Abstract
Many cancer cells exhibit an altered metabolic phenotype, in which glutamine consumption is upregulated relative to healthy cells. This metabolic reprogramming often depends upon mitochondrial glutaminase activity, which converts glutamine to glutamate, a key precursor for biosynthetic and bioenergetic processes. Two isozymes of glutaminase exist, a kidney-type (GLS) and a liver-type enzyme (GLS2 or LGA). While a majority of studies have focused on GLS, here we summarize key findings on both glutaminases, describing their structure and function, their roles in cancer and pharmacological approaches to inhibiting their activities.
Keywords: : 968, BPTES, cancer, CB-839, glutaminase, inhibitor, metabolism, Warburg effect
Hanahan and Weinberg famously described the ‘Hallmarks of Cancer’ in 2000, listing six discrete capabilities shared by the majority of cancer cells which together could be thought of as ‘defining’ cancer [1]. In 2011, two additional hallmarks were added to this list, one of which is the deregulation of cellular bioenergetics [2]. In fact, the reprogramming of cellular metabolism was among the first described changes in transformed (cancerous) cells, reported nearly a century ago by Otto Warburg [3,4]. Warburg observed that cancer cells engage in the fermentation of glucose to lactic acid, rather than respiration to yield carbon dioxide, even under aerobic conditions (aerobic glycolysis, or the ‘Warburg effect’). Although Warburg proposed that altered metabolism was the cause of cancer, it is now known that in most cases it is a supporting outcome of transformation driven by other mechanisms. A key function of metabolic reprogramming in cancer cells is to provide biosynthetic intermediates to support proliferation [5,6]. In the glycolytic pathway, conversion of phosphoenolpyruvate to pyruvate is slowed by expression of the low activity pyruvate kinase M2 (PKM2) isoform. Consequently glycolytic intermediates accumulate, and can be diverted into biosynthetic pathways [7]. Notably, other rapidly proliferating cells (e.g., activated T-cells) also exhibit the Warburg effect [8–10].
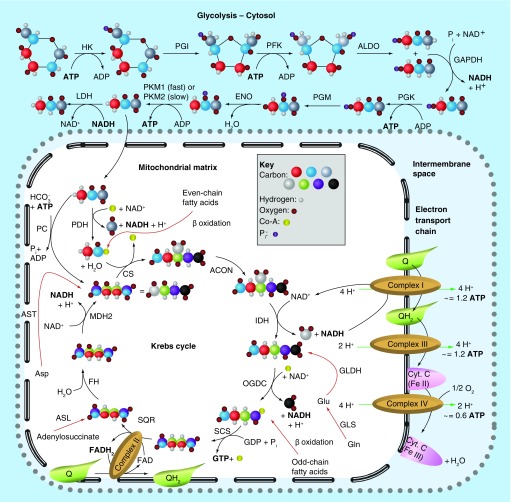
A central hub of cellular metabolism is the Krebs cycle [11]. It is initiated when one molecule of acetyl-CoA is synthesized from pyruvate; the acetyl-CoA subsequently fuels oxidative phosphorylation during one turn of the cycle. Combined, these steps generate one molecule of GTP, along with one FADH2 and four molecules of NADH, which may be used to generate ATP via the electron transport chain and ATP synthase (Figure 1). Oxidative phosphorylation efficiently generates ATP from glucose, with each glucose molecule yielding a theoretical maximum of 38 molecules of ATP or GTP, although consideration of alternate proton fates, such as membrane leakage or use in transporting pyruvate or phosphate into the matrix, reduces this to an estimated 29–30 molecules [12]. When oxygen is limited, cells instead ferment glucose to lactic acid. Fermentation is less efficient than oxidative phosphorylation in terms of ATP generation, such that only two net molecules of ATP are generated per molecule of glucose, during glycolysis [6]. A major carbon input for the Krebs cycle is the glucose metabolite pyruvate. However, the Krebs cycle continues to turn in most cancer cells, even when there is a loss of input from glucose-derived carbon [13,14]. This is accomplished by the utilization of alternate anaplerotic inputs, including carbon derived from glutamine, aspartate, adenylosuccinate and fatty acids (Figure 1, red arrows). The key enzyme which hydrolyzes glutamine to glutamate is glutaminase (EC 3.5.1.2, l-glutamine amidohydrolase), which is expressed in mammalian cells as two isozymes, kidney-type glutaminase (GLS) and liver-type glutaminase (LGA or GLS2). Of the two, GLS has recently been intensively studied, as it has been linked to the progression of a number of cancers, and thus targeted by extensive drug discovery efforts. LGA, in contrast, has alternately been suggested to be a tumor promoter or a tumor suppressor, and has been the subject of little drug discovery research. In this review, we examine the disparity in research efforts devoted to these enzymes. We focus on biochemical structure–function characterizations of GLS and LGA, as well as on studies implicating these enzymes in cancer progression. Finally, we summarize the drug discovery efforts targeting each protein.
Figure 1. . Schematic overview of cellular energetics.
Three primary metabolic pathways supply cells with biosynthetic intermediates and ATP: glycolysis (top), the Krebs cycle (left) and the electron transport chain (right and bottom). Glycolysis converts glucose to pyruvate, and consumes two molecules of ATP in the process, but splits glucose into two molecules (top right), further conversions of which eventually produce two ATPs each, for a net total of two ATPs. Two NADHs are also generated, which can fuel the electron transport chain. Pyruvate then enters the mitochondria where it is used to generate acetyl-CoA for the Krebs cycle, or exits the cell as lactate (regenerating NAD+ and allowing glycolysis to continue). The Krebs cycle turns once per acetyl-CoA molecule fed in, and so turns twice per molecule of glucose. Each turn of the Krebs cycle produces four molecules of NADH, one molecule of GTP and one molecule of FADH2. NADH enters the electron transport chain at Complex 1, which reduces membrane soluble ubiquinone (Q) to ubiquinol (QH2), and exports four protons to the intermembrane space (IMS). Complex III transfers electrons from QH2 to membrane-anchored cytochrome C, reducing the bound heme and simultaneously exports a further four protons to the IMS. Finally, Complex IV uses molecular oxygen to re-oxidize cytochrome C, and exports another 2 protons to the IMS per unit of NADH. Complex II (i.e., SQR) generates FADH2 to reduce Q to QH2, but without any proton transport. Every ten protons exported can ideally generate three units of ATP via ATP synthase (not shown), for a total of 34 ATP molecules (∼3 per NADH, ∼2 per FADH2 generated during cellular metabolism) and two GTP molecules per glucose molecule, but proton leakage and the use of protons to transport pyruvate and phosphate into the matrix reduce this number to approximately 29–31 ATP/GTP molecules formed per glucose molecule. The Krebs cycle can be supplied with intermediates from five sources in addition to glucose, shown with red arrows. Carbon atoms are represented by large spheres of different colors, and are given mixed colors to represent the point at which molecules gain symmetry and unique carbons become mixed; none of the pyruvate carbon atoms used to form citrate leave as CO2 during a single turn of the Krebs cycle. Hydrogens are shown only on carbon, to assist in tracing oxidation states. Anaplerotic reactions are shown as red arrows, and proton transport is shown by green arrows.
ACON: Aconitase; ALDO: Aldolase; ASL: Adenylosuccinate lyase; AST: Aspartate transaminase; CS: Citrate Synthase; ENO: Enolase; FH: Fumarase; GAPDH: Glyceraldehyde phosphate dehydrogenase; GLDH: Glutamate dehydrogenase; GLS: Glutaminase; HK: Hexokinase; IDH: Isocitrade dehydrogenase; IMS: Intermembrane space; LDH: Lactate dehydrogenase; MDH2: Malate dehydrogenase; OGDC: α-Ketoglutarate dehydrogenase; PC: Pyruvate carboxylase; PDH: Pyruvate dehydrogenase; PFK: Phosphofructokinase; PGI: Phosphoglucose isomerase; PGK: Phosphoglycerate kinase; PGM: Phosphoglycerate mutase; PKM1/PKM2: Pyruvate Kinase M1/M2; Q: Ubiquinone; QH2: Ubiquinol; SCS: Succinyl coenzyme A synthetase; SQR: Succinate dehydrogenase.
Glutaminases: early characterization
The existence of glutaminase was first proposed by Krebs, in 1935 [15]. Prior to this, it was known that glutamine could be hydrolyzed to glutamate through a mechanism similar to that used by asparaginase (ASPG) to hydrolyze asparagine to aspartate [16,17]. However, Krebs demonstrated that ASPG was not solely responsible for catalyzing the hydrolysis of glutamine. He then determined that different types of glutaminase were present in different tissues, based on their optimal activity at distinct pH values (the liver extracts being optimally active at higher pH values than the kidney extracts), and their ability to be inhibited by the product, glutamic acid (the catalytic rate of the kidney extracts was strongly inhibited by the addition of glutamic acid, while that of the liver extracts was barely affected). In the late 1940s, researchers at the National Cancer Institute detailed the mechanisms by which glutaminase enzymes become activated [18–21]. In particular, they determined that inorganic anions, such as phosphate, arsenate or sulfate, enhance glutaminase activity [19].
