Abstract
Aims:
To utilize previously reported lead SNPs for low-density lipoprotein cholesterol (LDL-c) levels to find additional loci of importance to statin response, and examine whether genetic predisposition to LDL-c levels associates with differential statin response.
Methods:
We investigated effects on statin response of 59 LDL-c SNPs, by combining summary level statistics from the Global Lipids Genetics and Genomic Investigation of Statin Therapy consortia.
Results:
Lead SNPs for APOE, SORT1 and NPC1L1 were associated with a decreased LDL-c response to statin treatment, as was overall genetic predisposition for increased LDL-c levels as quantified with 59 SNPs, with a 5.4% smaller statin response per standard deviation increase in genetically raised LDL-c levels.
Conclusion:
Genetic predisposition for increased LDL-c level may decrease efficacy of statin therapy.
Keywords: : MR–Egger, pharmacogenetics, statin therapy
HMG-CoA reductase inhibitors, also known as statins, have proven themselves as a highly effective treatment option in the management and prevention of cardiovascular disease, both in research and clinical settings [1,2]. Their effect is thought to primarily result from reducing low-density lipoprotein cholesterol (LDL-c) levels by up to 50% [3], thereby achieving a 20–30% reduction of cardiovascular events. However, substantial interindividual variability exists in the LDL-c response to statins, in part due to genetic factors, which influences their efficacy in reducing the occurrence of major adverse events.
Recently, through the largest pharmacogenomic meta-analysis for differential LDL-c response to statin therapy to date, the Genomic Investigation of Statin Therapy (GIST) consortium identified four loci (APOE, LPA, SORT1/CELSR2/PSRC1 and SLCO1B1) at a genome-wide significant level, whose effect on statin response was independent of off-treatment LDL-c levels [4]. With the exception of SLCO1B1, these loci have previously been independently reported to associate with LDL-c levels by the Global Lipids Genetics consortium (GLGC) [5]. As loci associated with LDL-c homeostasis are strong mechanistic candidates for differential LDL-c response to statin therapy, we performed a lookup of the previously reported lead SNPs for loci associated with LDL-c levels by the GLGC in the GIST consortium, to examine whether additional loci of importance to differential LDL-c statin response could be identified. Furthermore, we examined whether overall genetic predisposition to higher LDL-c levels (i.e., having more alleles associated with higher LDL-c levels) is associated with differential LDL-c response to statins, by combining summary level statistics from our GIST consortium with publicly available data from the GLGC for all lead SNPs through an inverse-variance weighted (IVW) approach.
Methods
Selection of SNPs associated with LDL-c levels
In the most recent and largest genome-wide association study (GWAS) for blood lipid levels, which examined up to 188,577 European-ancestry individuals, 157 nearly independent loci (r2 < 0.10) were found to associate with lipid levels at p-values lower than 5 × 10-8 [5]. Of the reported 157 lead SNPs, 60 were associated with LDL-c levels (Supplementary Table 1). Summary level data of the associations of these 60 lead SNPs with LDL-c levels were downloaded from the University of Michigan GLGC webpage [6]. Effects on lipid levels were reported in standard deviations. We excluded rs9411489 (ABO) from our analyses, as the genotype could not be imputed in our populations, and therefore included the remaining 59 lead SNPs in our analyses. To further isolate the effects on LDL-c levels from those of other lipids, we repeated all analyses with a restricted SNP list, excluding the 17 variants that also associated with either high-density lipoprotein cholesterol (HDL-c) or triglycerides (TG) levels at a genome-wide significant level. Of these, five associated solely with HDL-c, four solely with TG and eight with both lipid traits. As LDL-c is closely linked to total cholesterol, we did not exclude variants that also associated with total cholesterol at a genome-wide significant level. The restricted list therefore included the remaining 42 LDL-c-specific SNPs.
