Abstract
Aim:
Fungi are valuable resources for bioactive secondary metabolites. However, the chemical space of fungal secondary metabolites has been studied only on a limited basis. Herein, we report a comprehensive chemoinformatic analysis of a unique set of 207 fungal metabolites isolated and characterized in a USA National Cancer Institute funded drug discovery project.
Results:
Comparison of the molecular complexity of the 207 fungal metabolites with approved anticancer and nonanticancer drugs, compounds in clinical studies, general screening compounds and molecules Generally Recognized as Safe revealed that fungal metabolites have high degree of complexity. Molecular fingerprints showed that fungal metabolites are as structurally diverse as other natural products and have, in general, drug-like physicochemical properties.
Conclusion:
Fungal products represent promising candidates to expand the medicinally relevant chemical space. This work is a significant expansion of an analysis reported years ago for a smaller set of compounds (less than half of the ones included in the present work) from filamentous fungi using different structural properties.
Keywords: : chemical space, chemoinformatics, fungal metabolites, master key compound, molecular complexity, molecular fingerprint
Introduction
Natural products are important sources for new drugs and are also good lead compounds suitable for further optimization. The pharmaceutical industry has obtained many successful leads from natural sources. Approximately 45% of today's best selling drugs are either natural products or, more typically, their semisynthetic derivatives [1]. This could be because natural products have some important advantages over synthetic compounds: their biosynthesis involves repetitive interactions with enzymes, and for most of them, their actual biological function likely relies upon the ability to bind to proteins. This may explain why secondary metabolites exhibit advanced binding characteristics compared with synthetic compounds [2]. The chemical space of natural products (largely derived from plants), their synthetic derivatives and approved drugs from natural origin has been extensively explored. From such studies it has been concluded that, in general, natural products have larger size, greater 3D complexity, lower hydrophobicity, increased polarity and fewer aromatic rings than other compound libraries [3].
Most natural product-derived leads come from plant or microbial sources. There are also a few remarkable drugs from marine sources, such as the recently approved anticancer drug eribulin, a synthetic analog of halichondrin B isolated from the sponge Halichondria okadai [4]. In the microbial area the main sources have been terrestrial actinomycetes and fungi. From the latter, the secondary metabolite myriocin was the base for the development of fingolimod as the first oral orally active drug for multiple sclerosis.
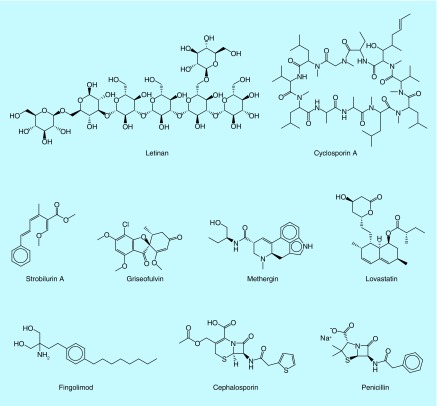
Out of the estimated 1.5–5.5 million species of fungi in the world, only approximately 75,000–100,000 species have been described, and yet, fungi from many different sources continue to be a valuable resource of bioactive secondary metabolites such as antibiotics (cephalosporin), immunosuppressants (cyclosporine A), cholesterol-lowering agents (statins), antifungals (echinocandin B), for the treatment of multiple sclerosis (fingolimod), anticancer drugs (lentinan), for the treatment of hemorrhage (methergin) and fungicides (strobilurins). Figure 1 shows remarkable examples of fungal metabolites (or synthetic analogues) used as drugs [5]. Since there are many unexplored fungal species, there is a high probability for discovering new lead structures. However, the chemical space of secondary metabolites isolated from fungi has been studied only on a limited basis.
Figure 1. . Representative examples of fungal metabolites as drugs.
An initial effort to explore the chemical space of fungal isolates was reported by El-Elimat et al. [6]. In that work, 105 compounds isolated from filamentous fungi, 75 from cyanobacteria and 163 compounds from tropical plants were compared with each other, and to 96 US FDA-approved anticancer drugs, using nine molecular descriptors. Data visualization was conducted using principal component analysis. It was concluded that anticancer drugs cover a large fraction of biologically active chemical space, and that the set of fungal metabolites had a high overlap with the chemical space of anticancer drugs. A reasonable explanation for the high overlap was that 59% of the anticancer drugs were either natural products or compounds derived and/or inspired by them. However, given the limited number of fungal metabolites that have been tested in anticancer assays, this was an encouraging finding. It was also noted that the secondary metabolites isolated from each source represented different areas of chemical space, and therefore, the collective study of each natural source has cumulative benefits toward probing chemical diversity.
Based on previous studies and as part of our continued effort to quantify the chemical diversity of fungal isolates [6], herein we expand the analysis to a larger set of 207 compounds described by our group [7–23] that represent twice the size of the dataset analyzed in 2012. In the current analysis, we employed molecular fingerprints as distinct molecular representations not analyzed in the previous study; also we emphasized the evaluation of molecular complexity that may have a significant impact in drug discovery endeavors, since this feature has been associated with target selectivity (and potential toxicity). As part of the analysis, we implemented a novel consensus measure of structural complexity. The chemical space and structural diversity of the fungal isolates were compared with anticancer and nonanticancer approved drugs, other reference datasets of pharmaceutical interest, and a unique dataset of Generally Recognized as Safe (GRAS) compounds used in the food industry [24].
Methods
General approach
The compound datasets were analyzed using validated chemoinformatics methods to characterize compound libraries [25–27]. Chemoinformatic-based analysis of compound libraries is part of the preliminary characterization of datasets used for drug discovery, providing key information to characterize molecular diversity and coverage of chemical space [28]. Of note, in the study published in 2012 for a smaller (105) set of compounds, nine molecular descriptors commonly used in compound characterizations were computed [6]. Herein, for the 207 fungal metabolites we included a number of previously computed properties plus a novel set of descriptors that capture different aspects of the molecules.