These early studies involved partially purified enzyme; over the following decades, a number of efforts were undertaken to obtain more pure preparations. Efforts pertaining to the kidney-type enzyme culminated in 1970, when Kvamme and colleagues reported a 10,000-fold purification of kidney-type glutaminase, via a series of centrifugation steps exploiting the tendency of the enzyme to transition between 150 and 2000 kDa molecular mass forms when placed in appropriate buffers [22–27]. It was not until 1987 that the first highly purified preparations of liver-type glutaminase were reported simultaneously by Heini and coworkers (400-fold purification) and by Smith and Watford (600-fold purification) [28,29]. Both investigators described the need to include protease inhibitors in their purification media, and Heini remarked upon the inherent instability of liver-type glutaminase compared with the kidney-type enzyme.
Glutaminases: structure & function
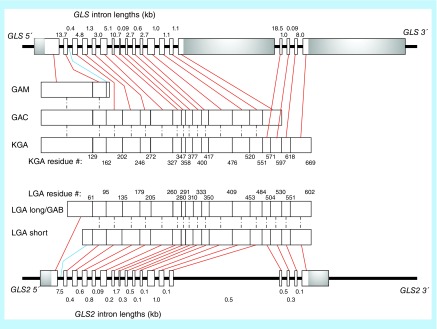
The ‘kidney-type’ and ‘liver-type’ glutaminases (GLS and LGA, respectively) are derived from distinct genes. GLS is encoded by the gene GLS, located on human chromosome 2, and exists as different splice variants (Figure 2). The first is a longer form called kidney glutaminase (KGA); its sequence in humans was deduced from cDNA isolated simultaneously in a massive cDNA sequencing effort and a more targeted study searching for analogs of the already discovered rat and pig KGA [30,31]. The latter effort also reported a shorter form called glutaminase C (GAC), which is identical to KGA except for the C-terminal region, and a third variant, GAM, which is significantly shorter than KGA or GAC, and exhibits no measurable catalytic activity [31]. Porter and colleagues described the gene GLS, which consists of 19 exons over 82 kb, and demonstrated the gene splicing which leads to GAC (exons 1–15) and KGA (exons 1–14 + 16–19) [32]. GAM appears to be formed by inclusion of intron 2–3 in the mRNA, and differs from KGA/GAC starting at residue 162. The GAM C-terminal sequence is ‘VSFYIFLS’, which is encoded by intron 2–3. Given the rarity of intron inclusion as a form of alternative splicing in mammals, it remains unclear if GAM was a result of a genetic defect in the cells from which its cDNA was isolated [33].
Figure 2. . Glutaminase gene and protein map.
The GLS gene is shown at the top, and the GLS2 gene is shown at the bottom. Proteins are drawn in the middle, with the shortest splice variants closest to the gene encoding them. Exons and protein segments encoded by them are represented by boxes, and are drawn to scale. Untranslated regions of 5′ and 3′ exons are shaded gray. Introns are represented by black lines and are not to scale; intron size is reported above or below each line. Protein segments are linked to encoding exons with red lines, and encoding introns with blue lines. Identical protein segments among splice variants are linked with dashed lines. The residue number at the end of each protein segment is labeled for the largest splice variant from each gene. Genes and proteins are aligned by homology. GLS and GLS2 show high homology in their exons, but GLS has substantially larger introns and untranslated regions of terminal exons. Data to generate this chart were taken from the RCSB Protein Data Bank, UniProtKB and Ensembl public data repositories. The smaller GLS2 isoform identified by Ota et al. is not included, due to uncertainty regarding its biological importance.
The N-termini of the GLS variants begin with a 16-residue sequence generally predicted to localize the proteins to the mitochondria, and indeed, there is near predictive and experimental consensus that GLS is localized to this organelle (Table 1 & Table 2) [34–45]. To predict GLS localization, we utilized five freely available predictive algorithms, one of which predicted localization based upon the N-terminal sequence only (TargetP) [42], one of which used the whole protein sequence (SCLPred) [44] and the remaining three of which made predictions based on whole-protein genetic ontology (GO) calculations [41,43,45]. SCLPred predicted both isoforms would localize to the cytosol, while three algorithms predicted both would localize to the mitochondria. The iLoc-Animal software predicts GAC to localize to mitochondria and KGA to localize to the cytoplasm, even though their N-termini are identical. Further, one report suggests that KGA resides in the cytoplasm of SK-BR-3 breast cancer cells, lending some support to the idea that more than just the N-termini of these proteins may determine their cellular localizations [37]. The KGA sequence deduced from the isolated cDNA has 669 residues, yielding a predicted molecular weight of 73.5 kDa. However, early work found that only 66 kDa and 68 kDa proteins were present in cell lysates. The Curthoys laboratory, using rat KGA cDNA, showed that these smaller proteins were due to the activity of matrix processing peptidase (MPP), a mitochondrial matrix enzyme that cleaves localization sequences. This results in the removal of the first 72 residues from rat KGA to yield a 66 kDa protein [46,47], although we note that a recent massive N-terminal sequencing analysis of the human mitochondrial proteome identified the first residue of processed human GLS to be residue 86 (with several later ‘first’ residues being observed as well) [48]. It is currently unclear if these different truncations are due to differences in the techniques used to detect them, or due to differences between rat and human GLS, which have nonidentical residues for 19 of the first 86 positions.
Table 1. . Predicted cellular localization of glutaminase isozymes.
| Algorithm |
Isoform |
Ref. | |||
|---|---|---|---|---|---|
| KGA | GAC | LGA_long | LGA_short | ||
| Hum-mPLoc 2.0 |
Mitochondria |
Mitochondria |
Mitochondria |
Mitochondria |
[41] |
| TargetP 1.1 |
Mitochondria |
Mitochondria |
Mitochondria |
Not mitochondria |
[42] |
| MultiLoc2 |
Mitochondria |
Mitochondria |
Mitochondria |
Cytosol |
[43] |
| SLCPred |
Cytosol |
Cytosol |
Cytosol |
Cytosol |
[44] |
| iLoc-Animal | Cytosol | Mitochondria | Cytosol | Cytosol | [45] |
Five cellular localization algorithms were queried with the full sequence of each major glutaminase isoform. Only Hum-mPloc 2.0 predicted that all four would localize to the mitochondria, and only SLCPred predicted that all four would localize to the cytosol.
GAC: Glutaminase; KGA: Kidney-type glutaminase; LGA: Liver-type glutaminase.
Table 2. . Experimental determinations of glutaminase localization in assorted tissues.
| Isoform | Tissue | Tumor? | Location | Technique | Ref. |
|---|---|---|---|---|---|
| GLS2† |
Liver |
N |
Mitochondrial |
OI |
[34] |
| GLS† |
Ascites |
Y |
Mitochondrial and lysosomal |
OI |
[35] |
| GLS† |
Kidney |
N |
Mitochondrial |
OI |
[36] |
| GAC |
Breast |
Y |
Mitochondrial |
IF, OI |
[37] |
| KGA |
Breast |
Y |
Cytosolic |
IF, OI |
[37] |
| GAC |
Prostate |
Y |
Mitochondrial |
OI |
[37] |
| KGA |
Prostate |
Y |
Cytosolic |
OI |
[37] |
| GAC |
Lung |
Y |
Mitochondrial |
OI |
[37] |
| KGA |
Lung |
Y |
Cytosolic |
OI |
[37] |
| GLS† |
Brain |
N |
Synaptosomal |
OI |
[38] |
| GLS† |
Kidney |
N |
Mitochondrial |
OI |
[38] |
| GLS† |
Kidney |
N |
Mitochondrial |
OI |
[39] |
| LGA |
Brain |
N |
Nuclear |
IF, OI |
[40] |
| KGA |
Brain |
N |
Mitochondrial |
IF, OI |
[40] |
| LGA(r) |
Insect cells |
N |
Mitochondrial and nuclear |
OI |
[51] |
| GLS2† |
Liver |
N |
Mitochondrial |
OI |
[56] |
| GLS2† |
Liver |
N |
Mitochondrial |
OI |
[57] |
| GAC(t) |
Breast |
Y |
Mitochondrial |
IF |
[60] |
| GAC |
Breast |
Y |
Mitochondrial |
OI |
[61] |
| LGA(t) | Lung | Y | Mitochondrial | IF | [62] |
Isoforms marked as ‘(r)’ or ‘(t)’ were being recombinantly expressed or ectopically transfected into host cells, respectively. Techniques are broadly characterized as those that viewed whole cells (IF), or those that isolated and characterized organelles (OI). A noncomprehensive tabulation of assorted experimental characterizations of glutaminase subcellular localization.
†These reports did not specifically state which glutaminase isoform was being studied and the isoform investigated was deduced from the tissue examined.
GAC: Glutaminase C; IF: Immunofluorescence; KGA: Kidney-type glutaminase; LGA: Liver-type glutaminase; OI: Organelle isolation.
LGA is encoded by the gene GLS2, located on human chromosome 12 and also exists as three transcriptional variants (Figure 2). The first cDNA for LGA was reported in 1997, when it was cloned from rat liver [49]. The first variant identified in human cells was cloned from the ZR-75 breast cancer cell line in 2000, and was termed ‘liver GA’; much of the field would ultimately refer to it as ‘LGA’ [50,51]. This variant became the accepted sequence of LGA, and represents the canonical sequence shown in the UniProtKB database. However, the same research group later renamed the ZR-75 sequence ‘GAB’, and hypothesized that a shorter LGA must also exist in humans, due to the different lengths of human GAB and rat LGA [52]. They subsequently demonstrated the existence of that shorter LGA transcript in human cells [53]. While some laboratories use this notation, most still refer to the canonical sequence as LGA.
