Abstract
The regulatory subunit of PI3K, p85α (encoded by PIK3R1), binds, stabilizes and inhibits the PI3K p110 catalytic subunit. Functional characterization of PIK3R1 mutations has identified not only hypomorphs with reduced inhibition of p110, but also hypomorphs and dominant negative mutants that disrupt a novel regulatory role of p85α on PTEN or neomorphs that activate unexpected signaling pathways. The diverse phenotypic spectrum of these PIK3R1 driver mutations underscores the need for different treatment strategies targeting tumors harboring these mutations. This article describes the functional consequences of the spectrum of PIK3R1 driver mutations and therapeutic liabilities they may engender.
Keywords: : MAPK, mutation, p85, PI3K, PIK3R1, targeted therapy
Tumorigenesis is a multistep process that involves accumulation of genomic aberrations resulting in signaling pathway deregulation. Cancer cells depend on one or more deregulated pathways to initiate and maintain malignant phenotypes. This concept of ‘oncogene addiction’ provides the rationale for targeted therapy blocking signaling pathways essential for maintenance of the malignant phenotype. Targeted cancer therapy, which interferes with the functional consequences of specific molecular abnormalities, has the potential to generate improved antitumor activity and lower systemic toxicity than conventional therapies. The identification of potential therapeutic targets is facilitated by comprehensive genomic profiling of tumors provided with robust functional genomics to characterize the functional consequences of genomic aberrations [1,2].
Aberrations of members along the PI3K pathway are among the most frequent driver events across different cancer lineages. The class I PI3Ks constitute a lipid kinase family that catalyze the production of phosphatidylinositol-3,4,5-triphosphate (PIP3) from phosphatidylinositol-4,5-bisphosphate (PIP2) at the cell membrane [3]. PIP3 is a lipid second messenger that recruits PH domain containing molecules including the proto-oncogene AKT for activation. The class IA PI3K is an obligate heterodimer composed of a catalytic subunit (p110) and a regulatory subunit (p85) [4]. The class 1A catalytic PI3Ks are comprised of three isoforms (p110α, p110β and p110δ). p110α and p110β are ubiquitously expressed whereas p110δ is more abundant in hematopoietic cells [5]. There are five regulatory isoforms which are recognized as p85-related regulatory subunits: p85α (and its splicing variants p55α and p50α), p85β and p55γ [6].
Although the catalytic reaction depends on the activity of p110, p85 plays a critical role in enabling the pathway activation by stabilizing p110 providing a pool of p110 that can be activated by intercellular signaling molecules. p110 molecules not associated with p85 are prone to thermo degradation [7]. The association with p85 also inhibits the activity of p110 but the inhibition is relieved upon RTK activation and recruitment of p85 to phosphotyrosine residues in the RTK or intracellular linker molecules [7,8]. Binding of the SH2 domains of p85 to phosphotyrosine relieves the inhibitory effect of p85 on p110. This process results in spatial regulation of PI3K activity. p85 therefore allows ligand-dependent activation of the PI3K pathway, which then contributes to widely divergent physiological processes including cell proliferation, survival, cell cycle progression, differentiation, protein synthesis, motility and metabolism [9–11].
The PI3K pathway in cancer: aberrations & therapeutic opportunities
In cancer, the PI3K pathway can be aberrantly activated through multiple mechanisms including cross-talk with other signaling pathways or ‘instrinsic’ hyperactivation due to genomic aberrations [12–17]. Common genomic aberrations across cancer types in the PI3K pathway include loss of the negative regulator PTEN via gene mutations, copy number loss or promoter hypermethylation and mutations or gene amplifications of PIK3CA (which encodes the p110α catalytic subunit). Specifically, the PIK3CA and PTEN genes are the 2nd and 3rd most frequently mutated gene respectively across 12 cancer types sequenced in The Cancer Genome Atlas (TCGA) [18]. PTEN is within the 9th most significantly deleted region whereas PIK3CA is within the 4th most amplified region across 11 cancer types [19]. There are genomic aberrations of other core members such as mutations in PIK3R1 (p85α regulatory subunit) or PIK3R2 (p85β regulatory subunit) and mutation or amplification of AKT isoforms (AKT1, AKT2 and AKT3) [18]. Aberrations are also detected in the upstream inputs (e.g., EGFR, FGFR) or downstream effectors of the pathway, such as MTOR, TSC1, TSC2 and RICTOR.
Being a critical player in tumorigenesis and the most frequently hyperactivated signaling pathway in cancer, pharmacological inhibition of the PI3K pathway is, therefore, considered to be among the most promising strategies for targeted cancer therapy [14,20]. Many inhibitors have been developed targeting components of the PI3K pathway and many of them have been introduced into clinical trials [21,22]. Since the majority of the components in the PI3K pathway are kinases, the pathway therefore presents a group of ‘druggable’ targets using small molecule inhibitors. Direct inhibition of the PI3K or inhibition of downstream kinases through a variety of small molecules with different mechanisms of action have been developed and entered clinical trials. These include small inhibitors targeting all PI3K isoforms, specific PI3K isoforms, dual PI3K/mammalian target of rapamycin (mTOR), mTOR, or AKT [23,24]. Representative first-generation inhibitors that have demonstrated activities in a range of tumor lineages include GDC-0941 (a pan-PI3K inhibitor) [25], NVP-BEZ235 (a dual PI3K/mTOR inhibitor), Rapalogs (mTOR inhibitors) [26,27] and MK–2206 (AKT inhibitor) [28]. Isoform-specific PI3K inhibitors offer the potential advantage of on-target blockade and minimizing off-target effects due to inhibition of other isoforms as in the case of pan-PI3K inhibitors. Encouragingly, GS-1101 (p110δ-specific, previously known as CAL-101) a p110δ-specific inhibitor has shown remarkable success in the clinic for the treatment of hematopoietic malignancies that express relatively high levels of p110δ [29]. In addition, an array of second generation and more potent agents targeting the PI3K pathway are in development or early clinical trials [30,31]. Blocking upstream signaling from RTKs provide an indirect approach to inhibiting the PI3K pathway. The first-in-class approaches that have been used in the clinic include trastuzumab (a monoclonal antibody that blocks the Erb2/HER2 receptor) [32] and imatinib (small molecule kinase inhibitor that inhibits the BCR-Abl, c-KIT, and PDGF-R tyrosine kinases) [33]. However, although, with the exception of GS-1101, PI3K pathway inhibitors have demonstrated limited activity as single agents in early trials, combinations with hormonal manipulation in breast cancer shows major promise. Thus it is likely that the potential of PI3K pathway inhibition will only be fulfilled by the implementation of rational drug combinations.