Datasets
Table 1 summarizes the datasets used, including the number of unique compounds after data curation. In total, 5494 compounds were analyzed. Prior to analysis, all datasets were curated using Molecular Operating Environment, version 2014.08 [29]. Molecules were washed using a protocol implemented in Molecular Operating Environment that involves removing salts and neutralizing the charges in the molecules. The largest fragments were kept, duplicates in each dataset were removed and all molecules with molecular weight (MW) over 1000 were excluded. The in-house library of fungal metabolites with 207 compounds (referred to hereafter as ‘FUNGI’) [7–23] was compared with the following five reference collections: 2249 compounds based on the Flavor and Extract Manufacturers Association of the United States GRAS list, updated to GRAS 27 (hereafter referred to as ‘GRAS’) [24,30]; FDA drugs obtained from DrugBank [31] containing: 76 drugs approved to treat cancer (hereafter referred to as ‘FDA-ONC’) and 1399 nononcological drugs (hereafter referred to as ‘FDA-NONC’); 713 drugs in clinical trials reported by the Therapeutic Target Database [32] (hereafter referred to as ‘CLINIC’); and 850 compounds from a commercial collection focused on epigenetic targets, available at Selleckchem (hereafter referred to as ‘GENERAL’) [33]. Supplementary Table 1 summarizes the number of duplicate molecules found in the initial datasets. The chemical structures of representative fungal metabolites analyzed in study are shown in Supplementary Figure 1. The full dataset of fungal metabolites is available upon request.
Table 1. . Compound datasets considered in this work.
| Dataset | Initial size | Unique compounds |
|---|---|---|
| Fungal isolates (FUNGI) |
224 |
207 |
| Approved anticancer drugs (FDA-ONC) |
82 |
76 |
| Approved nonanticancer drugs (FDA-NONC) |
1536 |
1399 |
| Drugs in clinical trials (CLINIC) |
832 |
713 |
| General screening collection (GENERAL) |
1094 |
850 |
| Generally Recognized as Safe (GRAS) | 2295 | 2249 |
Molecular representations
To reduce the dependence of chemical space with structure representation, we used different representations. These can be classified into three main groups: molecular complexity, structural fingerprints and physicochemical properties.
Molecular complexity
Three measures were used to compare the structural complexity of the datasets: fraction of sp3 carbon atoms (F-sp3), fraction of chiral centers (CCF) and globularity (GLOB). F-sp3 and CCF are complexity metrics that have been used broadly to measure MC [34,35]. A higher value of CCF means higher stereochemical complexity and a larger F-sp3 value indicates that the molecule is more likely to have a 3D structure, in other words, a nonplanar structure. 3D structures are preferred over planar structures with the hypothesis that molecules with out-of-plane substituents could adjust their molecular shape and increase receptor/ligand complementarity [34]. However, only considering sp3 carbons may not be enough to determine if a molecule has a 3D structure: F-sp3 does not account for 3D structures given by sp3-hybridized heteroatoms. However, GLOB does capture this structural feature. GLOB evaluates the resemblance of a compound to a sphere, a value equal to 1 represents a spherical molecule while a value equal to 0 represents a structure that is completely flat. F-sp3 was computed with Maya ChemTools [36], dividing the number of sp3 hybridized carbons by the total carbon count; CCF was calculated using Molecular Operating Environment software dividing the number of chiral centers (ChC) by the total carbon count. GLOB was calculated with Molecular Operating Environment software using the conformation with a low-energy conformation. The distributions of the F-sp3, GLOB and CCF values were analyzed using box plots. The statistical analysis of the distributions of F-sp3, GLOB and CCF was done with R Studio [37]. The statistical comparison of the complexity measures was made by the assessment of the homoscedasticity with a Shapiro test in which a normal distribution may be assumed if a p-value >0.05 is obtained. In order to do pairwise comparisons, a Kruskal–Nemenyi test was performed, for which a p-value <0.05 indicates that the null hypothesis must be accepted, meaning that there is not a statistically significant difference between the distributions in the datasets. All the statistics were generated with R Studio using the PMCMR package. In addition, we computed the mean complexity (MC) as the mean of F-sp3, GLOB and CCF values. MC represents a combined measure of MC. For each dataset, the distribution of MC values was obtained along with the corresponding summary statistics. In an attempt to determine whether differences could be observed when considering the total number of heavy atoms (HAs) or MW to calculate the fraction of sp3 carbon atoms, molecular globularity and CCF, we calculated six related fractions: sp3/MW, ChC/MW, sp3/HA, ChC/HA, GLOB/MW and GLOB/HA. An MC of the fractions normalized with MW (MCMW) and an MC for the fractions normalized with HA (MCHA) were calculated for each dataset. The statistical analysis between datasets was generated with R Studio as described for each metric of molecular complexity.
Structural fingerprints
Two structural fingerprints of different design: Molecular ACCess System (MACCS) keys (166-bits) [38] and extended connectivity fingerprints, ECFP4 (ECFPs) [39]. The distribution of the similarity values was analyzed by cumulative distribution functions (CDF) generated with MayaChem Tools and R Studio scripts.
Physicochemical properties
Six properties were calculated with Molecular Operating Environment software. Hydrogen bond donors (HBDs), hydrogen bond acceptors (HBAs), the octanol and/or water partition coefficient (SlogP) and MW; these physicochemical properties are associated with solubility and permeability. Topological polar surface area (TPSA), an important parameter related to solubility, permeability and transport of a compound; and number of rotatable bonds, which is an indicator of the flexibility of the molecules. For each database and property, a box plot was generated. The statistical comparison of the properties was made with R Studio as described in the ‘Methods’ section. To generate a visual representation of the chemical space based on physicochemical properties, a principal component analysis was performed with the six physicochemical properties using DataWarrior, version 4. 2. 2 [40].
