared with conventional treatment (ClinicalTrials.gov Identifier: NCT02930824). Patients who are being started on a PPI or who have continued symptoms on current PPI therapy are randomized to a genotype-supported strategy, with CYP2C19 genotype information integrated into treatment decisions, versus conventional management. We expect to complete data collection for the target population of 180 patients by mid-2017.
CYP2D6/CYP2C19–antidepressants
Within the pediatric psychiatry clinic, we are conducting a prospective randomized trial to examine the feasibility and acceptability of CYP2D6 and CYP2C19 testing to guide treatment of depression. Children and adolescents with major depression, anxiety or obsessive compulsive disorders are randomized to receive pharmacogenetic testing prior to starting or changing antidepressant medication or to usual treatment. Both clinician and parent understanding and acceptance of pharmacogenetic testing will be assessed, as will outcomes related to comfort with initiating medications and medication adherence.
Educational initiatives
The PMP's educational initiatives for providers incorporate principles of active learning and use of personal genotype information. Provider education for an implementation starts with a small group of key stakeholder clinicians and focuses on evidence review and developing the clinical recommendations for specific gene–drug pairs. We then expand educational efforts to include large provider groups (such as at departmental grand rounds presentations), trainees, nursing and administrative staff, to educate target audiences and support uptake and operationalizing of the clinical implementation. In addition, practice-based resources (e.g., genotype-guided dosing charts, patient education materials) are created for providers to assist with drug therapy changes and patient education within each implementation. On a broader scale, the PMP hosted the first UF Precision Medicine Conference in 2016 to reach providers across the nation, which will be offered again in 2017. To date, 249 providers have participated in PMP educational programs that included personal genotype evaluation.
The PMP has also developed educational offerings for post-graduate trainees and students. We currently offer one of only two accredited Post-graduate Year Two yearlong pharmacist training programs in pharmacogenetics, which has graduated five residents to date. Five additional postgraduate trainees have completed 4-week rotations in clinical pharmacogenetics. The PMP has also reached student pharmacists, with the development of a novel elective course currently in its fourth year that incorporates student genotype evaluation. Teaching approaches used in this course led to improved clinical pharmacogenetics knowledge and an increased student understanding of pharmacogenomics based on having undergone genotyping [13]. To date, 135 students have participated in these courses, including personal genotyping. Research efforts in this area are ongoing to further examine various teaching methods and use of personal genotype evaluation in broader multidisciplinary provider audiences.
Executive summary.
We have successfully implemented pharmacogenetic testing in a variety of inpatient and outpatient settings at University of Florida Health.
Gene–drug pairs implemented include CYP2C19–clopidogrel; TPMT–thiopurines; IFNL3–PEG–interferon; CYP2D6–opioids; CYP2D6/CYP2C19–antidepressants; and CYP2C19–proton pump inhibitors.
We have established pragmatic research studies to examine clinical outcomes with our pharmacogenetic implementations.
We recently showed improved outcomes among patients with a nonfunctional CYP2C19 allele who were prescribed an alternative antiplatelet agent versus clopidogrel.
We are collaborating with other medical centers in Florida to share and collectively disseminate data on implementation outcomes.
We offer novel educational programs to prepare current and future providers to manage pharmacotherapy in the age of genomics.
Our future plans include continued expansion of pharmacogenetic implementation across our medical center.
We are also working to overcome barriers to pre-emptive genotyping on a chip-based platform.
Footnotes
Financial & competing interests disclosure
This work was supported by the NIH grant U01 HG007269, as part of the NIH IGNITE network. Additional support provided by NIH U01 GM074492 and U01 HL105198 (both part of the NIH Pharmacogenomics Research Network), and by substantial institutional support from the University of Florida and its Clinical Translational Science Institute (UL1 TR000064 and UL1 TR001427). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Johnson JA, Elsey AR, Clare-Salzler MJ, Nessl D, Conlon M, Nelson DR. Institutional Profile: University of Florida and Shands Hospital Personalized Medicine Program: clinical implementation of pharmacogenetics. Pharmacogenomics. 2013;14(7):723–726. doi: 10.2217/pgs.13.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weitzel KW, Elsey AR, Langaee TY, et al. Clinical pharmacogenetics implementation: approaches, successes, and challenges. Am. J. Med. Genet. C Semin. Med. Genet. 2014;166C(1):56–67. doi: 10.1002/ajmg.c.31390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson JA, Burkley BM, Langaee TY, Clare-Salzler MJ, Klein TE, Altman RB. Implementing personalized medicine: development of a cost-effective customized pharmacogenetics genotyping array. Clin. Pharmacol. Ther. 2012;92(4):437–439. doi: 10.1038/clpt.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crews KR, Gaedigk A, Dunnenberger HM, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for codeine therapy in the context of cytochrome P450 2D6 (CYP2D6) genotype. Clin. Pharmacol. Ther. 2012;91(2):321–326. doi: 10.1038/clpt.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hicks JK, Bishop JR, Sangkuhl K, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin. Pharmacol. Ther. 2015;98(2):127–134. doi: 10.1002/cpt.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muir AJ, Gong L, Johnson SG, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for IFNL3 (IL28B) genotype and PEG interferon-alpha-based regimens. Clin. Pharmacol. Ther. 2014;95(2):141–146. doi: 10.1038/clpt.2013.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Relling MV, Gardner EE, Sandborn WJ, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing. Clin. Pharmacol. Ther. 2011;89(3):387–391. doi: 10.1038/clpt.2010.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yacyshyn BR, Thomson AB. The clinical importance of proton pump inhibitor pharmacokinetics. Digestion. 2002;66(2):67–78. doi: 10.1159/000065588. [DOI] [PubMed] [Google Scholar]
- 9.Sugimoto M, Furuta T. Efficacy of tailored Helicobacter pylori eradication therapy based on antibiotic susceptibility and CYP2C19 genotype. World J. Gastroenterol. 2014;20(21):6400–6411. doi: 10.3748/wjg.v20.i21.6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang HL, Li Y, Hu YF, Xie HG, Zhai SD. Effects of CYP2C19 loss-of-function variants on the eradication of H. pylori infection in patients treated with proton pump inhibitor-based triple therapy regimens: a meta-analysis of randomized clinical trials. PLoS ONE. 2013;8(4):e62162. doi: 10.1371/journal.pone.0062162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cavallari LH, Magvanjav O, Anderson RD, et al. Clinical implementation of CYP2C19 genotype guided antiplatelet therapy reduces cardiovascular events after PCI. Circulation. 2015 http://circ.ahajournals.org/content/132/Suppl_133/A11802.abstract?sid=cd11806bc11466--13451--11841a11808--18339--11503e71988d11882 [Google Scholar]
- 12.Cavallari LH, Denny JC, Lee CR, et al. Prospective clinical implementation of CYP2C19-genotype guided antiplatelet therapy after PCI: a multi-site investigation of MACE outcomes in a real-world setting. Circulation. 2016;134:e711–e712. [Google Scholar]
- 13.Weitzel KW, McDonough CW, Elsey AR, Burkley B, Cavallari LH, Johnson JA. Effects of using personal genotype data on student learning and attitudes in a pharmacogenomics course. Am. J. Pharm. Educ. 2016;80(7):122. doi: 10.5688/ajpe807122. [DOI] [PMC free article] [PubMed] [Google Scholar]