Unlike the GLS proteins KGA and GAC, which differ in their C-termini, the GLS2 proteins differ at their N-termini. The longer form is generated by transcription of all 18 exons, while the shorter variant is formed by an alternate promotor binding event at the 3′ end of the first GLS2 intron, and is thus encoded by the final 71 bases of intron 1–2 and the entirety of exons 2–18 (Figure 2). A third variant, much shorter than the others, and similar to GAM in lacking the glutaminase catalytic domain, was identified via another cDNA analysis project, but its existence has not yet been experimentally confirmed and it is as yet unclear if the isoform is an experimental artifact or if it has a biological role [54]. In contrast to GAM, there is no obvious way to generate this variant via gene splicing, and the ensemble database suggests that it (transcript ID ENST00000486896.5) is probably the result of nonsense mediated decay, further casting doubt onto its biological significance. The longer variant of LGA is predicted to localize to the mitochondria by three of the five algorithms we utilized, whereas the shorter isoform is predicted to be mitochondrial only by the Hum-mPLoc 2.0 software (Table 1) [41–45]. There is also uncertainty in the literature as to where LGA localizes (Table 2). Early reports consistently found LGA activity in mitochondria, but a study by Olalla and colleagues shows the protein to localize to the nucleus in mammalian brain tissue [40,55–57]. While this report is at odds with previous literature and the predicted localization of either LGA isoform, no study has yet been published which either disputes or supports these findings. The N-terminal residues of the longer LGA isoform are truncated following mitochondrial localization, but it is currently unclear if a similar truncation occurs for the shorter LGA variant [51,58].
By 1995, it was known that GLS exists naturally as both a dimer and a tetramer, that the tetramer is the active form, and that inorganic phosphate can promote the formation of tetramers from dimers [59]. At the time, LGA was thought to have a molecular mass of 58 kDa, and to have an active complex with a mass between 310 and 162 kDa, implying that the active form of LGA contained between 5 and 3 subunits, as analyzed by sucrose gradients, gel electrophoresis and HPLC. Given that we now know the molecular weight of LGA is approximately 66 kDa, these reported measurements apparently represent a tetramer and a dimer, similar to the oligomeric states reported for GLS.
The structures of both GLS and LGA have been determined by x-ray crystallography, although substantially more effort has been devoted to describing GLS than LGA. Table 3 shows the details of the currently available mammalian glutaminase crystal structures [37,63–68]. The constructs used for crystallography include the isolated glutaminase catalytic domain (e.g., 3CZD or 3VP1) and the entire biologically processed form of the enzyme (e.g., 3UNW or 5FI7), and structures have been determined with glutamine, glutamate or assorted inhibitors bound. However, only a single LGA structure (4BQM) has been released to date, and it represents a significantly truncated form of the enzyme. Neither the N- nor C-termini of GLS or LGA are resolved in any x-ray crystal structure to date. However, Cassago et al. have shown that GAC has greater catalytic activity than KGA (which differs only in the C-terminus), indicating that the C-terminus influences function [37]. Similarly, Campos-Sandoval et al. reported distinct catalytic properties for the longer and shorter LGA variants, which differ only at the N-terminus. In particular, they found that inhibition or activation of the recombinantly expressed longer isoform (GAB) by glutamate and ammonia, respectively, differed from what had previously been described for LGA isolated from rat liver [51,52]. However, the recombinant human GAB used in their study, which was isolated from insect cells, did not include the first approximately 40 residues, and as such it differed from LGA only by a few residues at the N-terminus [51]. Therefore, although this report might indicate that the N-terminus influences catalytic activity, it alternatively seems possible that the observed differences were species-specific, or due to impurities in the earlier preparations of rat liver LGA.
Table 3. . Crystal structures of mammalian glutaminase enzymes currently released via the Research Collaboration for Structural Bioinformatics Protein Databank.
| PDB ID | GA isoform | Residues used | Resolution (Å) | Ligand(s) | Deposition date | Ref. |
|---|---|---|---|---|---|---|
| 5JYO |
GLS |
221–533 |
2.1 |
CB-839 |
15/05/2016 |
[63] |
| 5JYP |
GLS |
221–533 |
2.74 |
Trans-CBTBP |
15/05/2016 |
[63] |
| 5HL1 |
GAC |
72–598 |
2.4 |
CB-839 |
14/01/2016 |
NA |
| 5FI2 |
GAC |
72–598 |
2.5 |
UPGL_00009 (reported as compound 7d in McDermott et al.) |
22/12/2015 |
[64] |
| 5FI6 |
GAC |
72–598 |
2.52 |
UPGL_00011 (reported as compound 7e in McDermott et al.) |
22/12/2015 |
[64] |
| 5FI7 |
GAC |
72–598 |
2.5 |
UPGL_00015 (reported as compound 14b in McDermott et al.) |
22/12/2015 |
[64] |
| 5I94 |
GAC |
72–598 |
2.98 |
UPGL_00019 (reported as compound 14d in McDermott et al.) |
19/02/2016 |
[64] |
| 5D3O |
GAC |
72–598 |
2.79 |
None |
06/08/2015 |
NA |
| 4O7D |
GLS |
221–531 |
2.3 |
DON |
24/12/2013 |
[65] |
| 4JKT |
GAC (m) |
128–555 |
2.77 |
BPTES |
11/03/2013 |
[66] |
| 4BQM |
LGA |
154–479 |
2.18 |
None |
31/05/2013 |
NA |
| 3VOY |
GLS |
221–533 |
2.2 |
None |
23/02/2013 |
[67] |
| 3VOZ |
GLS |
221–533 |
2.4 |
BPTES |
23/02/2012 |
[67] |
| 3VP0 |
GLS |
221–533 |
2.4 |
l-glutamine |
23/02/2012 |
[67] |
| 3VP1 |
GLS |
221–533 |
2.3 |
l-glutamate, BPTES |
23/02/2012 |
[67] |
| 3VP2 |
GLS |
221–533 |
2.7 |
BPTES derivative 2 |
23/02/2012 |
[67] |
| 3VP3 |
GLS |
221–533 |
2.7 |
BPTES derivative 3 |
23/02/2012 |
[67] |
| 3VP4 |
GLS |
221–533 |
2.45 |
BPTES derivative 4 |
23/02/2012 |
[67] |
| 3SS3 |
GAC (m) |
134–609 |
2.42 |
None |
07/07/2011 |
[37] |
| 3SS4 |
GAC (m) |
134–609 |
2.85 |
Phosphate |
07/07/2011 |
[37] |
| 3SS5 |
GAC (m) |
134–609 |
2.8 |
l-glutamate |
07/07/2011 |
[37] |
| 3UNW |
GAC |
71–598 |
2.56 |
l-glutamate |
16/11/2011 |
[68] |
| 3UO9 |
GAC |
71–598 |
2.3 |
l glutamate, BPTES |
16/11/2011 |
[68] |
| 3CZD | GLS | 221–533 | 2.4 | l-glutamate | 29/04/2008 | [67] |
Structures marked (m) are from mouse enzyme, all others are from human enzyme. Isoforms are specified only for structures that extended into the variable C-terminus (residue ≥550) of GLS. The only structure of liver glutaminase currently released is highlighted. References are provided where available, or marked ‘NA’ if the structure has been released, but not yet described formally in a scientific publication.
BPTES: Bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide; DON: 6-Diazo-5-oxo-l-norleucine; GAC: Glutaminase C; GLS: Kidney-type glutaminase; LGA: Liver-type glutaminase; NA: Not applicable.