Results & discussion
Structural complexity
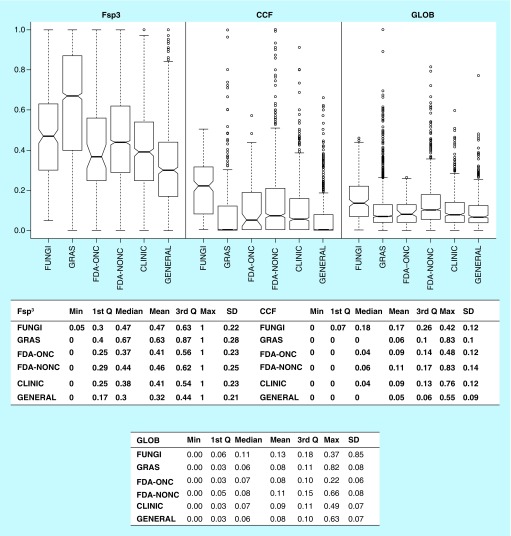
Many drug discovery efforts focus on compound libraries containing small molecules filtered using classical semi-empirical rules. Filtered molecules often interrogate a narrow range of medicinal chemical space but have failed to identify leads for many target classes [41]. Structural complexity is an attractive criterion for the drug discovery process. Indeed, previous studies have shown that increased structural complexity, as measured by simple metrics such as F-sp3 and CCF, is associated with increased probability to reach the market and 3D structures that might allow additional protein–ligand interactions not accessible to a flat aromatic ring [34]. Furthermore, preliminary experimental analyses have shown that increased structural complexity is related to target selectivity [35]. Therefore, we measured the distribution of the structural complexity of the 207 fungal metabolites using Fsp3 and CCF, previously studied metrics. GLOB was also included as an approach to obtain a more accurate description of the molecules shape and 3D structure. A consensus measure of complexity with these three descriptors was introduced and the results were compared with the reference collections. Of course, additional metrics to measure structural complexity could be used [42,43]. Figure 2 shows box plots of the distribution of CCF, GLOB and F-sp3 along with summary statistics. Supplementary Table 3 shows the Kruskal–Nemenyi test results for the complexity analysis.
Figure 2. . Molecular complexity of fungal isolates.
Box plots of the distributions of CCF, F-sp3 and globularity for the fungal isolates, molecules Generally Recognized as Safe, US FDA-approved drugs to treat cancer, FDA-approved drugs, drugs in clinical trials and compounds from a screening collection. Summary statistics are shown below each box plot. See text for details.
1st Q: First quartile; 3rd Q: Third quartile; CCF: Fraction of chiral centers; CLINIC: Drugs in clinical trials; FDA-NONC: FDA-approved drugs; FDA-ONC: FDA-approved drugs to treat cancer; FUNGI: Fungal isolates; F-sp3: Fraction of sp3 carbon atoms; GENERAL: Compounds from a screening collection; GLOB: Fraction of globularity; GRAS: Generally Recognized as Safe.
The CCF and F-sp3 values for the FDA-NONC compounds (that represent a larger fraction of all the FDA-approved compounds) are larger and statistically different from the CLINIC set (Figure 2 & Supplementary Table 2). In general, considering Fsp3 and CCF, the FDA-approved drugs and CLINIC are more complex than the GENERAL screening collection. Indeed, considering the FDA-approved drugs as a single compound set, the relative order of structural complexity decreases in the order Drugs (both FDA categories) > CLINIC > GENERAL which is in good agreement with previously reported data [34]. Interestingly, 3D measured with GLOB shows that FDA-ONC, GENERAL and CLINIC are not statistically different.
Impressively, FUNGI showed larger and statistically different CCF values than all reference datasets, including FDA-approved drugs and in particular FDA-ONC. FUNGI had similar F-sp3 and GLOB values as FDA-NONC, and both datasets exhibited higher values than FDA-ONC. Compared with GENERAL, FUNGI showed larger values of F-sp3, GLOB and CCF, demonstrating their increased structural complexity as compared with molecules typically used in high-throughput screening.
Interestingly, GRAS compounds had the largest structural complexity as measured by the distribution of F-sp3 values (Figure 2). This intriguing result and the inherent safety of GRAS chemicals for human consumption (at given doses) is in agreement with the general notion that large structure complexity can be associated with selectivity, and possibly, less toxicity [34]. The hypothesis of a putative correlation between compound safety and structural complexity is being fully evaluated in our group and will be reported in a separate work (see the ‘Future perspective’ section). An unexpected finding was that, even though GRAS has the highest F-sp3 values, it also has one of the lowest GLOB values. This is in agreement with the hypothesis that sp3-hybridized heteroatoms should be considered to study the complexity and 3D structure of molecules.
Combined measure of complexity
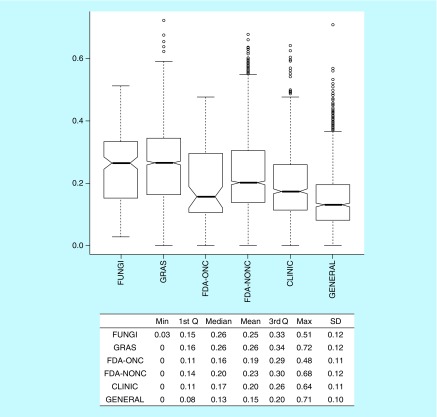
A simple aggregated measure of MC was obtained by computing the mean of F-sp3, GLOB and CCF (see the ‘Methods’ section). Figure 3 shows the distribution of the MC values for each dataset. Supplementary Table 2 summarizes the Kruskal–Nemenyi test results. According to this aggregated measure of complexity, FUNGI were statistically more complex than CLINIC, GENERAL and FDA-approved drugs, in particular FDA-ONC. Interestingly, according to MC, GRAS was not statistically different from FUNGI (Supplementary Table 2). Consistent with other metrics, the GENERAL set was the least complex. Finally, on average, the FDA-NONC and the FDA-ONC datasets showed a different degree of complexity as captured by MC values. This suggests that complexity among approved drugs can be significantly different depending on the therapeutic indication.