Description of pharmacogenetic meta-analysis
The GIST consortium included six randomized controlled statin trials (ASCOT, CARDS, CAP, PRINCE, PROSPER and TNT) and ten prospective, population-based studies (AGES, ARIC, BioVU, CHS, FHS, GoDARTS I, GoDARTS II, Health ABC, HVH and MESA) for the first stage, comprised of up to 18,596 statin recipients. In addition, 246 SNPs with p < 5 × 10-4 were further investigated in three additional studies (HPS, JUPITER, Rotterdam Study), contributing up to 22,318 additional statin-treated subjects to the meta-analysis. Of the 59 lead SNPs for LDL-c levels reported by the GLGC, only one (rs4420638, APOE) was included among these 246 SNPs. The GWAS was performed on the difference between natural log-transformed on- and off-treatment LDL-c levels, adjusting for the natural log-transformed off-treatment LDL-c level to control for possible mediation through off-treatment genetic effects. The beta of the corresponding regression therefore represents the fraction of differential LDL lowering in carriers versus noncarriers of each SNP. For observational studies, this meant that subjects with missing on- or off-treatment measurements were excluded, with the exception of the GoDARTS cohorts for which off-treatment LDL-c levels were imputed. In addition, analyses in the observational studies were, if available, additionally adjusted for statin dose through the use of the natural logarithm of the equivalent dose taken from the literature. Details on included studies, genotyping and GWAS analyses have been described previously [4].
Lookup of single SNPs
We performed a lookup of all 59 candidate LDL-c markers within the pharmacogenetic meta-analysis performed by the GIST consortium, assessing their effect on differential LDL-c response to statin therapy adjusted for off-treatment LDL-c values. Adjusted unstandardized beta-coefficients are given for the LDL-c-increasing alleles reported by the GLGC. Multiple testing was taken into account by means of a Bonferroni-corrected p-value threshold of 8.5 × 10-4 (i.e., 0.05/59).
Summary data methods for overall effect of LDL-c predisposition
Next, we investigated whether overall genetic predisposition for LDL-c levels was associated with statin response, making use of summary level data from both the GLGC and GIST consortia. All analyses were carried out separately for the full (n = 59) and restricted (n = 42) SNP lists. Analogous to pooling estimates from different studies in conventional meta-analysis using the IVW method, we pooled the causal estimates from the different genetic variants, defined as the ratio of each SNP's per-allele effect on response to statin therapy to its per-allele effect on LDL-c levels. The average of these ratio estimates was weighted by the inverse of the variance of the per-allele effect on response to statin therapy and can be visualized as a regression line constrained to pass through the origin [7,8]. As this approach may be biased by the inclusion of genetic variants violating the underlying assumptions of instrumental variable (IV) methods [9], most notably by the presence of unbalanced pleiotropic effects on phenotypes other than LDL-c, we performed two additional analyses that should be considered as sensitivity analyses for Mendelian randomization (MR) investigations with multiple genetic variants [10].
We first employed the recently published MR–Egger method [11] that provides a formal test of the presence of directional (i.e., unbalanced) pleiotropy from separate genetic variants by introducing an intercept term to the IVW method and determining whether this term deviates significantly from zero. Based on the Egger test [12], which assesses the presence of small study bias in meta-analysis, this intercept term can be interpreted as the average pleiotropic effect across the genetic variants. After taking these effects into account, the Egger-regression slope reflects the strength of any residual dose-response relationship. Under the assumption that the strength of the association of each variant with LDL-c levels is independent of the pleiotropic effects of the variant (i.e., not via LDL-c), MR–Egger regression gives a valid causal effect estimate even when all the genetic variants are invalid IVs [11].
Second, we calculated the weighted median estimator, defined as the 50% weighted percentile of the distribution of causal estimates given weights proportional to the inverse of their variance [10]. As the median of any distribution is less susceptible to outliers, this method provides a consistent causal estimate under the assumption that over 50% of the weight in the analysis is due to valid instruments. We also provide the penalized weighted median estimate, which severely limits the contribution of heterogeneous (i.e., outlying) variants, which are more likely to represent invalid IVs. This penalty is based on the heterogeneity between estimates as quantified by Cochran's Q-statistic. We considered p-values of 0.05 or smaller statistically significant for these summary data methods.
Finally, to examine whether the use of epidemiological cohort data by the GIST consortium might have introduced imprecision to the causal estimates, we repeated the summary data methods while solely including the data from the randomized controlled trials participating in the first-stage GIST meta-analysis. All analyses were performed with R software version 3.1.1. [13], utilizing the R code provided by the corresponding methodology papers on MR–Egger and median-based methods [10,11].