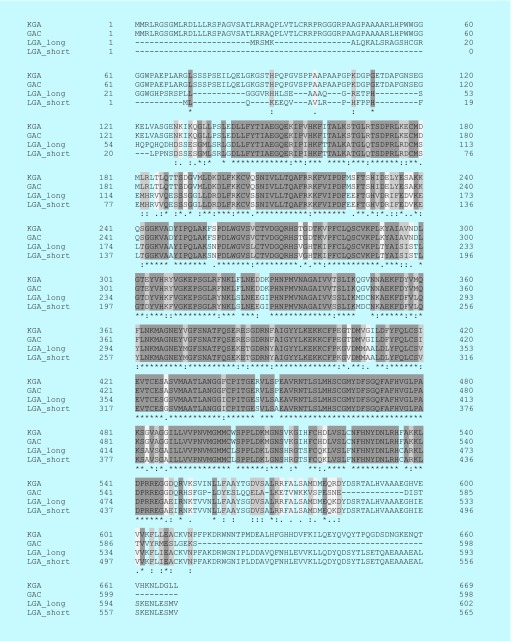
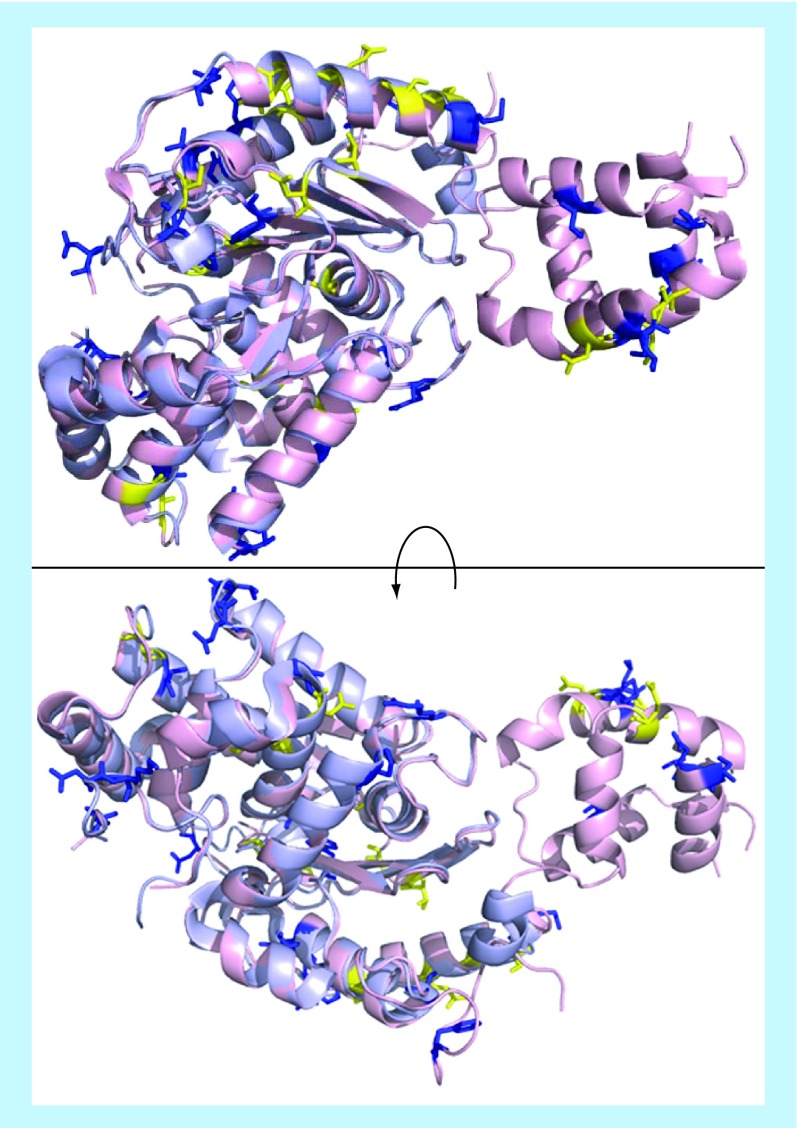
Both glutaminase isozymes are activated by phosphate; LGA is activated by lower concentrations of phosphate than GLS, but phosphate is ultimately able to increase GLS catalytic activity by a greater magnitude than for LGA [37,59]. Further, LGA is activated by ammonia, whereas ammonia inhibits GLS enzymes [57,69]. The mechanisms underlying these activating and inhibitory events are not known. As shown in Figure 3, the amino acid sequences of the major GLS and LGA variants align closely, with most of the variation occurring in their C- and N-termini. Similarly, overlaying the GLS and LGA crystal structures (Figure 4) shows that the catalytic domains fold in a nearly identical fashion, and that there are few concentrated regions of residues which differ significantly. Thus, it remains unclear what structural features lead to the differences in the allosteric regulation of these enzymes.
Figure 3. . Alignment of the amino acid sequences of the major isoforms of glutaminase and liver-type glutaminase.
Sequences were aligned with Clustal Omega via UniProt.org. Darker regions are identical, while lighter regions have greater differences between the isoforms. Each residue position is also marked with the Clustal alignment scores, “*”, “:”, “.”, or “ ”, which represent positions with identical residues, residues with strongly similar properties, residues with weakly similar properties or residues which are entirely dissimilar, respectively. Most of the differences lie in the N- and C-termini of the enzymes.
Figure 4. . Glutaminase C (light pink, crystal structure 5D3O) and liver-type glutaminase (light blue, crystal structure 4BQM) were aligned in PyMol.
Residues which are weakly similar or entirely dissimilar (Clustal alignment score of “.” or “ “, Figure 3) are drawn as sticks and shown in yellow and blue, respectively.
Recent advances in understanding glutaminase function have focused largely on GLS. In a study by Møller et al., the structures of the N- and C-termini of GAC were determined via small angle x-ray scattering (Figure 5A) and multi-angle light scattering measurements. This study also showed that GAC could exist not only as a dimer or tetramer, but also as an elongated higher order oligomer, with oligomerization increasing as a function of phosphate concentration [70]. This latter result was also reported by Ferreira and coworkers, who demonstrated the filament-like nature of the higher order GAC oligomer via electron microscopy [66]. This report also indicated the critical nature of a short loop, from Leu 316 to Leu 321 of the human enzyme, which the authors called a ‘gating loop’ (Figure 5B). This loop plays a critical role in GAC catalysis, and mutation of Lys 320 to alanine creates a constitutively activated GAC, which catalyzes glutamine hydrolysis even in the absence of phosphate or other inorganic anions. A recent study by Li et al. showed that it is not necessary to form higher order oligomers for GAC to be fully active. In fact, a dimeric GAC species, resulting from two mutations (K311Q and D286K) which render the enzyme unable to form a tetramer, can have full catalytic activity in a K320A background [60]. The same study shows that residue Tyr 249 acts as a ‘lid’, influencing substrate access or product release from the catalytic pocket (Figure 5B), and provides a mechanism by which phosphate may interact with the critical loop identified by Ferreira to enhance GAC catalytic activity. As such, this loop might better be referred to as an ‘activation loop’, as it seems to control catalytic activity rather than substrate access to the binding site. Of note, all of these structural studies were conducted with recombinantly expressed enzymes. In Li et al., we showed that wild-type GAC and constitutively active GAC provide a nearly identical growth advantage for NIH-3T3 cells stably expressing the tumor promotor onco-Dbl, while Ferreira reported just 10% enhancement in growth rate of MDA-MB-231 cells following selection for stable expression of GAC-K320A [60,66].
Figure 5. . Structural and mechanistic insights into glutaminase activity.
(A) The small angle x-ray scattering envelope calculated for tetrameric glutaminase C (GAC) shows the N-terminal residues (green) extending away from the catalytic domain (blue), and the C-terminal residues (magenta) coiling close to the catalytic domain. The N- and C-terminal residues have not been resolved to date by x-ray crystallography. Figure adapted with permission from [70] © PLoS ONE (2013). (B) The tetrameric form of GAC (crystal structure 3UO9). The recently identified ‘gating’ or ‘activation’ loop is shown as sticks, colored by chain, at the center of the structure. As is common for x-ray structures of GAC, not all of the four loops are fully resolved. The tyrosine 249 ‘lid’ is shown as blue spheres, and glutamate in the catalytic site is shown as green spheres.
Kidney-type glutaminase: implications in cancer
The role of kidney-type glutaminase in cancer has been heavily investigated [7,71–77]. The Cancer Genome Atlas (TCGA) pan-cancer gene expression data show high expression of GLS in acute myeloid leukemia, adrenocortical cancer, triple-negative breast cancer, colorectal cancer, kidney clear or papillary cell carcinoma, lung adenocarcinoma, melanoma, mesothelioma, pancreatic cancer, sarcoma and thyroid cancer [78,79]. Of these, GLS activity has been shown to be important for growth of acute myeloid leukemia [80–82], breast cancer [61,64,83,84], colorectal cancer [85], kidney cancer [86,87], lung cancer [88], melanoma [89,90] and pancreatic cancer cells [91] in studies that utilized small-molecule inhibitors or gene-silencing approaches in cell lines. Further, glioblastoma cell lines have been found to be sensitive to glutaminase inhibitors in vitro, despite having relatively low GLS expression according to TCGA data [80,92]. Combined, these results demonstrate that GLS is critical for the growth of a wide variety of cancer cell lines, and that inhibition of the enzyme is a potentially valuable therapeutic strategy.
The product of the glutaminase reaction, glutamate, can be converted to α-ketoglutarate for use in the Krebs cycle [73,93], contributes to nonessential amino acid synthesis, and is also required for production of glutathione, a key scavenger of reactive oxygen species (ROS) [94]. Although controlled levels of ROS can stimulate cell proliferation, excessive levels lead to cell stress and death [95]. Genetic silencing of GLS can lead to decreased glutathione levels [96,97], as can treatment of cells with a small-molecule inhibitor of GLS [13]. A novel role of GLS in cancer is in the formation of microvesicles or oncosomes, nontraditional secretory vesicles which mediate signaling between cancer cells and their environment [98]. Production of microvesicles by cultured cancer cells can be slowed or halted via glutaminase inhibition [99].