Figure 3. . Box plot and summary statistics of the mean of F-sp3, fraction of chiral centers and globularity or mean complexity.
See text for details.
1st Q: First quartile; 3rd Q: Third quartile; CCF: Fraction of chiral centers; F-sp3: Fraction of sp3 carbon atoms; GLOB: Fraction of globularity.
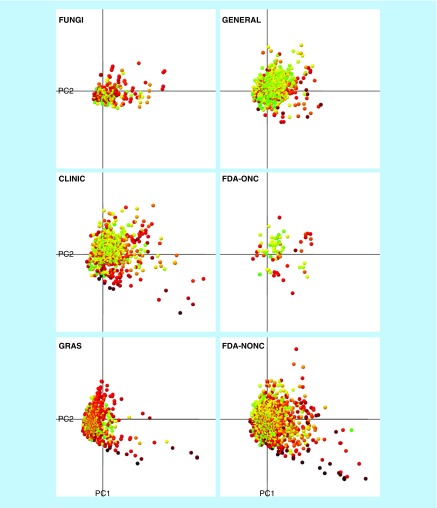
Figure 4 shows the mean of Fsp3 and FCC values mapped on a visual representation of the chemical space generated with principal component analysis of six physicochemical properties: HBA, HBD, SlogP, TPSA, MW and RB (see the ‘Profile of physicochemical properties’ section). A color scale was implemented to highlight each data point using MC in which the most complex compounds were marked red, the moderately complex compounds yellow and the less complex compounds were marked green. Based on the colors, the GRAS database contains more yellow-to-red colored molecules when compared with GENERAL where most of the molecules are green. This visualization was in line with the MC metric (in Figure 3) in which GENERAL was the least complex collection. The FUNGI dataset contained fewer molecules than GENERAL (Table 1), but based on the color scale, most of the molecules were orange-to-red indicating high complexity. The distribution of each dataset in the chemical space based on the physicochemical properties is discussed in the ‘Profile of physicochemical properties’ section.
Figure 4. . Visualization of the molecular complexity mapped on a representation of the chemical space generated with principal component analysis of six physicochemical properties.
The first two principal components recover 85% of the variance. Data points are colored by the mean of Fsp3 and FCC using a continuous color scale from green (less complex) to red (more complex). Each panel corresponds to the visualization of single datasets.
CLINIC: Drugs in clinical trials; FCC: Fraction of chiral centers; FDA-NONC: FDA-approved drugs; FDA-ONC: FDA-approved drugs to treat cancer; FUNGI: Fungal isolates; F-sp3: Fraction of sp3 carbon atoms; GENERAL: Compounds from a screening collection; GLOB: Fraction of globularity; GRAS: Generally Recognized as Safe; PC: Principal component.
FUNGI and the reference sets were compared using the MCHA and MCMW metrics (see the ‘Methods’ section). The goal was to explore the effects of different approaches to normalize the number of sp3 carbon atoms and ChC on the relative order of MC of the datasets. The corresponding box plots and summary statistics of the distributions of MCHA and MCMW are in Supplementary Figure 2. Overall, considering the different normalization methods, similar conclusions were obtained regarding the relative MC of FUNGI compared with the reference datasets. GRAS showed an increased MC, and was statistically more complex than the other datasets.
The analysis of MC supported the hypothesis that fungal metabolites are remarkable candidates to expand the medicinally relevant chemical space [41]. The profile of stereochemical complexity and 3D character (nonflat structures) makes fungal metabolites attractive compounds as drug sources with the potential to be selective and likely to succeed as future drugs [34]. Of note this is the first study that quantifies directly the structural complexity of fungal metabolites.
Structural diversity using molecular fingerprints
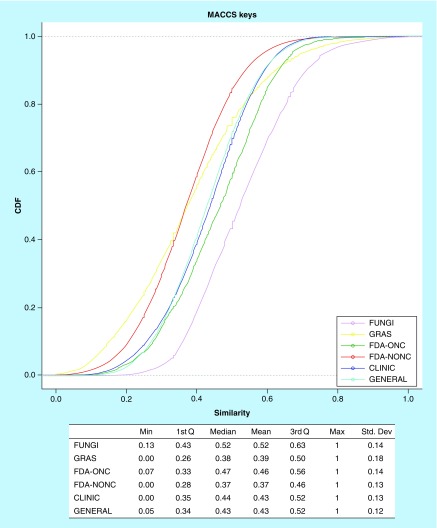
As discussed in the ‘Introduction’ section, this is the first report that addresses in a quantitative manner the structural diversity of fungal metabolites using molecular fingerprints. The structural diversity of each dataset was also measured computing all possible pair-wise comparisons with the Tanimoto coefficient [44,45] and two fingerprint-based structure representations: MACCS keys and ECFPs, previously described in the ‘Methods’ section. The Tanimoto coefficient measures the ratio of the set of structural features that two compounds have in common with respect to the total structural features of both compounds (as codified by the corresponding fingerprint representation). Figure 5 shows the CDF curves (along with summary statistics) of the pair-wise similarity values for each dataset using MACCS keys. The CDF curves of ECFPs are shown in Supplementary Figure 3.
Figure 5. . Molecular diversity of the fungal metabolite dataset and reference collections.