In addition to in vitro or in vivo evidence, clinical studies have suggested that inhibition of the PI3K pathway may be more effective in cancer patients with genomic aberration in the pathway [34]. Molecular aberrations in cancer genes can be utilized as ‘signals’ of potential resistance or sensitivity to specific targeted therapies. Further, the specific mutation present in a cancer gene can have different functional consequences as well as determine therapeutic sensitivity. For example, cancer patients with PIK3CA H1047R mutation showed better response to inhibitors targeting PI3K, mTOR or AKT than individuals with wild-type (WT) PIK3CA or other PIK3CA mutations [35]. PTEN aberrations including loss of PTEN protein and/or PTEN mutation were also associated with higher efficacy in PI3K pathway targeting [36]. Due to the surge in high-throughput sequencing of cancer genomes and the increased availability of genomic data, incorporation of the genomic information into cancer patient management is increasingly feasible. The key to successful translation into clinical benefit for cancer patients relies on understanding the functions of mutations as well as the therapeutic liabilities that these specific mutations engender.
PIK3R1 is one of the cancer genes in the PI3K pathway
There has been much effort in characterizing the functional consequence of PIK3R1 mutations recently. The PIK3R1 gene is the 12th most commonly mutated gene across cancer lineages and several mutation hotspot regions are detected, providing justification for exploring approaches to target the functional consequences of PIK3R1 mutations in cancer patients. Somatic mutations in PIK3R1 are particularly prevalent in endometrial cancer (20 and 34% in our and TCGA datasets, respectively [37,38]), glioma (11%; TCGA [39]) and colon cancer (4.5–10%; TCGA [40] and Genentech [41]). One major effort is to distinguish a ‘driver’ mutation that functionally contributes to tumor behavior from biologically neutral ‘passenger’ mutations resulting from instability of cancer genomes. A subset of PIK3R1 mutations detected are deemed functional and targeting these driver mutations, as compared with passenger mutations, has the potential to benefit cancer patients. In this article, we review the mechanisms by which different p85α driver mutations exert their oncogenic effects and the biological consequences of these drivers. The PIK3R1 driver mutations functionally characterized so far can be categorized as hypomorph (loss-of-function mutant), dominant negative that inhibits the activity of WT p85α, or neomorph (gain-of-novel-function mutant that has completely distinct activity from WT p85α). Here we also discuss preclinical data that may shed light on the therapeutic strategies in targeting PIK3R1 mutated cancers. Although p85α is not directly druggable, the driver mutations activate downstream signaling pathways and these downstream effectors represent potential therapeutic targets.
Functional & therapeutic relevance of PIK3R1 driver aberrations in cancer
Hypomorphic mutants with reduced inhibition of p110
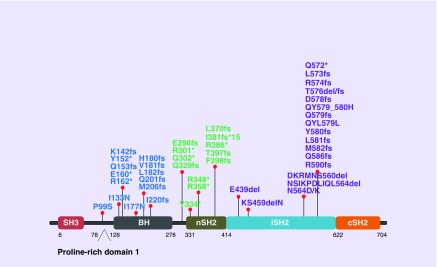
p85α itself has no kinase activity [42] and its functions are mediated by interaction with protein partners with the most well known binding partner being p110. The nSH2 and iSH2 domains of p85α (see Figure 1 for the domain structure of p85α) bind the helical and C2 domain of p110, respectively, to inhibit the catalytic activity of the p110 isoforms [43–45]. Two obvious hotspot regions of PIK3R1 mutations locate within the iSH2 domain (Figure 1). These hotspot mutations can be found in multiple cancer lineages, and they represent the first group of PIK3R1 mutations that were functionally characterized. The underlying mechanism of these PIK3R1 driver mutations appears to depend on disrupting the iSH2–C2 contact interface, thereby relieving the inhibitory effect on p110 [44,46]. These mutants retain the ability to bind and stabilize p110, leading to hyperactivation of the PI3K pathway in cancer. This subset of p85α mutants can be referred to as hypomorph.
Figure 1. . PIK3R1 mutations that are functionally verified or potentially functional based on locations.
Blue coded mutations potentially involved in p85α homodimerization and/or PTEN binding. Green coded mutations are neomorphs that activate MAPK signaling pathways. Purple coded mutations disrupt inhibition of p110 by p85α.
These p85α hypomorphic mutations include point mutations (D560Y, N564D), small deletions (E439del, KS459delN, DKRMNS560del, T576del, QYL579L) and frameshift or non-sense mutations (Q572*, R574fs) resulting in translation of truncation proteins [44,46–48]. No change in the binding activity between the p85α mutants and p110α or p110β has been detected. In vitro lipid activity assays showed that p110α coupled to these mutants had higher kinase activity than WT p85α-bound p110α. Several postulates based on the crystal structures and computational modeling of the p85α-p110α heterodimer have been proposed to explain the loss of the inhibitory effect. The mutants KS459delN and DKRMNS560del most likely disrupt the α-helical structural conformation of the iSH2 domain that mediates the inhibitory interaction with the C2 domain of p110 [46]. Truncation of p85α after residue 571 may destabilize a contact between the iSH2 domain and the C2 domain of p110α [49]. The p85α amino acid residues D560 and N564 of the iSH2 domain are within the hydrogen bonding distance of N345 of the C2 domain of p110α. This interaction is required for inhibition of p110α and the observation that the p110α-N345 mutant is also oncogenic provides support for this mechanism [44,47]. Consistent with the predicted mechanism of action, the effects of these hypomorphic p85α mutants were not additive with that of p110α-N345 [44,50].
The p85α iSH2 domain mutants lead to constitutive activation of PI3K pathway as demonstrated by increase in phosphorylation of downstream effectors including AKT. The PI3K pathway activation was associated with an increase in transforming activity of multiple cell models expressing these mutants. For example, in glioma models, three p85α driver mutants (DKRMNS560del, R574fs and T576del) significantly promoted anchorage-independent growth potential in soft agar in vitro and tumorigenicity in intracranial xenografts in vivo [48]. R574fs and T576del promoted IL3-independent growth of the IL3-dependent Ba/F3 cells [37]. D560Y and N564D induced anchorage-independent growth of NIH 3T3 cells and leukemia-like disease in mice [44,47]. Further, KS459delN, DKRMNS560del, R574fs and T576del increased the transformation potential of chicken embryo fibroblasts [46].