One of the most important drivers of oncogenesis is K-Ras, and a number of studies have connected K-Ras to glutaminase activation. Gaglio and coworkers took a metabolomics approach to show that NIH-3T3 cells transformed with oncogenic K-Ras exhibited dramatically altered metabolic flux, with an increase in flux through GLS accompanied, oddly, by a decrease in GLS expression [100]. Son et al. showed that in pancreatic ductal adenocarcinoma (PDAC), K-Ras suppresses glutamate dehydrogenase (GLDH) and activates aspartate transaminase (GOT1) to form α-ketoglutarate downstream of glutaminase [91]. This leads to a dependence upon GLS, as GOT1 is required in PDAC for redox homeostasis. This was also observed by Weinberg and coworkers, who noted that inhibition of GLS in PDAC could be rescued by addition of glutamate (which can be used to form ROS scavengers) but not by α-ketoglutarate [101]. K-Ras driven non-small-cell lung cancer cells have also been found to require GLS activity, although no strong correlation was detected between GLS expression, oncogene expression and resistance to glutamine withdrawal [88]. However, a recent study by Davidson et al. showed that Ras-driven, non-small-cell lung tumors are far less affected by glutaminase inhibition by the GLS inhibitor CB-839, or by CRISPR/Cas-9 based genetic silencing of GLS, than tumor-derived cell lines grown in culture [102]. The authors concluded that tumor microenvironment was responsible for reduced glutamine dependence in whole tumors relative to cultured cells, consistent with a separate report that lung cancer metabolism varies greatly with microenvironment [103]. Although Davidson et al. addressed GLS activity, TCGA data suggest that in most lung cancers, GLS and GLS2 are both expressed. It will therefore be of interest to determine whether simultaneous inhibition of both glutaminase isozymes is able to suppress lung tumor growth.
A number of other mechanisms regulate GLS expression at multiple levels, and GLS is upregulated by several proteins that are involved in cell cycle progression. The GLS transcript has a pH-responsive element, which results in increased expression at acidic pH [104–106]. The transcription factor c-Myc indirectly upregulates GLS in some contexts by repressing the expression of miRNA-23 [96,107], which is also repressed by NF-κB [108]. The transcription factor c-Jun can directly bind to the GLS gene promoter and enhance expression, and other recent reports have shown that the transcriptional coactivator PGC-1α, and loss of the tumor suppressor Rb, increase GLS transcription and translation, respectively [109–111]. Of further note are recently suggested roles for members of the Sirtuin family of proteins in the regulation of GLS. These enzymes act as deacetylases, desuccinylases and demalonylases, and at least two members of the Sirtuin family, SirT5 and SirT6, have been implicated in GLS regulation. Sebastian et al. showed that SirT6 represses MYC, and, by extension, the expression of GLS [112]. A more direct interaction was demonstrated by Poletta and colleagues, who used immunoprecipitation assays to show that GLS binds directly to SirT5 [113]. They additionally showed that GLS was succinylated in the absence of SirT5, and suggested that the desuccinylation activity of SirT5 acted to regulate GLS activity.
The GAC splice variant is more frequently upregulated in cancer cells than KGA [61]. A recent report by Redis and colleagues shows that the long, noncoding RNA colon cancer-associated transcript 2 (CCAT2) plays a role in this process, at least in colon cancer [114]. CCAT2 exists as two alleles: a G form, associated with greater predisposition for colon cancer, and a T form. The G allele strongly binds to CFIm25 (Cleavage Factor 1, small subunit), and this complex binds the 14th intron of GLS, resulting in preferential splicing of the GAC (exon 15 containing) isoform.
Liver-type glutaminase: implications in cancer
While GLS has been consistently tied to cancer progression, the case has been less clear for LGA. TCGA data suggest that GLS2 is relatively highly expressed in cancers of the bladder, breast (receptor positive), cervix, colon, kidney, liver, lung, ovary, paraganglia, prostate, rectum, thyroid and thymus [78,79]. However, it has yet to be verified as a pharmacological target in these cancers; moreover, there is still uncertainty over whether LGA functions as a tumor promotor or suppressor. In 2009, Szeliga and coworkers demonstrated that transfection of T98G glioblastoma cells with LGA reduces their ability to form colonies in soft agar [115]. The same group later demonstrated that transfection of the same cells with LGA increases their susceptibility to alkylating agents, by downregulating the DNA repair gene MGMT [116]. However, Lee et al. showed that LGA inhibition or genetic silencing inhibited the growth of both A549 lung cancer cells and HepG2 hepatoma cells [58]. Xiang et al. demonstrated that knock-downs of GLS2 greatly reduced the radiation resistance of the HeLa cervical cancer cell line, with a corresponding reduction in the levels of NADH and glutathione [117]. However, LGA overexpression was reported to synergize with oxidative stress in killing glioma cells in another study [97]. Consistent with the conflicting reports of the role of LGA in cancer, its expression can be upregulated both by the tumor suppressor p53 [62,118,119] and by the proto-oncoproteins c-Myc (reported in activated CD4+ T cells) and n-Myc [120,121].
Kidney-type glutaminase: drug discovery
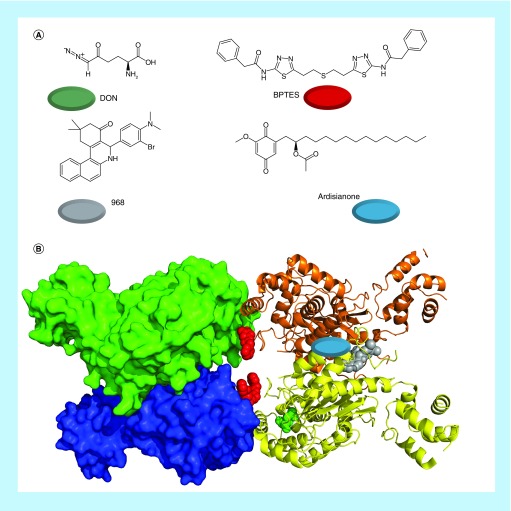
For most of the 80 years since glutaminase was discovered, the only chemical inhibitor of note was DON (6-diazo-5-oxo-l-norleucine), which is a glutamine mimetic and irreversibly binds to the catalytic serine of both glutaminase isozymes (Figure 6A) [122,123]. While other small molecules were utilized to inhibit GLS (particularly membrane-impermeable molecules for determining subcellular localization), DON was the most extensively used inhibitor until the last decade [39]. DON has a number of other targets, and offers little therapeutic potential due to excessive toxicity [123]. More recently, two novel inhibitors of glutaminase have been described. In 2007, Robinson et al. reported the molecule BPTES, and in 2010 Wang et al. reported the chemically distinct molecule 968 (Figure 6A) [61,124].
Figure 6. . The most important classes of glutaminase inhibitors described in recent years.
(A) The nonselective inhibitor DON, the kidney-type glutamase and liver-type glutaminase inhibitor 968, the kidney-type glutaminase selective inhibitor BPTES and the liver-type glutaminase selective inhibitor ardisianone. The colored circles match the colors of the inhibitors below. (B) Tetrameric kidney-type glutamase (crystal structure 4JKT) with 968 docked to it (gray, on right). BPTES (red, in center) is overlayed from crystal structure 3UO9, as is DON (green, in yellow subunit) from crystal structure 4O7D. The described binding site for ardisianone on liver-type glutaminase is shown with a blue oval (right-hand subunits).
BPTES: Bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide; DON: 6-Diazo-5-oxo-l-norleucine.
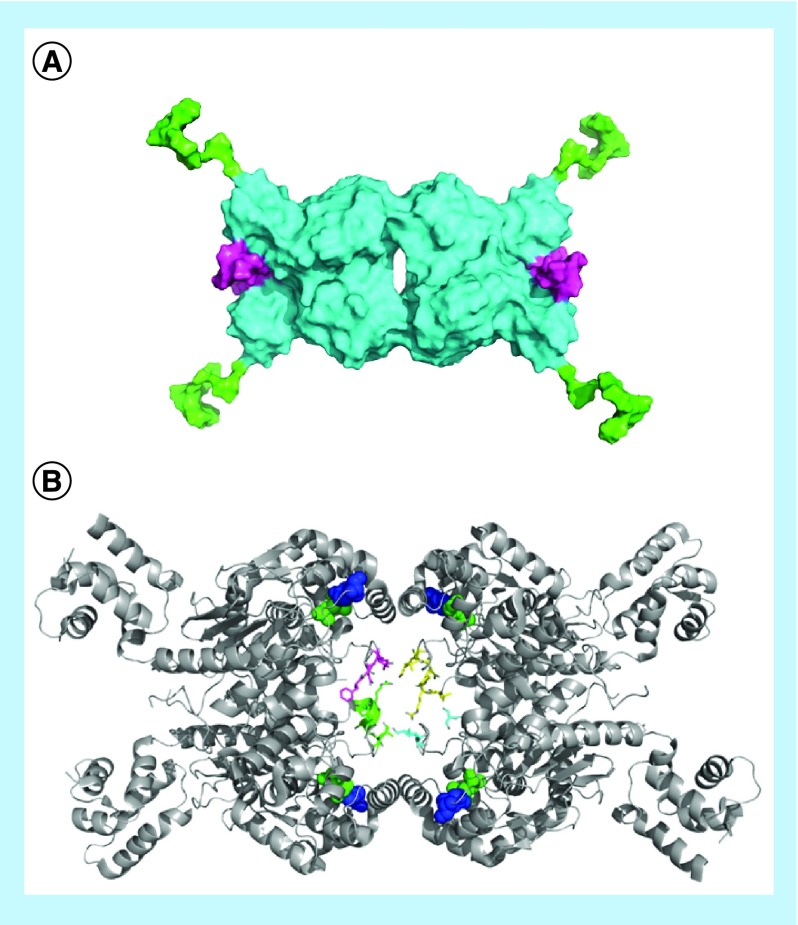
BPTES is a long, highly flexible and C2-symmetrical molecule which binds in a 1:2 ratio with GLS and sits at the dimer–dimer binding interface, where it stabilizes an inactive tetramer (Figure 6B) [67,124]. It has been suggested to have approximately 1000-fold greater affinity for GLS than for LGA, however to date, the only studies showing this with purified recombinant enzymes have used mutants of GLS which resemble LGA at the BPTES binding site, rather than using full-length LGA [68,123]. BPTES has been the subject of significant research efforts since its discovery, aided by the early reports of crystal structures of the molecule in complex with GLS. Research has substantially focused on enhancing inhibitor affinity and improving drug-like properties.