This figure shows the cumulative distribution functions of all pair-wise similarity comparisons using the Tanimoto coefficient using Molecular ACCess System keys. The table summarizes the summary statistics of the cumulative distribution functions. The FUNGI set was less diverse than GRAS, US FDA, CLINIC and GENERAL but is as diverse as other natural product datasets reported in the literature [41,46].
1st Q: First quartile; 3rd Q: Third quartile; CLINIC: Drugs in clinical trials; FDA-NONC: FDA-approved drugs; FDA-ONC: FDA-approved drugs to treat cancer; FUNGI: Fungal isolates; F-sp3: Fraction of sp3 carbon atoms; GENERAL: Compounds from a screening collection; GLOB: Fraction of globularity; GRAS: Generally Recognized as Safe; MACCS: Molecular ACCess System.
Analysis of the CDF indicated that the FUNGI set was diverse with, for instance, median MACCS/Tanimoto similarity of 0.52 (and median of 0.143 with ECFP). Despite the fact that the FUNGI set showed the lowest structural diversity as compared with the other collections studied in this work (e.g., the intralibrary similarity was higher, Figure 5), their diversity was comparable (calculated using the same metrics) to other natural products such as molecules from traditional Chinese medicine or a dataset of commercially available natural products for high-throughput screening [41,46].
Out of the datasets compared in this work, FDA-NONC was the most diverse followed by GRAS and compounds in the GENERAL and CLINIC datasets. Overall, the similarity values for the reference datasets were consistent with the values reported in the literature. The FDA-ONC set had higher similarity values compared with the values reported for approved drugs [41]. This result can be explained because FDA anticancer drugs are focused on fewer molecular targets and occupy a narrower (focused) region of the drug-like chemical space.
Profile of physicochemical properties
In addition to measuring structural complexity and molecular diversity using structural fingerprints, we also obtained the physiochemical profile of the FUNGI set. This is because in library selection and design it is important to have a balance between the structural diversity and physicochemical drug likeness [47]. As mentioned in the ‘methods’ section, six physicochemical properties of pharmaceutical relevance were analyzed for the 207 fungal metabolites and the reference datasets. Box plots and summary statistics of the six properties for all collections are shown in Supplementary Figure 4 & Supplementary Table 1 .
TPSA is an important parameter for the permeability, solubility and transport of compounds [48]. The FUNGI set had comparable values of TPSA with FDA-ONC. Indeed, it has been suggested that TPSA is one of the key physicochemical properties that confer the drug-likeness character to natural products [49].
Regarding molecular flexibility as measured by RB, the FUNGI set was similar to GRAS, and both sets were less flexible than FDA-approved drugs, CLINIC and GENERAL datasets. The fungal metabolites analyzed in this work were not statistically different from FDA-ONC in terms of HBA, HBD and MW. This result is in line with the conclusions obtained in the previous work for a smaller set of fungal metabolites described in the ‘Introduction’ section. [6].
It is noteworthy that there was a statistical difference between the datasets with FDA-approved drugs; on average FDA-ONC drugs had higher values than FDA-NONC for most of the computed properties. In other words, there is a considerable difference in the physicochemical properties of drugs used to treat cancer and drugs approved for other indications.
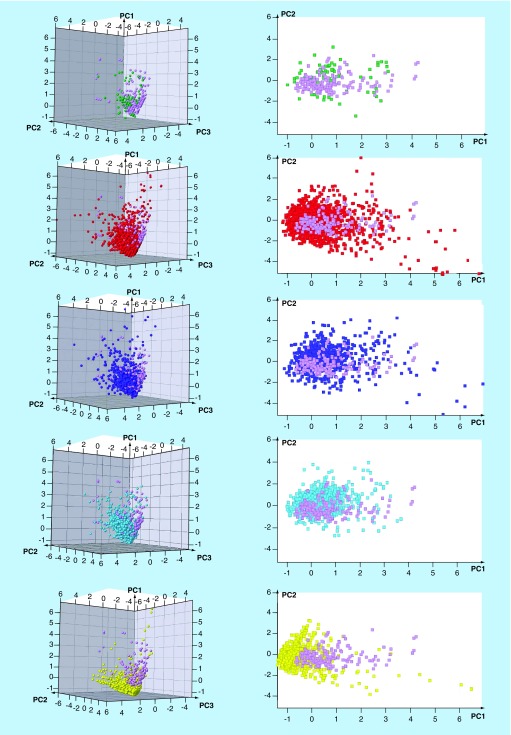
2D and 3D visual representations of the chemical space based on the six properties are shown in Figure 6. The first two principal components retrieved 85.2% of the variance, whereas 93.9% was recovered by using the first three principal components. Supplementary Table 5 summarizes the loadings for the six properties. Supplementary Table 5 indicates that HBA and HBD had the highest contributions to the first principal component, SlogP had the highest contribution to the second principal component and RB had the highest contribution to the third principal component. Figure 6 shows that fungal metabolites cover similar regions of the property space of FDA-approved drugs, CLINIC and GENERAL, particularly FDA-ONC. This result was consistent with the conclusions obtained in our previous work [6]. Interestingly, most of the outliers were either molecules with many ChC, or highly saturated molecules. As expected, most of the molecules in FDA-ONC, GENERAL and CLINIC occupy similar regions of the chemical space, indicating that these sets have, in general, comparable physicochemical properties (as quantitatively captured by the box plots in Supplementary Figure 4).
Figure 6. . 3D and 2D visual representation of the chemical space of 207 fungal isolates considered in this work.
The visual representation was generated with a principal component analysis of six physicochemical properties: molecular weight, hydrogen bond donors, hydrogen bond acceptors, the octanol and/or water partition coefficient, topological polar surface area and number of rotatable bonds. The first two principal components capture 85.2% of the variance and the first three principal components capture 93.9% of the variance. FUNGI: purple; GRAS: yellow; FDA-ONC: green; FDA-NONC: red; CLINIC: blue; GENERAL: turquoise.