Results
Lookup of single SNPs
After correction for multiple testing, three SNPs were found to have attained a statistically significant association with LDL-c response to statins (all p < 8.5 × 10-4, Table 1). The results indicate that carriers of these SNPs have a smaller LDL-c response to statin therapy when compared with noncarriers. The magnitudes of these per-allele proportional decreases were 2.5% (APOE, 95% CI: 1.8–3.1), 1.5% (SORT1, 95% CI: 0.9–2.1) and 1.8% (NPC1L1, 95% CI: 0.8–2.7), respectively. When restricting the SNP list to those 42 variants primarily associated with LDL-c, which did not include the lead SNPs for APOE and SORT1, NPC1L1 was the sole statistically significant finding (p = 2.1 × 10-4), also after adjusting the Bonferroni-corrected p-value threshold to 1.2 × 10-3 (i.e., 0.05/42).
Table 1. . Candidate markers significantly associated with low-density lipoprotein cholesterol response to statin therapy.
| SNP | Locus | Chr |
Global Lipids Genetics Consortium |
Genomic Investigation of Statin Therapy consortium |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| EA | EA freq. (1000G) | Beta (SE)† | p-value | Other lipids | EA freq. | Beta (SE)‡ | p-value | |||
| rs4420638 |
APOE |
19 |
G |
0.19 |
0.225 (0.008) |
2 × 10-178 |
HDL-c, triglycerides, TC |
0.17 |
0.025 (0.003) |
3.9 × 10-15 |
| rs629301 |
SORT1 |
1 |
T |
0.79 |
0.167 (0.005) |
5 × 10-241 |
HDL-c, TC |
0.77 |
0.015 (0.003) |
9.4 × 10-7 |
| rs2072183 | NPC1L1 | 7 | C | 0.24 | 0.039 (0.004) | 7 × 10-16 | TC | 0.24 | 0.018 (0.005) | 2.1 × 10-4 |
Listed variants are those with p-values smaller than the Bonferroni-corrected threshold of 8.5 × 10-4 (i.e., 0.05/59) for the association with statin response.
†Beta for effect on low-density lipoprotein cholesterol (LDL-c) levels, in standard deviations.
‡Beta for difference between the natural log-transformed on- and off-treatment LDL-c levels adjusted for natural log-transformed off-treatment LDL-c, age-, sex- and study-specific covariates. A negative beta indicates a better statin response, a positive beta indicates a worse statin response.
Chr: Chromosome; EA: Effect allele for increased low-density lipoprotein cholesterol levels from the Global Lipids Genetics consortium; HDL-c: High-density lipoprotein cholesterol; freq: Frequency; SE: Standard error; TC: Total cholesterol.
Summary results for overall effect of LDL-c predisposition
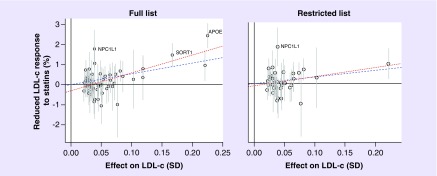
As shown in Figure 1 and Table 2, the conventional IVW method revealed strong evidence that overall genetic predisposition for higher LDL-c levels associates with a decreased LDL-c response to statin therapy. For the full list (all LDL-c-associated variants), this amounted to a 5.4% (95% CI: 4.2–6.7; p = 8.4 × 10-12) smaller response per standard deviation increase in genetically raised LDL-c levels. Despite the effect being slightly reduced, the direction of the association was similar for the restricted list (excluding HDL-c- and TG-associated variants), showing a 3.2% (95% CI: 1.2–5.1; p = 2.1 × 10-3) decreased response per standard deviation increase in genetically raised LDL-c levels.
Figure 1. . Scatter plots of the genetic associations with low-density lipoprotein cholesterol against genetic associations with differential low-density lipoprotein cholesterol response to statin therapy, both plotted as per-allele effects.