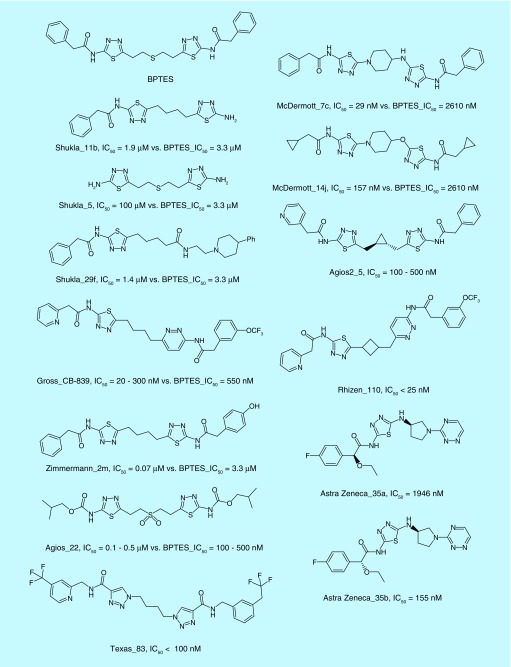
The first major structure–activity relationship (SAR) study around BPTES was reported in 2012 by Shukla et al., and focused on alteration of the flexible chain in the center of the molecule and desymmetrization via replacement of one or both phenyl groups at the ends of the molecule [125]. The key findings were that the central linker could be replaced by a four carbon chain, and that one, but not both, phenyl groups could be removed to enhance solubility without significantly reducing potency (Figure 7, Shukla_11b, Shukla_5). The authors also found a variety of terminal substituents that could be tolerated, but did not find any that dramatically improved potency (Figure 7, Shukla_29f). Calithera Biosciences (CA, USA) subsequently described several hundred BPTES derivatives, one of which, CB-839, is currently considered the best-in-class GLS inhibitor, and is undergoing clinical trials for several indications [84,126]. Development of CB-839 expanded upon the studies of Shukla and colleagues, replacing one thiadiazole ring and both phenyl rings, and was the first GLS inhibitor reported to have a low nanomolar potency. However, CB-839 is short-lived in mice, and dosing orally with 200 mg drug/kg mouse bodyweight twice per day resulted in a concentration of 2 μM drug in plasma (∼67 μg/kg) [83]. Despite this issue, the drug was exceptionally effective under this treatment regimen, inhibiting the growth of patient-derived xenografts by approximately 60% and almost entirely blocking growth when combined with the common chemotherapeutic paclitaxel. Further, the drug exhibits an as-yet-unexplained requirement to be incubated for up to 4 h with GLS before it exhibits maximum potency. Very similar series of compounds were described in a report from Zimmermann and coworkers, and in patents from Agios (MA, USA) and the University of Texas, with representative examples shown in Figure 7 [127–132].
Figure 7. . Bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide, and a selection of potent inhibitors derived from its scaffold, are shown.
Inhibitors are named by first author for academic publications, or patent assignee for patents, and then by the compound code within the relevant publication. IC50 values are reported for each compound. Because assay conditions varied between investigators, the value for bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide (BPTES) in their assay system is also reported where available.
BPTES: Bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide.
A second major focus has been on replacement of the flexible linker portion of BPTES with assorted cyclic scaffolds. Recently, we described a series of heterocyclic compounds with potencies comparable to CB-839, while exhibiting greater resistance to liver microsome metabolism [64]. Similar strategies have also been followed by Agios (MA, USA), Calithera Biosciences (CA, USA), Rhizen Pharmaceuticals (PA, USA) and AstraZeneca (DE, USA) [133–137]. Representative molecules from each of these studies are shown in Figure 7. In total, several thousand derivatives of BPTES have been reported, and the chemical space, while not exhausted, has been well investigated. However, several issues remain unclear, particularly pertaining to the role played by the terminal rings of these molecules, which in crystal structures have very high B-factors, suggesting that they do not bind tightly to the protein, even while the structure–activity relationship (SAR) reports demonstrate their importance for inhibitory potency.
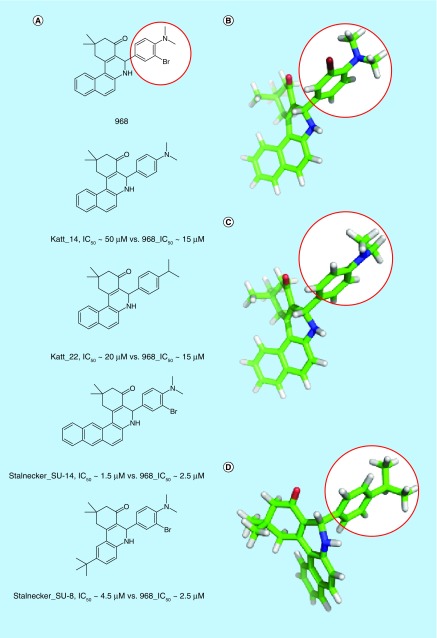
968 is an allosteric regulator of GAC, and has been suggested to bind to a cavity where two GAC monomers form a dimer (Figure 6B) [61,83,138]. The original report of 968 determined that the ‘hot-spot’ ring of the molecule (Figure 8A & B, circled in red) was essential for activity. We now understand that this ring requires a large, antiplanar group at the para position for significant inhibitory potency (e.g., 968 vs Katt_14 vs Katt_22 in Figure 8B–D), and the naphthyl portion of the molecule can be significantly altered without affecting inhibitory activity (e.g., Figure 8A, Stalnecker_SU-14 or Stalnecker_SU-8). Notably, one study indicates that 968 inhibits LGA as potently as it inhibits GAC, and prevents LGA-dependent ferroptosis in several cell systems [139]. Of further note is that 968 is not able to inhibit recombinantly expressed GAC which has been pre-activated via exposure to phosphate [61]. It has shown efficacy in both cell culture and animal models, however, which we hypothesize represents either a rapid binding to newly synthesized protein or a low overall activation state of GLS in the cell at any given time.
Figure 8. . 968 and select potent derivatives.
(A) Important compounds that help to elucidate the 968 structure–activity relationship. Compounds are labeled by first author of the reporting literature, and compound code within that manuscript. Because different assays were used for the two relevant studies, IC50 values are given for each value and for 968 as reported within that study. (B–D) 968 (B), Katt_14 (C) and Katt_22 (D), energy minimized in the MMFF94 forcefield. 968 and Katt_22, both potent compounds, have ‘hot-spot’ rings (circled in red) with para-substituents perpendicular to the plane of the ring. Katt_14, a weak inhibitor, has a ‘hot-spot’ ring with the para-substituent parallel to the plane of the ring. Similar orientations were determined following ab initio calculations at the 6–31+G* level.
Liver-type glutaminase: drug discovery
In contrast to GLS, little drug-discovery effort has been directed toward LGA, and only a single study describes LGA-selective inhibitors. Lee et al. screened an assortment of natural products, and found that several alkyl benzoquinones isolated from Ardisia virens or Ardisia kusukuensis had submicromolar potency against recombinantly expressed LGA, while exhibiting up to tenfold higher affinity for LGA than for KGA [59]. Further, they provided evidence that these molecules bind to a novel site on LGA, distinct from the equivalent binding sites for BPTES, 968 or DON on GLS (Figure 6). Finally, they showed that their primary molecule, AV-1, inhibited the growth of HepG2 hepatoma and A549 lung carcinoma cells. Notably, AV-1, better known as ardisianone, is a major product of many species of Ardisia, which have been used for centuries in traditional Chinese medicine. The compound has been shown to lead to mitochondrial failure, apoptosis and downregulation of survivin, an important prosurvival protein in many cancers [140–142] and it is possible that it might have additional cellular targets besides LGA.