FDA-NONC compounds showed a broader coverage of the property space. Interestingly, GRAS molecules occupy a well-defined and distinct area of the space that was associated with the smallest molecules. In fact, based on the distribution of the physicochemical properties and visual representation of the chemical space, GRAS molecules were, overall, smaller molecules than the other reference collections. However, GRAS compounds have comparable SlogP values. Of note, the octanol/water partition coefficient is one of the most important drug-like physicochemical properties [49]. Fungal metabolites share pharmaceutical important physicochemical properties with the approved drugs. Even though natural products do not necessarily follow all Lipinski's Rule of Five [49], in general, the fungal isolates studied in this work fulfilled those rules.
Conclusion
In this work, we quantified for the first time the structural complexity of 207 fungal natural products previously isolated and characterized during our investigation into cytotoxic fungal metabolites conducted over the past decade. It was concluded that the chemical structures of fungal metabolites are more complex than FDA-approved drugs, compounds undergoing clinical trials and general screening molecules. This distinct feature of fungal metabolites, combined with similar structural diversity compared with other natural products datasets, quantified using several molecular fingerprints and drug-like physicochemical properties make fungal metabolites an attractive source to cover novel regions of the medicinally relevant chemical space. During the course of this study, differences in the structural complexity and physicochemical profile of FDA-approved anticancer and nonanticancer drugs were evaluated. Interestingly, GRAS compounds had the highest MC profile as measured by the fraction of sp3 carbon atoms. This result led to the hypothesis that MC could be related to compound safety, in other words, low toxicity. However, considering the mean of F-sp3, GLOB and CCF as a simple measure of complexity, a similar profile of MC of fungal metabolites and GRAS molecules was observed. This result suggested that both types of structures have the potential to present similar target selectivity profiles. Taking all results together, it can be concluded that the fungal metabolites are attractive sources of leads, likely because they combine high complexity and structural diversity with drug-like physicochemical properties.
Future perspective
The long-term goal of this study is to contribute to the systematic identification of bioactive compounds from fungi. This plan is in agreement with an effort to synergize natural product-based and computer-aided drug discovery [50]. One of the first steps to systematically identify active molecules in compound libraries is to characterize the chemical space. However, only few reports of efforts to characterize the chemical space of fungal metabolites have been published. To our knowledge, this is the second computational study that expands the findings of our work published 4 years ago for a smaller set of fungal metabolites (with half of the molecules reported) using different metrics and structure representations. Therefore, it is expected that this study will continue to stimulate the research community, including our own groups, by expanding our knowledge of the chemical space of fungal metabolites. Moreover, the findings of this Short Communication led to hypotheses that warrant further computational and experimental exploration. At least three follow-up studies can be envisioned: experimental testing of fungal metabolites across different molecular targets. In addition to uncovering new bioactive leads, results of the screening will test the hypothesis that the large structural complexity of fungal metabolites is associated with target selectivity. Computational and experimental assessment of the putative association between compound toxicity with MC, and target fishing, in other words, computational prediction of molecular targets of fungal metabolites followed by rigorous experimental validation.
Executive summary.
Fungal metabolites have high structural complexity, large structural diversity (comparable to other natural products) and drug-like physicochemical properties. Therefore, they are attractive compounds to expand the medicinally relevant chemical space.
Findings of this work led to the hypothesis that fungal metabolites have, in general, a promising potential to be selective if tested across diverse molecular targets. Future systematic experimental screening will test this hypothesis.
Generally Recognized as Safe compounds had larger structural complexity than approved drugs and fungal metabolites. Hence, it is hypothesized that molecular complexity can be associated with compound safety, in other words, low toxicity. This hypothesis is currently being tested by our group.
Notable differences were found between the chemical space of approved oncological and nononcological drugs.
Supplementary Material
Footnotes
Financial & competing interests disclosure
The authors thank the Universidad Nacional Autónoma de México (UNAM) for grant PAPIME No. PE200116, and the institutional program Nuevas Alternativas de Tratamiento para Enfermedades Infecciosas (NUATEI) of the Instituto de Investigaciones Biomédicas (IIB) UNAM for financial support, and the Consejo Nacional de Ciencia y Tecnología (CONACyT) for grant 236564. FDP-M is grateful to CONACyT for the fellowship 660465/576637. FD Prieto-Martínez is grateful to CONACyT for the fellowship No. 660465/576637. The isolation of fungal metabolites from the Mycosynthetix library via researchers at UNCG was funded by grant P01 CA125066 from the National Cancer Institute/National Institutes of Health, Bethesda, MD, USA. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Newman DJ, Cragg GM. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016;79(3):629–661. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- 2.Muller-Kuhrt L. Putting nature back into drug discovery. Nat. Biotech. 2003;21(6):602–602. doi: 10.1038/nbt0603-602. [DOI] [PubMed] [Google Scholar]
- 3.Stratton CF, Newman DJ, Tan DS. Cheminformatic comparison of approved drugs from natural product versus synthetic origins. Bioorg. Med. Chem. Lett. 2015;25(21):4802–4807. doi: 10.1016/j.bmcl.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okouneva T, Azarenko O, Wilson L, et al. Inhibition of centromere dynamics by Eribulin (E7389) during mitotic metaphase. Mol. Cancer Ther. 2008;7(7):2003–2011. doi: 10.1158/1535-7163.MCT-08-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strader CR, Pearce CJ, Oberlies NH. Fingolimod (FTY720): a recently approved multiple sclerosis drug based on a fungal secondary metabolite. J. Nat. Prod. 2011;74(4):900–907. doi: 10.1021/np2000528. [DOI] [PubMed] [Google Scholar]
- 6.El-Elimat T, Zhang X, Jarjoura D, et al. Chemical diversity of metabolites from fungi, cyanobacteria, and plants relative to FDA-approved anticancer agents. ACS Med. Chem. Lett. 2012;3(8):645–649. doi: 10.1021/ml300105s. [DOI] [PMC free article] [PubMed] [Google Scholar]; • First study of the computational analysis of fungal metabolites.