Given the activation of PI3K pathway signaling by p85α hypomorphic mutants, it may not be surprising that cells harboring these mutants become addicted to the PI3K pathway and are, therefore, sensitive to inhibitors targeting the pathway. Ba/F3 cells expressing DKRMNS560del, R574fs, or T576del were more sensitive to an AKT allosteric inhibitor MK-2206 than cells expressing WT p85α [48]. Another independent study showed that KS459delN or DKRMNS560del rendered chicken embryo fibroblasts sensitive to the mTOR inhibitor rapamycin [46]. Interestingly, these cells were also sensitive to inhibition of p110α (by the isoform specific inhibitor A66), but not sensitive to inhibition of p110β (TGX-221) or p110δ (IC87114) [46]. This suggested p110α is the major class 1A p110 isoform that mediates the phenotypic changes induced by the p85α mutants at least in this particular cell type. p110δ is unlikely involved in the PI3K signaling in this cell type because its expression was undetectable [46]. Although binding between p110β with WT or the mutated p85α could be detected, the lack of response to p110β inhibition can be due to several potential mechanisms. It is possible that the interaction of p85α mutants with p110α and p110β are different and that the effects of p110α on the PI3K pathway are dominant over p110β when both isoforms are expressed. Moreover, the mechanisms of upstream activation are different between the two isoforms. Unlike p110α, p110β is activated downstream of both receptor tyrosine kinases and G-protein coupled receptor signaling [51,52]. p110α/p85α is more responsive to phosphorylated RTK than p110β/p85α [45]. Further, p110β is more tightly regulated than p110α [45,53]. Apart from the inhibitory contacts mediated by the p85α nSH2 and iSH2 domains on both p110α and p110β, a third inhibitory interaction that involves contact between the cSH2 domain of p85α and the kinase domain of p110β has been suggested [45,53]. Therefore, mutation of amino acid residues at the cSH2 interacting interface may be required to relieve the inhibition of p110β. Along this line, it has been shown that engineered p85α mutants at the cSH2 domain (Y677A or E675A) relieved cSH2-mediated inhibition of p110β [53]. The kinase domain/cSH2 contact exists also in p110δ but not in p110α [45]. This indicates that p85α and most likely other p85 isoforms have unique regulatory interactions with different class IA p110 isoforms. As the development of p110 isoform specific inhibitors emerge, identification of genomic markers that predict the effect of different p85α mutations on response will be important for successful application of the isoform-selective therapeutic strategies in the clinic. Moreover, it would be interesting to investigate whether p110 mutants that disrupt the p85α inhibitory contact, such as p110α-N345, confer similar sensitivity to the same group of inhibitors as these hypomorphic iSH2 p85α mutants. Further, p110 isoforms have been shown to contribute to blood platelet activation which is essential for coagulation [54]. If p110 isoform specific inhibitors are given to mutant-carrying cancer patients, bleeding risk upon blockage of platelet activation may have to be considered.
Dominant negative & hypomorphic mutants that disrupt p85α homodimerization &/or PTEN binding
Approximately a third of PIK3R1 mutations are found outside the iSH2 domain. It is highly likely that these mutants are involved in altering p85α functions independent of p110 because these N-terminal domains (see Figure 1 for the domain structure of p85α) do not bind p110. Indeed, WT p85α has demonstrated PI3K-independent activities by interaction with multiple signaling molecules through its N-terminal domains. For example, the BH domain of p85α interacts with Cdc42 of the Rho family of small GTPases to regulate cytokinesis [55]. A direct interaction between the BH domain of p85α and PTEN has also been demonstrated contributing to increased PTEN activity [56,57]. Thus p85α has both positive (p110) and negative (PTEN) effects on the PI3K pathway allowing tight spatial and temporal regulation of pathway activity. The SH3 domain of p85α binds to a proline-rich motif in the Na+, K+-ATPase α subunit suppressing cell motility by reorganizing the actin cytoskeleton [58,59]. Intriguingly, p85α itself has two proline-rich motifs and one of them contains a canonical class I SH3-interacting sequence, suggesting that there may be intramolecular or intermolecular interaction between the SH3 and the proline-rich domains of p85α monomers or dimers, respectively. Moreover, p85α is shown to be in excess of p110α in fibroblasts [60], indicating that the amount of endogenous p85α is more than p110α in certain cell context and resulting in a pool of p85α that is not bound to p110α. p85α, if in excess of p110α, could compete with the p85α-p110α complex for binding to phosphorylated insulin receptor substrate or to other phosphotyrosine-containing proteins [61], suggesting that p110α-unbound p85α could act as a negative regulator of PI3K signaling and potentially act as a tumor suppressor.
We have recently proposed an alternative but compatible mechanistic model of how p110-free p85α negatively regulates the PI3K pathway. p85α homodimers were first reported in 1999 [62]. We have extended this finding by demonstrating that p110α-unbound p85α homodimerizes through multiple intermolecular interactions between two p85α monomers [37,63]. The SH3 domain of one p85α protomer interacted with the first proline-rich motif (residues 93–99; proline-rich domain 1, Figure 1) of another p85α protomer in a canonical SH3/proline-rich domain interaction, constituting a major homodimerization interface. Additional interactions through BH:BH domain dimerization also contributed to stabilizing the homodimer. Intriguingly, the p85α homodimer competed with the E3 ligase of PTEN WWP2 for binding to the PTEN phosphatase domain and protected PTEN against WWP2-mediated proteosomal degradation [63]. Further, p85α homodimers also enhanced the lipid phosphatase activity and membrane association of PTEN providing increased access to its targets. Indeed, Pik3r1 knockout mice display decreased PTEN protein levels and increased PI3K pathway activity [64]. p110α-unbound homodimeric p85α, therefore, indirectly downregulates PI3K signaling through positive regulation of PTEN.
Of significant clinical importance, we have identified cancer patient-derived p85α mutants that disrupt p85α homodimerization and PTEN binding resulting in decreased PTEN levels, indicating the relevance of the p85α homodimer in cancer biology. One of these mutants E160* leads to premature stop codon and a truncated protein lacking the BH domain and other C terminal domains. E160* binds WT p85α resulting in decreased WT p85α dimers [37]. Consistent with the lack of the BH domain that binds PTEN, E160* could not bind PTEN and further, the binding between PTEN and WT p85α was decreased in the presence of E160* [37]. E160*, therefore, represents a dominant negative mutant inhibiting p85α homodimerization. As a result, WT p85α does not interact with PTEN and does not protect PTEN from ubiquitination. I177, which is located at the BH:BH p85α homodimer interface, is highly conserved across species. Intriguingly, a naturally occurring I177N mutation decreased homodimer formation and decreased PTEN binding [63] with a consequent deceased PTEN protein levels and increased phosphorylated AKT. Thus I177N is a hypomorph due to weakened ability to form homodimers and a decreased ability to stabilize PTEN compared with WT p85α. Strikingly, another cancer patient-derived p85α point mutation hypomorph located at the PTEN interacting interface has been discovered. Without an impact on p85α homodimer formation, the mutant I133N led to decreased PTEN binding, increase in PTEN ubiquitination and activation of the PI3K pathway [63]. There are multiple other patient mutations in regions potentially involved in p85α homodimerization [37,40] for example, other truncation mutants around E160 (Figure 1). It is very likely that these mutants share the same oncogenic mechanism as E160*. The proline residue (residue 99) in the first proline-rich motif, which is involved in p85α homodimerization, is a site of recurrent mutation.