Conclusion
Many cancer cells develop a reliance upon metabolism of the common nutrient glutamine for their bioenergetic and biosynthetic needs. The glutaminase enzymes are the key players in facilitating the use of glutamine as an energy source and biosynthetic precursor. These enzymes are derived from two independent genes, each of which encodes at least two unique proteins. While advances have been made in discovering links between glutaminase expression and signaling pathways such as those involving KRas, c-Myc, or c-Jun, the question of how specific isozymes of glutaminase are differentially regulated in assorted tissues has not yet been answered. Similarly, while the isozymes have been well characterized in isolated systems, and recent crystallographic advances have demonstrated the importance of features such as the ‘gating’ or ‘activation’ loop, much remains to be investigated pertaining to both basic mechanistic details and the functional differences between the assorted glutaminases in a cellular context. Although these important enzymes are not yet fully understood, their roles in cancer have been well studied, particularly for the isozyme GAC, and substantial efforts have been devoted towards development of small, non-toxic inhibitors of this enzyme, one of which, CB-839, is currently in clinical trials. It is likely that glutaminase enzymes will continue to be studied in the coming years as both a fundamental player in the metabolism of rapidly proliferating cells and as a potentially important target for cancer therapeutics.
Future perspective
In the 80 years since its discovery by Krebs, glutaminase has been intensely studied with an increasing focus on its role in cancer. However, the kidney isozyme of glutaminase has garnered substantially more attention than the liver isozyme, even though the latter likely also plays important roles in cancer. Thus far, virtually all preclinical studies have focused on the BPTES class of molecules. In the next 5 to 10 years, we expect further studies around these molecules; however, we anticipate new scaffolds to be developed as well Further, insight into the mechanism of glutaminase activation is growing, and this will open the door both to further mechanistic studies and potentially to new pharmaceutical approaches. A major question is the importance of LGA in tumors, and whether its expression could lead to resistance to GLS-specific inhibitors. The recent finding that inhibition of GLS alone is insufficient to halt progression of some tumors makes it especially important for investigators to further characterize LGA, and its possible roles in cancer cell metabolism and tumorigenesis (Box 1).
Box 1. . A note on terminology.
Glutaminases (EC 3.5.1.2) have been denoted by a number of names and symbols historically. The Human Genome Organization (HUGO) has approved GLS and GLS2 as the gene symbols for the two human isoforms, and these are primarily used in the literature (although GLS1 is sometimes used in place of GLS, and is considered an acceptable substitute by HUGO). The protein names and symbols are less consistent. The two isozymes together have historically been referred to as phosphate-activated glutaminase, glutaminase I, GA or simply GLS. The GLS-derived protein is usually referred to as ‘kidney-type glutaminase’, but in older literature is sometimes referred to as ‘brain-type glutaminase’. The GLS2-derived protein is almost always referred to as ‘liver-type glutaminase’, although this term was used in older literature to describe an unrelated enzymatic complex, sometimes referred to as the glutaminase II pathway, consisting of glutamine transaminase and ω-amidase (which, contrary to its name, actually catalyzes a deamidation reaction). Further, some literature refers to ‘phosphate-independent glutaminases’, which in at least some cases are γ-glutamyl transpeptidases, in other words, transglutaminases. Regarding symbols, GLS-derived protein is designated GLS or KGA, although the latter can refer to all GLS-derived protein or to a particular splice variant. The other GLS splice variants are GAC and GAM. GLS2-derived protein is called GLS2 or liver-type glutaminase (LGA), but the latter can also refer to either of two GLS2 transcripts, with the longer form sometimes being called GAB instead.
In this review, we use GLS to refer to all protein derived from GLS, whereas KGA, GAC and GAM refer to the specific splice variants. Since GAB has not seen wide adoption in the literature, we refer to the protein derived from GLS2 as LGA, and refer to ‘LGA long’ or ‘LGA short’ forms when it is pertinent to differentiate between the two.
Executive summary.
Structure & function
GLS and liver-type glutaminase (LGA) isoforms have high homology, and fold similarly in their catalytic domains.
GLS isoforms differ in their C-termini, LGA isoforms differ in their N-termini.
GLS and LGA isoforms have different responses to allosteric regulators.
Role in cancer
GLS, and particularly glutaminase C, has been implicated in the progression of a diverse array of cancers.
LGA has been shown to promote the proliferation of some cancer cells, and to suppress the proliferation of others.
Drug discovery
Two small molecule inhibitors of GLS were recently identified, bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide (BPTES) and 968.
The BPTES class of inhibitors has been extensively investigated, with the current best-in-class molecule CB-839 undergoing clinical trials.
BPTES class molecules are generally selective for GLS over LGA.
The 968 class of inhibitors is less well investigated, but evidence suggests that they inhibit GLS and LGA with similar potency.
Natural products isolated from species of Ardisia are selective for LGA over GLS, but may have significant off-target effects as well.
Acknowledgements
The authors would like to thank C Westmiller for her excellent secretarial assistance in assembling this manuscript.
Footnotes
Financial & competing interests disclosure
This work was supported by funding from the NIH (GM040654, GM047458, CA201402). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J. Gen. Physiol. 1927;8:519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 5.DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci. Adv. 2016;2(5):e1600200. doi: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koppenol WH, Bounds PL, Dang CV. Otto Warburg's contributions to current concepts of cancer metabolism. Nat. Rev. Cancer. 2011;11(5):325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 8.Pearce EL. Metabolism in T cell activation and differentiation. Curr. Opin. Immunol. 2010;22(3):314–320. doi: 10.1016/j.coi.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pearce EL, Poffenberger MC, Chang CH, Jones RG. Fueling immunity: insights into metabolism and lymphocyte function. Science. 2013;342(6155):1242454. doi: 10.1126/science.1242454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rathmell JC. Metabolism and autophagy in the immune system: immunometabolism comes of age. Immunol. Rev. 2012;249(1):5–13. doi: 10.1111/j.1600-065X.2012.01158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krebs HA, Johnson WA. The role of citric acid in intermediate metabolism in animal tissues. Enzymologia. 1937;4:148–156. doi: 10.4159/harvard.9780674366701.c143. [DOI] [PubMed] [Google Scholar]
- 12.Rich PR. The molecular machinery of Keilin's respiratory chain. Biochem. Soc. Trans. 2003;31(6):1095–1105. doi: 10.1042/bst0311095. [DOI] [PubMed] [Google Scholar]
- 13.Le A, Lane AN, Hamaker M, et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012;15(1):110–121. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan J, Kamphorst JJ, Mathew R, et al. Glutamine-driven oxidative phosphorylation is a major ATP source in transformed mammalian cells in both normoxia and hypoxia. Mol. Syst. Biol. 2013;9(1):712. doi: 10.1038/msb.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krebs HA. Metabolism of amino-acids. Biochem. J. 1935;29(8):1951–1969. doi: 10.1042/bj0291951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geddes WF, Hunter A. Observations upon the enzyme asparaginase. J. Biol. Chem. 1928;77(1):197–229. [Google Scholar]
- 17.Grassmann W, Mayr O. Zur Kenntnis der Hefeasparaginase. Hoppe-Seyler’ s Zeitschrift für Physiol. Chemie. 1933;214(5–6):185–210. [Google Scholar]
- 18.Errera M, Greenstein JP. Phosphate-activated glutaminase in kidney and other tissues. J. Biol. Chem. 1949;178(1):495–502. [PubMed] [Google Scholar]
- 19.Carter CE, Greenstein JP. Acceleration of enzymatic desamidation of glutamine by several inorganic anions. J. Natl Cancer Inst. 1947;7(6):433–436. [PubMed] [Google Scholar]
- 20.Greenstein JP, Price VE. α-Keto acid-activated glutaminase and asparaginase. J. Biol. Chem. 1949;178(2):695–705. [PubMed] [Google Scholar]
- 21.Greenstein JP, Leuthardt FM. Effect of added phosphate on glutamine desamidation in tumors. J. Natl Cancer Inst. 1948;8(4):161–162. [PubMed] [Google Scholar]
- 22.Kvamme E, Tveit B, Svenneby G. Glutaminase from pig renal cortex: I. purification and general properties. J. Biol. Chem. 1970;245(8):1871–1877. [PubMed] [Google Scholar]
- 23.Svenneby G, Torgner IA, Kvamme E. Purification of phosphate-dependent pig brain glutaminase. J. Neurochem. 1973;20(4):1217–1224. doi: 10.1111/j.1471-4159.1973.tb00090.x. [DOI] [PubMed] [Google Scholar]