- 7.Ayers S, Ehrmann BM, Adcock AF, et al. Peptaibols from two unidentified fungi of the order Hypocreales with cytotoxic, antibiotic, and anthelmintic activities. J. Pep. Sci. 2012;18(8):500–510. doi: 10.1002/psc.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ayers S, Ehrmann BM, Adcock AF, et al. Thielavin B methyl ester: a cytotoxic benzoate trimer from an unidentified fungus (MSX 55526) from the order Sordariales. Tetrahedron Lett. 2011;52(44):5733–5735. doi: 10.1016/j.tetlet.2011.08.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ayers S, Graf TN, Adcock AF, et al. Resorcylic acid lactones with cytotoxic and NF-kB inhibitory activities and their structure–activity relationships. J. Nat. Prod. 2011;74(5):1126–1131. doi: 10.1021/np200062x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ayers S, Graf TN, Adcock AF, et al. Cytotoxic xanthone-anthraquinone heterodimers from an unidentified fungus of the order Hypocreales (MSX 17022) J. Antibiot. 2012;65:3–8. doi: 10.1038/ja.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ayers S, Graf TN, Adcock AF, et al. Obionin B: an O-pyranonaphthoquinone decaketide from an unidentified fungus (MSX 63619) from the Order Pleosporales. Tetrahedron Lett. 2011;52(40):5128–5230. doi: 10.1016/j.tetlet.2011.07.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El-Elimat T, Figueroa M, Ehrmann BM, Cech NB, Pearce CJ, Oberlies NH. High-resolution MS, MS/MS, and UV database of fungal secondary metabolites as a dereplication protocol for bioactive natural products. J. Nat. Prod. 2013;76(9):1709–1716. doi: 10.1021/np4004307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El-Elimat T, Figueroa M, Raja HA, et al. Waol A, trans-dihydrowaol A, and cis-dihydrowaol A: polyketide-derived gamma-lactones from a Volutella species. Tetrahedron Lett. 2013;54(32):4300–4302. doi: 10.1016/j.tetlet.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El-Elimat T, Figueroa M, Raja HA, et al. Benzoquinones and terphenyl compounds as phosphodiesterase-4B inhibitors from a fungus of the Order Chaetothyriales (MSX 47445) J. Nat. Prod. 2013;76(3):382–387. doi: 10.1021/np300749w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El-Elimat T, Figueroa M, Raja HA, et al. Biosynthetically distinct cytotoxic polyketides from Setophoma terrestris . Eur. J. Org. Chem. 2015;2015(1):109–121. doi: 10.1002/ejoc.201402984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El-Elimat T, Raja HA, Figueroa M, et al. Sorbicillinoid analogs with cytotoxic and selective anti-Aspergillus activities from Scytalidium album . J. Antibiot. 2015;68(3):191–196. doi: 10.1038/ja.2014.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Figueroa M, Graf TN, Ayers S, et al. Cytotoxic epipolythiodioxopiperazine alkaloids from filamentous fungi of the Bionectriaceae. J. Antibiot. 2012;65(11):559–564. doi: 10.1038/ja.2012.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Figueroa M, Raja H, Falkinham JO, et al. Peptaibols, tetramic acid derivatives, isocoumarins, and sesquiterpenes from a Bionectria sp. (MSX 47401) J. Nat. Prod. 2013;76(6):1007–1015. doi: 10.1021/np3008842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sy-Cordero AA, Figueroa M, Raja HA, et al. Spiroscytalin, a new tetramic acid and other metabolites of mixed biogenesis from Scytalidium cuboideum . Tetrahedron. 2015;71(47):8899–8904. doi: 10.1016/j.tet.2015.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sy-Cordero AA, Graf TN, Adcock AF, et al. Cyclodepsipeptides, sesquiterpenoids, and other cytotoxic metabolites from the filamentous fungus Trichothecium sp. (MSX 51320) J. Nat. Prod. 2011;74(10):2137–2142. doi: 10.1021/np2004243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sy-Cordero AA, Graf TN, Wani MC, Kroll DJ, Pearce CJ, Oberlies NH. Dereplication of macrocyclic trichothecenes from extracts of filamentous fungi through UV and NMR profiles. J. Antibiot. 2010;63(9):539–544. doi: 10.1038/ja.2010.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sy-Cordero AA, Pearce CJ, Oberlies NH. Revisiting the enniatins: a review of their isolation, biosynthesis, structure determination, and biological activities. J. Antibiot. 2012;65(11):541–549. doi: 10.1038/ja.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaur A, Raja HA, Darveaux BA, et al. New diketopiperazine dimer from a filamentous fungal isolate of Aspergillus sydowii . Magn. Reson. Chem. 2015;53(8):616–619. doi: 10.1002/mrc.4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burdock GA, Carabin IG, Griffiths JC. The importance of GRAS to the functional food and nutraceutical industries. Toxicology. 2006;221(1):17–27. doi: 10.1016/j.tox.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 25.Feher M, Schmidt JM. Property distributions: differences between drugs, natural products, and molecules from combinatorial chemistry. J. Chem. Inf. Comput. Sci. 2003;43(1):218–227. doi: 10.1021/ci0200467. [DOI] [PubMed] [Google Scholar]
- 26.Fernandez-De Gortari E, Medina-Franco JL. Epigenetic relevant chemical space: a chemoinformatic characterization of inhibitors of DNA methyltransferases. RSC Adv. 2015;5(106):87465–87476. [Google Scholar]
- 27.Singh N, Guha R, Giulianotti MA, Pinilla C, Houghten RA, Medina-Franco JL. Chemoinformatic analysis of combinatorial libraries, drugs, natural products, and molecular libraries small molecule repository. J. Chem. Inf. Model. 2009;49(4):1010–1024. doi: 10.1021/ci800426u. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Chemoinformatic analysis of natural products using multiple structure representations.