Similar to the mutants within the SH2 domains described above, these p85α homodimer and/or PTEN destabilizing mutants lead to activation of the downstream PI3K pathway, therefore implicating PI3K pathway inhibitors as potential therapies for tumor cells carrying these mutants. Indeed, a drug sensitivity screen with a 145-compound library revealed that E160* rendered BaF/3 cells more sensitive to mTOR inhibitors (rapamycin and temsirolimus), but not to pan-PI3K inhibitors (GSK2126458A and GDC0941) [65]. The cells were not sensitive to RTK inhibitors such as the EGFR inhibitors lapatinib and erlotinib [65]. Observations from studies by others suggest that PI3K pathway activation confers resistance to RTK inhibition and that dual blockade of PI3K pathway and RTK may improve the therapeutic outcome [66–68]. Interestingly, along this line, E160* conferred BaF/3 cells sensitivity to the inhibitor PP121 which is a dual inhibitor of RTKs (Abl, Src, VEGFR–2 and PDGFR) and PI3K family kinases (p110α, DNA-PK and mTOR). The efficacy of such combined inhibition definitely needs further confirmation using more inhibitors in more cell models.
A previous study has shown that subtle decreases in PTEN expression may have profound effects in promoting tumor-suppressor ‘quasi-insufficiency’ and increased susceptibility to cancer [69]. With the discovery of PTEN-destabilizing p85α mutants, it may be important to investigate whether cells with the PTEN-destabilizing p85α mutants display the same drug sensitivity profile as PTEN-deficient cells. The preclinical and clinical drug sensitivity data of PTEN-deficient cancer cells available so far are conflicting, for example, to AKT inhibitor [70,71] or mTOR inhibitor [72–74]. This is probably due to the differences in genetic background in these cell models under investigation with an isogenic cell panel potentially providing a better model for the comparison. Further, p110β, but not p110α, is suggested to be the dominant PI3K isoform mediating PI3K signaling in PTEN-deficient tumors [75,76]. Some studies demonstrated that the p110β-selective inhibitor was more effective than p110α-specific inhibitors in PTEN-deficient tumors in at least some lineages [75,77–79]. The therapeutic liability engendered by the PTEN-destabilizing mutants to PI3K isoform specific inhibitors would be another interesting area for investigation.
Neomorphic mutants that activate the MAPK signaling pathways
Perhaps the most exciting discovery among all functionally characterized p85α mutants is the identification of a neomorph that activates the MAPK pathway in addition to the PI3K pathway. R348* is the most recurrent PIK3R1 mutation in endometrial cancers (9.6 and 6.9% of all PIK3R1 mutations in our and TCGA datasets, respectively) and colon cancers (13.3%; TCGA) [37,40]. Multiple patient-derived truncation mutations within close distance from R348 have also been detected including a Q329fs in renal papillary cell carcinoma and a L370fs in ovarian cancer [40] (Figure 1). This high occurrence frequency provides justification for investigating whether these mutations can be biomarker of drug response and the underlying mechanism of sensitivity.
Consistent with an activation of AKT, R348* engendered sensitivity to an AKT inhibitor MK2206 [65]. In contrast to E160*, R348* had no effect on sensitivity to rapamcyin. While both E160* and R348* do not stabilize the PTEN protein, the activation of signaling downstream of AKT (including phosphorylation of S6, p70SK6 and GSK3) was more robust in cells with R348* than E160* mutations [65]. The distinct drug sensitivity profiles could, therefore, be due to difference in activation mechanisms or degree of activation of the pathway and/or involvement of different feedback machinery along the PI3K pathway axis. The exact mechanism by which R348* activates the PI3K pathway warrants further investigation. Preliminary data suggested that the activation requires the activities of Cdc42 and Rac1 [80].
Surprisingly, BaF/3, endometrial or ovarian cancer cells expressing R348* or L370fs displayed sensitivity to multiple MEK (PD0325901, AZD6244, PD98059, CI1040 and hypothemycin) and JNK (SP600125, BI78D3 and AEG3482) inhibitors [65]. The sensitivity to these MAPK inhibitors was unique to R348*- or L370fs-expressing cells because WT p85α, E160* and other known p85α driver mutants (DKRMNS560del, R574fs and T576del) had no effect on sensitivity towards these inhibitors, highlighting the neomorphic activities of R348* and L370fs [65]. Xenograft models with endometrial or ovarian cancer cells stably expressing the neomorphs displayed significantly increased tumor growth and the tumors were highly sensitive to MEK and JNK inhibitors. Activation of the pathways appears to underlie pattern of sensitivity to the inhibitors because the neomorphs were associated with phosphorylation of components along the ERK and JNK pathways. Higher phosphorylation of nuclear ERK and JNK could also be observed by immunohistochemical staining in endometrial cancer patients with R348* compared with those without the mutation [65]. Importantly, the activation of MAPK by the neomorphs was independent of the convention role of p85α in PI3K signaling and this ties in perfectly to the fact that R348* lacks an iSH2 domain that mediates association with p110. The neomorphic activity is expected to be applicable to multiple patient-derived truncating and other mutations in PIK3R1 within close proximity of R348 or L370 (Figure 1).
The ability of the neomorphs to activate the MAPK pathways underscores the need to functionally characterize each patients specific driver mutation. The identification of neomorphs argues that patient stratification for treatment approaches needs to be based on the specific mutations present in the cancer gene rather than on the mutated cancer gene alone because of the potential unique functional consequences of individual mutations. The treatment strategy for tumors with the p85α neomorphs may be similar to that of KRAS driven tumors or tumors with co-mutations in both MAPK and PI3K pathways. Activated MAPK pathway in Ras mutant tumors mediates resistance to therapy targeting PI3K pathway [81]. Conversely, PI3K pathway has also been implicated in mediating resistance to MEK inhibitors [82,83]. Combined inhibition of PI3K and MAPK signaling may be required to achieve maximal therapeutic response.
Reduced PIK3R1 expression also confers drug sensitivity
PIK3R1 was significantly decreased in a subset of human cancer tissues including ovarian, prostate, liver, lung, breast, kidney, compared with control tissues [64,84]. Reduced expression of PIK3R1 mRNA and p85α protein was associated with poorer metastasis-free survival of breast cancer patients [84]. Further, low PIK3R1 mRNA level was a signature of high-risk stage I nonsmall cell lung cancers compared with their low-risk counterparts [85]. Likewise, reduced PIK3R1 mRNA expression was associated with migration and invasion of breast cancer cell line [86]. In mouse models of hepatocellular cancer, liver specific-Pik3r1 knockout with 80–90% decrease in p85α protein expression led to PI3K pathway activation and conferred spontaneous tumor development [64]. Consistent with a role of p85α in PTEN stabilization, these Pik3r1 knockout tumors had decreased PTEN expression. All these findings clearly suggest that reduced p85α expression promotes tumorigenesis at least in certain tumor types. One plausible underlying mechanism could be decrease in the amount of p110–unbound p85α, which is tumor suppressive as described earlier. However, it is important to note that in vitro knockdown of PIK3R1 resulted in decreased proliferation, migration and invasion in glioblastoma multiforme cells [87], suggesting that increased or decreased p85α levels could have similar effects on tumor progression depending on the context.