In addition, 95% CIs are presented for the genetic associations with statin response. The dashed and dotted line correspond to the inverse-variance weighted and Mendelian randomization–Egger estimators, respectively, and are shown for the full (59 SNPs) and restricted (42 SNPs) lists with a positive slope reflecting a worse statin response.
LDL-c: Low-density lipoprotein cholesterol; SD: Standard deviation.
Table 2. . Inverse-variance weighted, Mendelian randomization–Egger and median-based estimators for the association between low-density lipoprotein cholesterol levels and proportional low-density lipoprotein cholesterol response to statin therapy.
| Analysis method |
Full list of 59 variants |
42 variants primarily associated with LDL-c |
||
|---|---|---|---|---|
| Beta (SE) | p-value | Beta (SE) | p-value | |
| Inverse-variance weighted |
0.054 (0.006) |
8.4 × 10-12* |
0.032 (0.010) |
2.1 × 10-3* |
| MR–Egger: slope |
0.089 (0.010) |
1.0 × 10-11* |
0.044 (0.018) |
1.7 × 10-2* |
| MR–Egger: intercept |
-0.003 (0.001) |
7.6 × 10-5* |
-0.001 (0.001) |
0.40 |
| Weighted median |
0.070 (0.011) |
4.2 × 10-8* |
0.043 (0.015) |
6.4 × 10-3* |
| Penalized weighted median | 0.051 (0.011) | 2.8 × 10-5* | 0.043 (0.015) | 6.8 × 10-3* |
Beta's (SE) given as differential LDL-c response to statin therapy per standard deviation increase in LDL-c levels. The MR–Egger intercept term provides a formal test of directional pleiotropy.
*Statistically significant results, using a p-value threshold of 0.05.
LDL-c: Low-density lipoprotein cholesterol; MR: Mendelian randomization; SE: Standard error.
Results from both sensitivity analyses were largely consistent with those seen for the IVW approach, with regard to magnitude and direction of the association, especially for the restricted SNP list (Table 2). The MR–Egger results indicated the presence of unbalanced pleiotropy for the full list of variants, as the intercept deviated significantly from zero (p = 7.6 × 10-5), which was not present when analyses were restricted to those variants primarily associated with LDL-c (p = 0.40). Though inconclusive, further attempts to disentangle the influence of HDL-c- and TG-associated variants suggested that the variants associated with HDL-c were especially influential with regard to possible unbalanced pleiotropic effects on statin response, as their exclusion led to the greatest decrease in the MR–Egger intercept term (Supplementary Table 2). Of the median-based methods, the penalized estimator was the most consistent with the IVW estimate, for both SNP lists. As shown in Supplementary Table 3, there was large homogeneity between the causal estimates obtained from the full sample and when restricting the analyses to the data from the randomized controlled trials participating in the first-stage GIST meta-analysis.
Discussion
Within the present study, we aimed to examine whether additional loci of importance to LDL-c response to statin therapy could be identified by focusing our efforts on previously reported lead SNPs explaining variation in LDL-c levels. In addition to reconfirming the previously described associations of APOE and SORT1 with LDL-c response to statin therapy, we found suggestive evidence that NPC1L1 is of importance to statin pharmacogenetics. Of note, our previously reported association of LPA with statin response was not among these results, reflecting the different lead SNP reported by the GLGC, which also explains why the association with statin response was not genome-wide significant for SORT1. Consistent with the results for the individual lead SNPs, we found strong evidence that overall genetic predisposition for higher LDL-c levels is associated with a decreased LDL-c statin response, and robustly quantified this association using summary level data from the largest and most recent GWA studies on lipid levels and LDL-c response to statin therapy. In addition, MR–Egger and median-based estimators showed largely consistent results, both in direction and magnitude, thereby strengthening the findings of the IVW approach.
Localized to gastrointestinal tract epithelial cells as well as hepatocytes, the NPC1L1 protein is a key regulator of cholesterol absorption [14] and is the drug target of ezetimibe [15]. Shown to associate with interindividual variation in response to ezetimibe treatment [16,17], genetic variation in NPC1L1 has also been previously linked to LDL-c response to statin therapy in smaller studies. In 37 men with central obesity, Chan et al. found that subjects with the NPC1L1 2/2 haplotype had a greater reduction in LDL-c levels than non-2/2 haplotype subjects, independent of their higher baseline LDL-c levels [18]. Moreover, in the PROSPER trial, the NPC1L1 -133A>G variant was found to associate with greater 6-month change in lipid levels in pravastatin-treated individuals and also with higher baseline LDL-c levels, which were not adjusted for in the analyses [19].