- 28.Lucas X, Grüning BA, Bleher S, Günther S. The purchasable chemical space: a detailed picture. J. Chem. Inf. Mod. 2015;55(5):915–924. doi: 10.1021/acs.jcim.5b00116. [DOI] [PubMed] [Google Scholar]
- 29.Molecular operating environment (MOE), version 2014.08, Chemical Computing Group Inc. Montreal, Quebec, Canada: www.chemcomp.com [Google Scholar]
- 30.Medina-Franco JL, Martínez-Mayorga K, Peppard TL, Del Rio A. Chemoinformatic analysis of GRAS (Generally Recognized as Safe) flavor chemicals and natural products. PLoS ONE. 2012;7(11):e50798. doi: 10.1371/journal.pone.0050798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Law V, Knox C, Djoumbou Y, et al. Drugbank 4.0: shedding new light on drug metabolism. Nucl. Acids Res. 2014;42(D1):D1091–D1097. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu F, Shi Z, Qin C, et al. Therapeutic target database update 2012: a resource for facilitating target-oriented drug discovery. Nucleic Acids Res. 2012;40:D1128–D1136. doi: 10.1093/nar/gkr797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Selleckchem. www.selleckchem.com
- 34.Lovering F, Bikker J, Humblet C. Escape from flatland: increasing saturation as an approach to improving clinical success. J. Med. Chem. 2009;52(21):6752–6756. doi: 10.1021/jm901241e. [DOI] [PubMed] [Google Scholar]; •• Comprehensive discussion of the association between molecular complexity and clinical success.
- 35.Clemons PA, Bodycombe NE, Carrinski HA, et al. Small molecules of different origins have distinct distributions of structural complexity that correlate with protein-binding profiles. Proc. Natl Acad. Sci. USA. 2010;107(44):18787–18792. doi: 10.1073/pnas.1012741107. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Experimental assessment of the relationship between molecular complexity and target selectivity.
- 36.Maya ChemTools. www.mayachemtools.org/
- 37.Team RDC. R Foundation for Statistical Computing; Vienna, Austria: 2011. www.gbif.org/resource/81287 [Google Scholar]
- 38.Symyx Software; San Ramon, CA, USA: MACCS structural keys. [Google Scholar]
- 39.Rogers D, Hahn M. Extended-connectivity fingerprints. J. Chem. Inf. Model. 2010;50(5):742–754. doi: 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- 40.Sander T, Freyss J, Von Korff M, Rufener C. Datawarrior: an open-source program for chemistry aware data visualization and analysis. J. Chem. Inf. Model. 2015;55(2):460–473. doi: 10.1021/ci500588j. [DOI] [PubMed] [Google Scholar]
- 41.López-Vallejo F, Giulianotti MA, Houghten RA, Medina-Franco JL. Expanding the medicinally relevant chemical space with compound libraries. Drug Discovery Today. 2012;17(13–14):718–726. doi: 10.1016/j.drudis.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 42.Allu TK, Oprea TI. Rapid evaluation of synthetic and molecular complexity for in silico chemistry. J. Chem. Inf. Model. 2005;45(5):1237–1243. doi: 10.1021/ci0501387. [DOI] [PubMed] [Google Scholar]
- 43.Böttcher T. An additive definition of molecular complexity. J. Chem. Inf. Model. 2016;56(3):462–470. doi: 10.1021/acs.jcim.5b00723. [DOI] [PubMed] [Google Scholar]
- 44.Jaccard P. Etude comparative de la distribution florale dans une portion des alpes et des jura. Bull. Soc. Vaudoise Sci. Nat. 1901;37:547–579. [Google Scholar]
- 45.Medina-Franco JL, Maggiora GM. Molecular similarity analysis. In: Bajorath J, editor. Chemoinformatics for Drug Discovery. John Wiley & Sons; NJ, USA: 2014. pp. 343–399. [Google Scholar]
- 46.Yongye AB, Waddell J, Medina-Franco JL. Molecular scaffold analysis of natural products databases in the public domain. Chem. Biol. Drug Des. 2012;80(5):717–724. doi: 10.1111/cbdd.12011. [DOI] [PubMed] [Google Scholar]
- 47.Medina-Franco JL, Martinez-Mayorga K, Meurice N. Balancing novelty with confined chemical space in modern drug discovery. Expert Opin. Drug Discov. 2014;9(2):151–165. doi: 10.1517/17460441.2014.872624. [DOI] [PubMed] [Google Scholar]
- 48.Bergström CA. In silico predictions of drug solubility and permeability: two rate-limiting barriers to oral drug absorption. Basic Clin. Pharmacol. Toxicol. 2005;96(3):156–161. doi: 10.1111/j.1742-7843.2005.pto960303.x. [DOI] [PubMed] [Google Scholar]
- 49.Ganesan A. The impact of natural products upon modern drug discovery. Curr. Opin. Chem. Biol. 2008;12(3):306–317. doi: 10.1016/j.cbpa.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 50.Medina-Franco JL. Discovery and development of lead compounds from natural sources using computational approaches. In: Mukherjee P, editor. Evidence-Based Validation of Herbal Medicine. Elsevier; Amsterdam, The Netherlands: 2015. pp. 455–475. [Google Scholar]; • Recent review of natural product-based drug discovery driven by computational methods.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.