Loss-of-function RNAi-based screen showed that silencing PIK3R1 enhanced the sensitivity of breast cancer cell lines to rapamycin [88], implicating a negative role of PIK3R1 in PI3K pathway activation. Intriguingly, the histone deacetylase inhibitor (HDACi) decreased the expression of PIK3R1 mRNA, suggesting the use of combination of HDACi and rapamycin as a potential treatment. The rapamycin/HDACi combination led to decreased PI3K pathway signaling, induced G0/G1 cell cycle arrest as well as enhanced cleavage of PARP and caspase-3 [88]. As another level of evidence, the effect of the rapamycin/HDACi combination of growth of breast cancer cells injected into the mammary fat pad of female SCID mice was associated with decreased p85α protein expression and PI3K pathway inhibition [88]. Another independent study showed that PIK3R1 was overexpressed in cisplatin-resistant ovarian cancer cell lines and knockdown of PIK3R1 resensitized these cell lines to platinum treatment, indicating the potential involvement of p85α in acquired cisplatin resistance [89]. The PI3K pathway has been shown to be a mediator of platinum resistance [90–92]. For example, AKT activation conferred resistance to caspase-independent cisplatin-induced apoptosis by inhibiting the apoptosis-inducing factor-associated pathway [91]. However, the mechanism of how p85α mediates resistance to chemotherapy and whether this involves the canonical PI3K signaling pathway remain to be investigated.
Conclusion
Different driver aberrations of the same gene may be functional through divergent oncogenic mechanisms and influence distinct downstream signaling processes. PIK3R1 can be hailed as the ‘poster child’ for this concept. Although the p85α driver mutants characterized thus far all activate the PI3K pathway, the mutants may lead to activation of different nodes along the axis or elicit different feedback mechanisms therefore engendering sensitivity to inhibitors of different nodes in the pathway. The mutants may also activate unpredicted pathways, for example the MAPK pathway. Clearly, tailored therapeutic approaches will be required to target these individual PIK3R1 mutations.
Future perspective
The key challenge for successful implementation of targeted therapy is to identify the appropriate patient populations most likely to benefit from the treatment. Currently, many targeted therapeutics are entering clinical evaluation and even more compounds are under development. The development and validation of biomarkers that can identify the sensitive or resistant populations appears to be much slower than the pace of new drug development. Indeed, only a small subset of targeted cancer therapies are administered based on the genetic background of the tumors [93–95]. Aggressive efforts in functional characterization of drivers are needed to speed up the translation of genomic data into the clinic, in order to identify additional biomarkers and to devise better therapeutic strategies.
The PIK3R1 gene is currently included in the sequencing panel of multiple clinical laboratories for the analysis of cancer patient tumor samples, further highlighting the importance of understanding of the biological significance of the genomic alterations of the gene. In addition to small molecule inhibitors that target specific kinases along the PI3K pathway, the identification of interacting partners of p85α and cancer patient-derived mutants that target these interactions may allow development of potential alternative treatment approaches. For instance, pharmacological compounds or small peptides that specifically target the interacting interfaces between p85α and p110 or PTEN may be a new avenue of targeting tumors harboring specific PIK3R1 mutations.
Executive summary.
Functionally characterized p85α mutants are hypomorphs, dominant negative mutants or neomorphs.
PIK3R1 aberrations including gene mutations or downregulation of gene expression could potentially be a biomarker of responsiveness to inhibitors targeting different nodes along the PI3K pathway or the MAPK pathway.
Detailed functional characterization and identification of underlying mechanism of driver mutations is imperative to deliver optimal therapy to cancer patients.
Footnotes
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Hoadley KA, Yau C, Wolf DM, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158(4):929–944. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weinstein JN, Collisson EA, Mills GB, et al. The Cancer Genome Atlas Pan-Cancer Analysis Project. Nat. Genet. 2013;45(10):1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296(5573):1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 4.Geering B, Cutillas PR, Nock G, Gharbi SI, Vanhaesebroeck B. Class IA phosphoinositide 3-kinases are obligate p85–p110 heterodimers. Proc. Natl Acad. Sci. USA. 2007;104(19):7809–7814. doi: 10.1073/pnas.0700373104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fruman DA, Cantley LC. Phosphoinositide 3-kinase in immunological systems. Semin. Immunol. 2002;14(1):7–18. doi: 10.1006/smim.2001.0337. [DOI] [PubMed] [Google Scholar]
- 6.Fruman DA, Meyers RE, Cantley LC. Phosphoinositide kinases. Annu. Rev. Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- 7.Yu J, Zhang Y, Mcilroy J, Rordorf-Nikolic T, Orr GA, Backer JM. Regulation of the p85/p110 phosphatidylinositol 3′-kinase: stabilization and inhibition of the p110alpha catalytic subunit by the p85 regulatory subunit. Mol. Cell. Biol. 1998;18(3):1379–1387. doi: 10.1128/mcb.18.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cuevas BD, Lu Y, Mao M, et al. Tyrosine phosphorylation of p85 relieves its inhibitory activity on phosphatidylinositol 3-kinase. J. Biol. Chem. 2001;276(29):27455–27461. doi: 10.1074/jbc.M100556200. [DOI] [PubMed] [Google Scholar]
- 9.Cully M, You H, Levine AJ, Mak TW. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat. Rev. Cancer. 2006;6(3):184–192. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- 10.Foukas LC, Okkenhaug K. Gene-targeting reveals physiological roles and complex regulation of the phosphoinositide 3-kinases. Arch. Biochem. Biophys. 2003;414(1):13–18. doi: 10.1016/s0003-9861(03)00177-2. [DOI] [PubMed] [Google Scholar]
- 11.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat. Rev. Genet. 2006;7(8):606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 12.Brugge J, Hung MC, Mills GB. A new mutational AKTivation in the PI3K pathway. Cancer Cell. 2007;12(2):104–107. doi: 10.1016/j.ccr.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 13.Shayesteh L, Lu Y, Kuo WL, et al. PIK3CA is implicated as an oncogene in ovarian cancer. Nat. Genet. 1999;21(1):99–102. doi: 10.1038/5042. [DOI] [PubMed] [Google Scholar]