In contrast, our findings are unlikely to be explained by differences in off-treatment LDL-c levels, as these were statistically accounted for in the GIST meta-analysis. Rather, the genetic associations with LDL-c levels reflect lifelong effects on lipid metabolism, which we now show may influence the efficacy of clinical interventions later in life. Unfortunately, our use of summary level data precludes providing more detailed mechanistic insights, though there exists some evidence that statin therapy efficacy interacts with cholesterol synthesis and absorption, possibly in part through changes in intestinal expression of NPC1L1 [20,21].
While the MR–Egger test did not show evidence for directional pleiotropy after excluding variants associated with HDL-c or TG at a genome-wide significant level, it is possible that the remaining variants are not solely of importance to LDL-c homeostasis, as meaningful subthreshold associations may exist for HDL-c or TG. Similarly, we cannot be certain that the associations with HDL-c and TG of the excluded genetic variants reflected true biological pleiotropy, or merely downstream effects of LDL-c on other phenotypic traits (lipid or otherwise), which are specifically the effects of interest in MR investigations [22]. However, by creating a restricted list we attempted to isolate variants more specific to LDL-c levels, as has previously been done when constructing genetic risk scores consisting of large numbers of genetic variants [23]. In line with this, the consistency of the different methods for the restricted score indicates that this score is less likely to contain invalid instruments. Furthermore, the relatively large difference in mean estimates between the MR–Egger and median-weighted methods for the full list of variants possibly reflects violation of MR–Egger's underlying assumptions, as variants associated with LDL-c levels might be proportionally associated with HDL-c and TG levels.
As we included summary level data from partially overlapping data sources, our findings may have been influenced by weak IV bias [24]. More specifically, of the ten prospective, population-based studies that contributed to the first-stage meta-analysis of GIST, six (AGES, ARIC, CHS, FHS, Go-DARTS I and Go-DARTS II) also contributed to the GLGC meta-analysis. With the exception of rs4420638 (APOE), which was validated in additional populations in the second-stage meta-analysis of GIST, this means that up to 43% of GIST participants included in the first-stage meta-analysis were possibly also included in the GLGC analyses. However, the median F-statistic of our instruments for LDL-c levels was 58.35 (interquartile range: 42.51–118.59), making it unlikely to have substantially influenced our results, as instruments with F-statistics over 10 are generally considered sufficiently strong [25]. The homogeneity between the causal estimates obtained from the full GIST sample and those generated when solely including data from the GIST randomized controlled trials strengthens this claim, as these trials were not included in the GLGC meta-analysis.
Conclusion
In summary, we found that three lead SNPs (for APOE, SORT1 and NPC1L1) were associated with smaller LDL-c response to statin treatment after taking multiple testing into account, thereby identifying one new locus of importance to statin response, namely NPC1L1. In addition, our findings indicate that individuals with overall genetic predisposition for high LDL-c levels are less likely to respond well to statins.
Future perspective
To date, pharmacogenetic research on statin therapy has identified genetic variants with only modest effect sizes and therefore limited clinical utility [26]. Recently, Leusink et al. generated a genetic risk score based on previously reported genetic variants of importance to statin response in ABCG2, LPA and APOE. However, the small effect size (roughly 2% of average LDL-c reduction) suggests that the applicability of this score in clinical practice would be limited [26]. While the main aim of our study was to examine whether overall predisposition to LDL-c level associates with statin response, our results suggest that risk stratification based on a LDL-c genetic risk score might identify individuals most likely to benefit from combination therapy of statin and nonstatin lipid-lowering medication, as genetic predisposition to higher LDL-c levels may not affect their efficacy to the same degree. However, as the various summary method effect estimates observed in our study varied between 3 and 9% reduced LDL-c response to statin therapy per standard deviation increase in genetically raised LDL-c levels, clinical utility will likely be limited. If genetic information becomes available, large experimental studies such as the recently completed IMPROVE-IT trial [27] would be most suited to determine possible clinical significance. In addition, pharmacogenetic studies of nonstatin LDL-lowering therapies should also consider examining the role of genetic predisposition for higher LDL-c levels. Finally, it would be of great interest for future studies to examine whether NPC1L1-dependent compensatory mechanisms to lipid-lowering treatment exist, which could add to the rationale behind combination therapy with ezetimibe.
Executive summary.
Background
There exists substantial interindividual variation in low-density lipoprotein cholesterol (LDL-c) response to statin treatment, in part due to genetic factors. Several genetic loci have been found to associate with differential LDL-c response to statins, independent of off-treatment LDL-c levels.
The majority of these loci have additionally been found to associate with LDL-c levels. LDL-c level-associated loci may therefore represent strong candidates for pharmacogenetic studies on statin therapy.
Methods
To identify additional loci of importance to statin response, we performed a lookup of 59 lead SNPs for LDL-c levels in the pharmacogenetic meta-analysis of the Genomic Investigation of Statin Therapy consortium.
We further examined whether overall genetic predisposition for higher LDL-c levels associates with statin response, by combining summary statistics from the Global Lipids Genetics and Genomic Investigation of Statin Therapy consortia for 59 lead SNPs for LDL-c levels from the Global Lipids Genetics consortium, through an inverse-variance weighted approach. Mendelian randomization–Egger regression and median-based methods were then performed as sensitivity analyses.
Results: main findings
Lead SNPs for APOE, SORT1 and NPC1L1 were associated with diminished statin response, as was overall genetic predisposition for increased LDL-c level.
Supplementary Material
Acknowledgements
The authors thank the Global lipids Genetic consortium for making the result files of their meta-analysis available. In addition, the authors wish to express our gratitude to all studies participating in the Genomic Investigation of Statin Therapy consortium.
Footnotes
Financial & competing interests disclosure
BM Psaty serves on the Data and Safety Monitoring Board of a clinical trial funded by the manufacturer, and on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. DI Chasman received research support for independent genetic analysis in JUPITER from AstraZeneca. JW Jukema is an Established Clinical Investigator of The Netherlands Heart Foundation (grant no. 2001 D 032). RM Krauss serves on the Merck Global Atherosclerosis Advisory Board. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Cholesterol Treatment Trialist’ (CTT) Collaborators. Mihaylova B, Emberson J, et al. The effects of lowering LDL cholesterol with statin therapy in people at low risk of vascular disease: meta-analysis of individual data from 27 randomised trials. Lancet. 2012;380(9841):581–590. doi: 10.1016/S0140-6736(12)60367-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cholesterol Treatment Trialists’ (CTT) Collaboration. Baigent C, Blackwell L, et al. Efficacy and safety of more intensive lowering of LDL cholesterol: a meta-analysis of data from 170,000 participants in 26 randomised trials. Lancet. 2010;376(9753):1670–1681. doi: 10.1016/S0140-6736(10)61350-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davidson MH, Toth PP. Comparative effects of lipid-lowering therapies. Prog. Cardiovasc. Dis. 2004;47(2):73–104. doi: 10.1016/j.pcad.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Postmus I, Trompet S, Deshmukh HA, et al. Pharmacogenetic meta-analysis of genome-wide association studies of LDL cholesterol response to statins. Nat. Commun. 2014;5:5068. doi: 10.1038/ncomms6068. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Largest pharmacogenetic meta-analysis on lipoprotein cholesterol response to statin therapy.
- 5.Global Lipids Genetics Consortium. Willer CJ, Schmidt EM, et al. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013;45(11):1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Largest genome-wide association study on blood lipid levels.
- 6.Center for Statistical Genetics, University of Michigan. http://csg.sph.umich.edu//abecasis/public/lipids2013/
- 7.Johnson T. Efficient calculation for multi-SNP genetic risk scores. 2012. https://cran.r-project.org/web/packages/gtx/vignettes/ashg2012.pdf The Comprehensive R Archive Network.