- 14.Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat. Rev. Drug Discov. 2005;4(12):988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- 15.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27(41):5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendoza MC, Er EE, Blenis J. The Ras–ERK and PI3K–mTOR pathways: cross-talk and compensation. Trends Biochem. Sci. 2011;36(6):320–328. doi: 10.1016/j.tibs.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zabransky DJ, Park BH. Estrogen receptor and receptor tyrosine kinase signaling: use of combinatorial hormone and epidermal growth factor receptor/human epidermal growth factor receptor 2-targeted therapies for breast cancer. J. Clin. Oncol. 2014;32(10):1084–1086. doi: 10.1200/JCO.2013.53.5070. [DOI] [PubMed] [Google Scholar]
- 18.Kandoth C, Mclellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zack TI, Schumacher SE, Carter SL, et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 2013;45(10):1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agarwal R, Carey M, Hennessy B, Mills GB. PI3K pathway-directed therapeutic strategies in cancer. Curr. Opin. Invest. Drugs. 2010;11(6):615–628. [PubMed] [Google Scholar]
- 21.Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat. Rev. Drug Discov. 2009;8(8):627–644. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodon J, Dienstmann R, Serra V, Tabernero J. Development of PI3K inhibitors: lessons learned from early clinical trials. Nat. Rev. Clin. Oncol. 2013;10(3):143–153. doi: 10.1038/nrclinonc.2013.10. [DOI] [PubMed] [Google Scholar]
- 23.Thorpe LM, Yuzugullu H, Zhao JJ. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat. Rev. Cancer. 2015;15(1):7–24. doi: 10.1038/nrc3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janku F., Jr Molecular targets for cancer therapy in the PI3K/AKT/mTOR pathway. Pharmacol. Ther. 2014;142(2):164–175. doi: 10.1016/j.pharmthera.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 25.Wagner AJVHDH, Lorusso PM, Tibes R, Mazina KE, Ware JA. 45th Annual Meeting of the American Society of Clinical Oncology. Orlando, FL: 29 May–2 June 2009. A first-in-human Phase I study to evaluate the pan-PI3K inhibitor GDC-0941 administered QD or BID in patients with advanced solid tumors. Presented at. [Google Scholar]
- 26.Battelli C, Cho DC. mTOR inhibitors in renal cell carcinoma. Therapy. 2011;8(4):359–367. doi: 10.2217/thy.11.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fasolo A, Sessa C. Targeting mTOR pathways in human malignancies. Curr. Pharm. Des. 2012;18(19):2766–2777. doi: 10.2174/138161212800626210. [DOI] [PubMed] [Google Scholar]
- 28.Pal SK, Reckamp K, Yu H, Figlin RA. Akt inhibitors in clinical development for the treatment of cancer. Expert Opin. Invest. Drugs. 2010;19(11):1355–1366. doi: 10.1517/13543784.2010.520701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macias-Perez IM, Flinn IW. G-1101: a delta-specific PI3K inhibitor in chronic lymphocytic leukemia. Curr. Hematol. Malig. Rep. 2013;8(1):22–27. doi: 10.1007/s11899-012-0142-1. [DOI] [PubMed] [Google Scholar]
- 30.Benjamin D, Colombi M, Moroni C, Hall MN. Rapamycin passes the torch: a new generation of mTOR inhibitors. Nat. Rev. Drug Discov. 2011;10(11):868–880. doi: 10.1038/nrd3531. [DOI] [PubMed] [Google Scholar]
- 31.Massacesi C, Di Tomaso E, Fretault N, Hirawat S. Challenges in the clinical development of PI3K inhibitors. Ann. NY Acad. Sci. 2013;1280:19–23. doi: 10.1111/nyas.12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nahta R, Esteva FJ. Trastuzumab: triumphs and tribulations. Oncogene. 2007;26(25):3637–3643. doi: 10.1038/sj.onc.1210379. [DOI] [PubMed] [Google Scholar]
- 33.Druker BJ. Imatinib as a paradigm of targeted therapies. Adv. Cancer Res. 2004;91:1–30. doi: 10.1016/S0065-230X(04)91001-9. [DOI] [PubMed] [Google Scholar]
- 34.Martini M, Ciraolo E, Gulluni F, Hirsch E. Targeting PI3K in cancer: any good news? Front. Oncol. 2013;3:108. doi: 10.3389/fonc.2013.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janku F, Wheler JJ, Naing A, et al. PIK3CA mutation H1047R is associated with response to PI3K/AKT/mTOR signaling pathway inhibitors in early-phase clinical trials. Cancer Res. 2013;73(1):276–284. doi: 10.1158/0008-5472.CAN-12-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janku F, Hong DS, Fu S, et al. Assessing PIK3CA and PTEN in early-phase trials with PI3K/AKT/mTOR inhibitors. Cell Rep. 2014;6(2):377–387. doi: 10.1016/j.celrep.2013.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheung LW, Hennessy BT, Li J, et al. High frequency of PIK3R1 and PIK3R2 mutations in endometrial cancer elucidates a novel mechanism for regulation of PTEN protein stability. Cancer Discov. 2011;1(2):170–185. doi: 10.1158/2159-8290.CD-11-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kandoth C, Schultz N, Cherniack AD, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497(7447):67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brennan CW, Verhaak RG, Mckenna A, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seshagiri S, Stawiski EW, Durinck S, et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488(7413):660–664. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Otsu M, Hiles I, Gout I, et al. Characterization of two 85 kd proteins that associate with receptor tyrosine kinases, middle-T/pp60c-src complexes, and PI3-kinase. Cell. 1991;65(1):91–104. doi: 10.1016/0092-8674(91)90411-q. [DOI] [PubMed] [Google Scholar]
- 43.Miled N, Yan Y, Hon WC, et al. Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science. 2007;317(5835):239–242. doi: 10.1126/science.1135394. [DOI] [PubMed] [Google Scholar]
- 44.Wu H, Shekar SC, Flinn RJ, et al. Regulation of Class IA PI 3-kinases: C2 domain-iSH2 domain contacts inhibit p85/p110alpha and are disrupted in oncogenic p85 mutants. Proc. Natl Acad. Sci. USA. 2009;106(48):20258–20263. doi: 10.1073/pnas.0902369106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burke JE, Williams RL. Dynamic steps in receptor tyrosine kinase mediated activation of class IA phosphoinositide 3-kinases (PI3K) captured by H/D exchange (HDX-MS) Adv. Biol. Regul. 2013;53(1):97–110. doi: 10.1016/j.jbior.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun M, Hillmann P, Hofmann BT, Hart JR, Vogt PK. Cancer-derived mutations in the regulatory subunit p85alpha of phosphoinositide 3-kinase function through the catalytic subunit p110alpha. Proc. Natl Acad. Sci. USA. 2010;107(35):15547–15552. doi: 10.1073/pnas.1009652107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jaiswal BS, Janakiraman V, Kljavin NM, et al. Somatic mutations in p85alpha promote tumorigenesis through class IA PI3K activation. Cancer Cell. 2009;16(6):463–474. doi: 10.1016/j.ccr.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quayle SN, Lee JY, Cheung LW, et al. Somatic mutations of PIK3R1 promote gliomagenesis. PLoS ONE. 2012;7(11):e49466. doi: 10.1371/journal.pone.0049466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang CH, Mandelker D, Schmidt-Kittler O, et al. The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations. Science. 2007;318(5857):1744–1748. doi: 10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- 50.Gymnopoulos M, Elsliger MA, Vogt PK. Rare cancer-specific mutations in PIK3CA show gain of function. Proc. Natl Acad. Sci. USA. 2007;104(13):5569–5574. doi: 10.1073/pnas.0701005104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guillermet-Guibert J, Bjorklof K, Salpekar A, et al. The p110beta isoform of phosphoinositide 3-kinase signals downstream of G protein-coupled receptors and is functionally redundant with p110gamma. Proc. Natl Acad. Sci. USA. 2008;105(24):8292–8297. doi: 10.1073/pnas.0707761105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maier U, Babich A, Nurnberg B. Roles of non-catalytic subunits in gbetagamma-induced activation of class I phosphoinositide 3-kinase isoforms beta and gamma. J. Biol. Chem. 1999;274(41):29311–29317. doi: 10.1074/jbc.274.41.29311. [DOI] [PubMed] [Google Scholar]
- 53.Zhang X, Vadas O, Perisic O, et al. Structure of lipid kinase p110beta/p85beta elucidates an unusual SH2-domain-mediated inhibitory mechanism. Mol. Cell. 2011;41(5):567–578. doi: 10.1016/j.molcel.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laurent PA, Severin S, Gratacap MP, Payrastre B. Class I PI 3-kinases signaling in platelet activation and thrombosis: PDK1/Akt/GSK3 axis and impact of PTEN and SHIP1. Adv. Biol. Regul. 2014;54:162–174. doi: 10.1016/j.jbior.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 55.Garcia Z, Silio V, Marques M, et al. A PI3K activity-independent function of p85 regulatory subunit in control of mammalian cytokinesis. EMBO J. 2006;25(20):4740–4751. doi: 10.1038/sj.emboj.7601324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chagpar RB, Links PH, Pastor MC, et al. Direct positive regulation of PTEN by the p85 subunit of phosphatidylinositol 3–kinase. Proc. Natl Acad. Sci. USA. 2010;107(12):5471–5476. doi: 10.1073/pnas.0908899107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rabinovsky R, Pochanard P, Mcnear C, et al. p85 Associates with unphosphorylated PTEN and the PTEN-associated complex. Mol. Cell. Biol. 2009;29(19):5377–5388. doi: 10.1128/MCB.01649-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yudowski GA, Efendiev R, Pedemonte CH, Katz AI, Berggren PO, Bertorello AM. Phosphoinositide-3 kinase binds to a proline-rich motif in the Na+, K+-ATPase alpha subunit and regulates its trafficking. Proc. Natl Acad. Sci. USA. 2000;97(12):6556–6561. doi: 10.1073/pnas.100128297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barwe SP, Anilkumar G, Moon SY, et al. Novel role for Na, K-ATPase in phosphatidylinositol 3-kinase signaling and suppression of cell motility. Mol. Biol. Cell. 2005;16(3):1082–1094. doi: 10.1091/mbc.E04-05-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ueki K, Fruman DA, Brachmann SM, Tseng YH, Cantley LC, Kahn CR. Molecular balance between the regulatory and catalytic subunits of phosphoinositide 3-kinase regulates cell signaling and survival. Mol. Cell. Biol. 2002;22(3):965–977. doi: 10.1128/MCB.22.3.965-977.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luo J, Field SJ, Lee JY, Engelman JA, Cantley LC. The p85 regulatory subunit of phosphoinositide 3-kinase down-regulates IRS-1 signaling via the formation of a sequestration complex. J. Cell Biol. 2005;170(3):455–464. doi: 10.1083/jcb.200503088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harpur AG, Layton MJ, Das P, et al. Intermolecular interactions of the p85alpha regulatory subunit of phosphatidylinositol 3-kinase. J. Biol. Chem. 1999;274(18):12323–12332. doi: 10.1074/jbc.274.18.12323. [DOI] [PubMed] [Google Scholar]
- 63.Cheung LW, et al. Regulation of the PI3K pathway through a p85α monomer-homodimer equilibrium. Elife. 2015;4:e06866. doi: 10.7554/eLife.06866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taniguchi CM, Winnay J, Kondo T, et al. The phosphoinositide 3-kinase regulatory subunit p85alpha can exert tumor suppressor properties through negative regulation of growth factor signaling. Cancer Res. 2010;70(13):5305–5315. doi: 10.1158/0008-5472.CAN-09-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cheung LW, Yu S, Zhang D, et al. Naturally occurring neomorphic PIK3R1 mutations activate the MAPK pathway, dictating therapeutic response to MAPK pathway inhibitors. Cancer Cell. 2014;26(4):479–494. doi: 10.1016/j.ccell.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ludovini V, Bianconi F, Pistola L, et al. Phosphoinositide-3-kinase catalytic alpha and KRAS mutations are important predictors of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in patients with advanced non-small cell lung cancer. J. Thorac. Oncol. 2011;6(4):707–715. doi: 10.1097/JTO.0b013e31820a3a6b. [DOI] [PubMed] [Google Scholar]
- 67.Fumarola C, Bonelli MA, Petronini PG, Alfieri RR. Targeting PI3K/AKT/mTOR pathway in non small cell lung cancer. Biochem. Pharmacol. 2014;90(3):197–207. doi: 10.1016/j.bcp.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 68.Engelman JA, Mukohara T, Zejnullahu K, et al. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J. Clin. Invest. 2006;116(10):2695–2706. doi: 10.1172/JCI28656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alimonti A, Carracedo A, Clohessy JG, et al. Subtle variations in PTEN dose determine cancer susceptibility. Nat. Genet. 2010;42(5):454–458. doi: 10.1038/ng.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davies BR, Greenwood H, Dudley P, et al. Preclinical pharmacology of AZD5363, an inhibitor of AKT: pharmacodynamics, antitumor activity, and correlation of monotherapy activity with genetic background. Mol. Cancer Ther. 2012;11(4):873–887. doi: 10.1158/1535-7163.MCT-11-0824-T. [DOI] [PubMed] [Google Scholar]