- 8.Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 2013;37(7):658–665. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenland S. An introduction to instrumental variables for epidemiologists. Int. J. Epidemiol. 2000;29(4):722–729. doi: 10.1093/ije/29.4.722. [DOI] [PubMed] [Google Scholar]
- 10.Bowden J, Davey Smith G, Haycock PC, et al. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 2016;40(4):304–314. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Methodology paper on median-based methods.
- 11.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015;44(2):512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Methodology paper on the Mendelian randomization–Egger regression method.
- 12.Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.R Core Team. R Foundation for Statistical Computing; Vienna, Austria: 2016. R: A language and environment for statistical computing.www.r-project.org/ [Google Scholar]
- 14.Jia L, Betters JL, Yu L. Niemann-pick C1-like 1 (NPC1L1) protein in intestinal and hepatic cholesterol transport. Annu. Rev. Physiol. 2011;73:239–259. doi: 10.1146/annurev-physiol-012110-142233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Calvo M, Lisnock J, Bull HG, et al. The target of ezetimibe is Niemann-Pick C1-Like 1 (NPC1L1) Proc. Natl Acad. Sci. USA. 2005;102(23):8132–8137. doi: 10.1073/pnas.0500269102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hegele RA, Guy J, Ban MR, et al. NPC1L1 haplotype is associated with inter-individual variation in plasma low-density lipoprotein response to ezetimibe. Lipids Health Dis. 2005;4:16. doi: 10.1186/1476-511X-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simon JS, Karnoub MC, Devlin DJ, et al. Sequence variation in NPC1L1 and association with improved LDL-cholesterol lowering in response to ezetimibe treatment. Genomics. 2005;86(6):648–656. doi: 10.1016/j.ygeno.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 18.Chan DC, Watts GF, Wang J, et al. Variation in Niemann-Pick C1-like 1 gene as a determinant of apolipoprotein B-100 kinetics and response to statin therapy in centrally obese men. Clin. Endocrinol. (Oxf.) 2008;69(1):45–51. doi: 10.1111/j.1365-2265.2007.03144.x. [DOI] [PubMed] [Google Scholar]
- 19.Polisecki E, Peter I, Simon JS, et al. Genetic variation at the NPC1L1 gene locus, plasma lipoproteins, and heart disease risk in the elderly. J. Lipid. Res. 2010;51(5):1201–1207. doi: 10.1194/jlr.P001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qi Y, Liu J, Ma C, et al. Association between cholesterol synthesis/absorption markers and effects of cholesterol lowering by atorvastatin among patients with high risk of coronary heart disease. J. Lipid. Res. 2013;54(11):3189–3197. doi: 10.1194/jlr.P040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tremblay AJ, Lamarche B, Lemelin V, et al. Atorvastatin increases intestinal expression of NPC1L1 in hyperlipidemic men. J. Lipid Res. 2011;52(3):558–565. doi: 10.1194/jlr.M011080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 2014;23(R1):R89–R98. doi: 10.1093/hmg/ddu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holmes MV, Asselbergs FW, Palmer TM, et al. Mendelian randomization of blood lipids for coronary heart disease. Eur. Heart J. 2014;36(9):539–550. doi: 10.1093/eurheartj/eht571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pierce BL, Burgess S. Efficient design for Mendelian randomization studies: subsample and 2-sample instrumental variable estimators. Am. J. Epidemiol. 2013;178(7):1177–1184. doi: 10.1093/aje/kwt084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stock JH, Yogo M. Testing for weak instruments in linear IV regression. In: Andrews DWK, Stock JH, editors. Identification and Inference for Econometric Models. Cambridge University Press; NY, USA: 2005. pp. 80–108. [Google Scholar]
- 26.Leusink M, Maitland-van der Zee AH, Ding B, et al. A genetic risk score is associated with statin-induced low-density lipoprotein cholesterol lowering. Pharmacogenomics. 2016;17(6):583–591. doi: 10.2217/pgs.16.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cannon CP, Blazing MA, Giugliano RP, et al. Ezetimibe added to statin therapy after acute coronary syndromes. N. Engl. J. Med. 2015;372(25):2387–2397. doi: 10.1056/NEJMoa1410489. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.