- 71.Li J, Davies BR, Han S, et al. The AKT inhibitor AZD5363 is selectively active in PI3KCA mutant gastric cancer, and sensitizes a patient-derived gastric cancer xenograft model with PTEN loss to Taxotere. J. Transl. Med. 2013;11:241. doi: 10.1186/1479-5876-11-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weigelt B, Warne PH, Lambros MB, Reis-Filho JS. Downward J. PI3K pathway dependencies in endometrioid endometrial cancer cell lines. Clin. Cancer. Res. 2013;19(13):3533–3544. doi: 10.1158/1078-0432.CCR-12-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oza AM, Elit L, Tsao MS, et al. Phase II study of temsirolimus in women with recurrent or metastatic endometrial cancer: a trial of the NCIC Clinical Trials Group. J. Clin. Oncol. 2011;29(24):3278–3285. doi: 10.1200/JCO.2010.34.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mackay HJ, Eisenhauer EA, Kamel-Reid S, et al. Molecular determinants of outcome with mammalian target of rapamycin inhibition in endometrial cancer. Cancer. 2014;120(4):603–610. doi: 10.1002/cncr.28414. [DOI] [PubMed] [Google Scholar]
- 75.Wee S, Wiederschain D, Maira SM, et al. PTEN-deficient cancers depend on PIK3CB. Proc. Natl Acad. Sci. USA. 2008;105(35):13057–13062. doi: 10.1073/pnas.0802655105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ni J, Liu Q, Xie S, et al. Functional characterization of an isoform-selective inhibitor of PI3K-p110beta as a potential anticancer agent. Cancer Discov. 2012;2(5):425–433. doi: 10.1158/2159-8290.CD-12-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Greshock J. AACR Precision Medicine Series: Synthetic Lethal Approaches to Cancer Vulnerabilities. Bellevue, WA, USA: 17–20 May 2013. Exploiting the synthetic lethal properties of selective PI3K-β inhibition in PTEN deficient cells with GSK2636771. Presented at. [Google Scholar]
- 78.Arkenau H-T, Mateo J, Lemech CR, Infante JR, et al. American Society of Clinical Oncology (ASCO) Annual Meeting 2014. Chicago, IL, USA: 30 May–3 June 2014. Phase I/II, first-in-human dose-escalation study of GSK2636771 in patients (pts) with PTEN-deficient advanced tumors. Presented at. [Google Scholar]
- 79.Edgar KA, Wallin JJ, Berry M, et al. Isoform-specific phosphoinositide 3-kinase inhibitors exert distinct effects in solid tumors. Cancer Res. 2010;70(3):1164–1172. doi: 10.1158/0008-5472.CAN-09-2525. [DOI] [PubMed] [Google Scholar]
- 80.Murga C, Zohar M, Teramoto H, Gutkind JS. Rac1 and RhoG promote cell survival by the activation of PI3K and Akt, independently of their ability to stimulate JNK and NF-kappaB . Oncogene. 2002;21(2):207–216. doi: 10.1038/sj.onc.1205036. [DOI] [PubMed] [Google Scholar]
- 81.Ihle NT, Lemos R, Jr, Wipf P, et al. Mutations in the phosphatidylinositol-3-kinase pathway predict for antitumor activity of the inhibitor PX-866 whereas oncogenic Ras is a dominant predictor for resistance. Cancer Res. 2009;69(1):143–150. doi: 10.1158/0008-5472.CAN-07-6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wee S, Jagani Z, Xiang KX, et al. PI3K pathway activation mediates resistance to MEK inhibitors in KRAS mutant cancers. Cancer Res. 2009;69(10):4286–4293. doi: 10.1158/0008-5472.CAN-08-4765. [DOI] [PubMed] [Google Scholar]
- 83.Balmanno K, Chell SD, Gillings AS, Hayat S, Cook SJ. Intrinsic resistance to the MEK1/2 inhibitor AZD6244 (ARRY-142886) is associated with weak ERK1/2 signalling and/or strong PI3K signalling in colorectal cancer cell lines. Int. J. Cancer. 2009;125(10):2332–2341. doi: 10.1002/ijc.24604. [DOI] [PubMed] [Google Scholar]
- 84.Cizkova M, Vacher S, Meseure D, et al. PIK3R1 underexpression is an independent prognostic marker in breast cancer. BMC Cancer. 2013;13:545. doi: 10.1186/1471-2407-13-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lu Y, Lemon W, Liu PY, et al. A gene expression signature predicts survival of patients with stage I non-small cell lung cancer. PLoS Med. 2006;3(12):e467. doi: 10.1371/journal.pmed.0030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Uchino M, Kojima H, Wada K, et al. Nuclear beta-catenin and CD44 upregulation characterize invasive cell populations in non-aggressive MCF-7 breast cancer cells. BMC Cancer. 2010;10:414. doi: 10.1186/1471-2407-10-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Weber GL, Parat MO, Binder ZA, Gallia GL, Riggins GJ. Abrogation of PIK3CA or PIK3R1 reduces proliferation, migration, and invasion in glioblastoma multiforme cells. Oncotarget. 2011;2(11):833–849. doi: 10.18632/oncotarget.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ou O, Huppi K, Chakka S, et al. Loss-of-function RNAi screens in breast cancer cells identify AURKB, PLK1, PIK3R1, MAPK12, PRKD2, and PTK6 as sensitizing targets of rapamycin activity. Cancer Lett. 2014;354(2):336–347. doi: 10.1016/j.canlet.2014.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stronach EA, Alfraidi A, Rama N, et al. HDAC4-regulated STAT1 activation mediates platinum resistance in ovarian cancer. Cancer Res. 2011;71(13):4412–4422. doi: 10.1158/0008-5472.CAN-10-4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hui RC, Francis RE, Guest SK, et al. Doxorubicin activates FOXO3a to induce the expression of multidrug resistance gene ABCB1 (MDR1) in K562 leukemic cells. Mol. Cancer Ther. 2008;7(3):670–678. doi: 10.1158/1535-7163.MCT-07-0397. [DOI] [PubMed] [Google Scholar]
- 91.Yang X, Fraser M, Abedini MR, Bai T, Tsang BK. Regulation of apoptosis-inducing factor-mediated, cisplatin-induced apoptosis by Akt. Br. J. Cancer. 2008;98(4):803–808. doi: 10.1038/sj.bjc.6604223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Madhunapantula SV, Sharma A, Robertson GP. PRAS40 deregulates apoptosis in malignant melanoma. Cancer Res. 2007;67(8):3626–3636. doi: 10.1158/0008-5472.CAN-06-4234. [DOI] [PubMed] [Google Scholar]
- 93.Martini M, Vecchione L, Siena S, Tejpar S, Bardelli A. Targeted therapies: how personal should we go? Nat. Rev. Clin. Oncol. 2012;9(2):87–97. doi: 10.1038/nrclinonc.2011.164. [DOI] [PubMed] [Google Scholar]
- 94.Dimasi JA, Feldman L, Seckler A, Wilson A. Trends in risks associated with new drug development: success rates for investigational drugs. Clin. Pharmacol. Ther. 2010;87(3):272–277. doi: 10.1038/clpt.2009.295. [DOI] [PubMed] [Google Scholar]
- 95.Mills GB. An emerging toolkit for targeted cancer therapies. Genome Res. 2012;22(2):177–182. doi: 10.1101/gr.136044.111. [DOI] [PMC free article] [PubMed] [Google Scholar]