- 24.Curthoys NP, Kuhlenschmidt T, Godfrey SS. Regulation of renal ammoniagenesis. Arch. Biochem. Biophys. 1976;174(1):82–89. [PubMed] [Google Scholar]
- 25.Haser WG, Shapiro RA, Curthoys NP. Comparison of the phosphate-dependent glutaminase obtained from rat brain and kidney. Biochem. J. 1985;229(2):399–408. doi: 10.1042/bj2290399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiu JF, Boeker EA. Cow brain glutaminase: partial purification and mechanism of action. Arch. Biochem. Biophys. 1979;196(2):493–500. doi: 10.1016/0003-9861(79)90301-1. [DOI] [PubMed] [Google Scholar]
- 27.Archibald RM. Preparation and assay of glutaminase for glutamine determinations. J. Biol. Chem. 1944;154(3):657–667. [Google Scholar]
- 28.Heini HG, Gebhardt R, Brecht A, Mecke D. Purification and characterization of rat liver glutaminase. Eur. J. Biochem. 1987;162(3):541–546. doi: 10.1111/j.1432-1033.1987.tb10673.x. [DOI] [PubMed] [Google Scholar]
- 29.Smith EM, Watford M. Rat hepatic glutaminase: purification and immunochemical characterization. Arch. Biochem. Biophys. 1988;260(2):740–751. doi: 10.1016/0003-9861(88)90504-8. [DOI] [PubMed] [Google Scholar]
- 30.Nagase T, Ishikawa K, Suyama M, et al. Prediction of the coding sequences of unidentified human genes. XII. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro . DNA Res. 1998;5(6):355–364. doi: 10.1093/dnares/5.6.355. [DOI] [PubMed] [Google Scholar]
- 31.Elgadi KM, Meguid RA, Qian M, Souba WW, Abcouwer SF. Cloning and analysis of unique human glutaminase isoforms generated by tissue-specific alternative splicing. Physiol. Genomics. 1999;1(2):51–62. doi: 10.1152/physiolgenomics.1999.1.2.51. [DOI] [PubMed] [Google Scholar]
- 32.Porter LD, Ibrahim H, Taylor L, Curthoys NP. Complexity and species variation of the kidney-type glutaminase gene. Physiol. Genomics. 2002;9(3):157–166. doi: 10.1152/physiolgenomics.00017.2002. [DOI] [PubMed] [Google Scholar]
- 33.Sammeth M, Foissac S, Guigó R. A general definition and nomenclature for alternative splicing events. PLoS Comput. Biol. 2008;4(8):e1000147. doi: 10.1371/journal.pcbi.1000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brosnan JT, Ewart HS, Squires SA. Hormonal control of hepatic glutaminase. Adv. Enzyme Regul. 1995;35:131–146. doi: 10.1016/0065-2571(94)00003-l. [DOI] [PubMed] [Google Scholar]
- 35.Kalra J, Brosnan JT. The subcellular localization of glutaminase isoenzymes in rat kidney cortex. J. Biol. Chem. 1974;249(10):3255–3260. [PubMed] [Google Scholar]
- 36.Aledo JC, de Pedro E, Gomez-Fabre PM, Nunez de Castro I, Marquez J. Submitochondrial localization and membrane topography of Ehrlich ascitic tumour cell glutaminase. Biochim. Biophys. Acta. 1997;1323(2):173–184. doi: 10.1016/s0005-2736(96)00189-7. [DOI] [PubMed] [Google Scholar]
- 37.Cassago A, Ferreira APS, Ferreira IM, et al. Mitochondrial localization and structure-based phosphate activation mechanism of Glutaminase C with implications for cancer metabolism. Proc. Natl Acad. Sci. USA. 2012;109(4):1092–1097. doi: 10.1073/pnas.1112495109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kvamme E, Torgner IA, Roberg B. Kinetics and localization of brain phosphate activated glutaminase. J. Neurosci. Res. 2001;66(5):951–958. doi: 10.1002/jnr.10041. [DOI] [PubMed] [Google Scholar]
- 39.Shapiro RA, Haser WG, Curthoys NP. The orientation of phosphate-dependent glutaminase on the inner membrane of rat renal mitochondria. Arch. Biochem. Biophys. 1985;243(1):1–7. doi: 10.1016/0003-9861(85)90767-2. [DOI] [PubMed] [Google Scholar]
- 40.Olalla L, Gutiérrez A, Campos JA, et al. Nuclear localization of L-type glutaminase in mammalian brain. J. Biol. Chem. 2002;277(41):38939–38944. doi: 10.1074/jbc.C200373200. [DOI] [PubMed] [Google Scholar]
- 41.Shen HB, Chou KC. A top-down approach to enhance the power of predicting human protein subcellular localization: Hum-mPLoc 2.0. Anal. Biochem. 2009;394(2):269–274. doi: 10.1016/j.ab.2009.07.046. [DOI] [PubMed] [Google Scholar]
- 42.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000;300(4):1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 43.Blum T, Briesemeister S, Kohlbacher O. MultiLoc2: integrating phylogeny and gene ontology terms improves subcellular protein localization prediction. BMC Bioinformatics. 2009;10(1):1–11. doi: 10.1186/1471-2105-10-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mooney C, Wang Y, Pollastri G. SCLpred: protein subcellular localization prediction by N-to-1 neural networks. Bioinformatics. 2011;27(20):2812–2819. doi: 10.1093/bioinformatics/btr494. [DOI] [PubMed] [Google Scholar]
- 45.Lin WZ, Fang JA, Xiao X, Chou KC. iLoc-Animal: a multi-label learning classifier for predicting subcellular localization of animal proteins. Mol. Biosyst. 2013;9(4):634–644. doi: 10.1039/c3mb25466f. [DOI] [PubMed] [Google Scholar]
- 46.Shapiro RA, Farrell L, Srinivasan M, Curthoys NP. Isolation, characterization, and in vitro expression of a cDNA that encodes the kidney isoenzyme of the mitochondrial glutaminase. J. Biol. Chem. 1991;266(28):18792–18796. [PubMed] [Google Scholar]
- 47.Srinivasan M, Kalousek F, Curthoys NP. In vitro characterization of the mitochondrial processing and the potential function of the 68-kDa subunit of renal glutaminase. J. Biol. Chem. 1995;270(3):1185–1190. doi: 10.1074/jbc.270.3.1185. [DOI] [PubMed] [Google Scholar]
- 48.Vaca Jacome AS, Rabilloud T, Schaeffer-Reiss C, et al. N-terminome analysis of the human mitochondrial proteome. Proteomics. 2015;15(14):2519–2524. doi: 10.1002/pmic.201400617. [DOI] [PubMed] [Google Scholar]
- 49.Chung-Bok MI, Vincent N, Jhala U, Watford M. Rat hepatic glutaminase: identification of the full coding sequence and characterization of a functional promoter. Biochem. J. 1997;324(1):193–200. doi: 10.1042/bj3240193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gómez-Fabre PM, Aledo JC, Del Castillo-Olivares A, et al. Molecular cloning, sequencing and expression studies of the human breast cancer cell glutaminase. Biochem. J. 2000;345(2):365–375. [PMC free article] [PubMed] [Google Scholar]
- 51.Campos-Sandoval JA, López de la Oliva AR, Lobo C, et al. Expression of functional human glutaminase in baculovirus system: affinity purification, kinetic and molecular characterization. Int. J. Biochem. Cell Biol. 2007;39(4):765–773. doi: 10.1016/j.biocel.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 52.de la Rosa V, Campos-Sandoval JA, Martín-Rufián M, et al. A novel glutaminase isoform in mammalian tissues. Neurochem. Int. 2009;55(1–3):76–84. doi: 10.1016/j.neuint.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 53.Martín-Rufián M, Tosina M, Campos-Sandoval JA, et al. Mammalian glutaminase Gls2 gene encodes two functional alternative transcripts by a surrogate promoter usage mechanism. PLoS ONE. 2012;7(6):e38380. doi: 10.1371/journal.pone.0038380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ota T, Suzuki Y, Nishikawa T, et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 2004;36(1):40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- 55.Häussinger D, Gerok W, Sies H. Regulation of flux through glutaminase and glutamine synthetase in isolated perfused rat liver. Biochim. Biophys. Acta. 1983;755(2):272–278. doi: 10.1016/0304-4165(83)90214-3. [DOI] [PubMed] [Google Scholar]
- 56.Meijer AJ. Channeling of ammonia from glutaminase to carbamoyl-phosphate synthetase in liver mitochondria. FEBS Lett. 1985;191(2):249–251. doi: 10.1016/0014-5793(85)80018-1. [DOI] [PubMed] [Google Scholar]
- 57.McGivan JD, Lacey JH, Joseph SK. Localization and some properties of phosphate-dependent glutaminase in disrupted liver mitochondria. Biochem. J. 1980;192(2):537–542. doi: 10.1042/bj1920537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee YZ, Yang CW, Chang HY, et al. Discovery of selective inhibitors of glutaminase-2, which inhibit mTORC1, activate autophagy and inhibit proliferation in cancer cells. Oncotarget. 2014;5(15):6087–6101. doi: 10.18632/oncotarget.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Although the compounds may have substantial off-targets, the natural products investigated in this manuscript by Lee and coworkers remain the only small molecules so far reported to have potent selectivity for liver-type glutaminase over kidney-type glutaminase.
- 59.Curthoys NP, Watford M. Regulation of glutaminase activity and glutamine metabolism. Annu. Rev. Nutr. 1995;15:133–159. doi: 10.1146/annurev.nu.15.070195.001025. [DOI] [PubMed] [Google Scholar]
- 60.Li Y, Erickson JW, Stalnecker CA, et al. Mechanistic basis of glutaminase activation: a key enzyme that promotes glutamine metabolism in cancer cells. J. Biol. Chem. 2016;291(40):20900–20910. doi: 10.1074/jbc.M116.720268. [DOI] [PMC free article] [PubMed] [Google Scholar]; • A number of key structural features of GLS are described which significantly impact its catalytic activities.
- 61.Wang JB, Erickson JW, Fuji R, et al. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell. 2010;18(3):207–219. doi: 10.1016/j.ccr.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; • The initial discovery of 968 by Wang et al. introduced one of two important inhibitors to the field.
- 62.Hu W, Zhang C, Wu R, Sun Y, Levine A, Feng Z. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc. Natl Acad. Sci. USA. 2010;107(16):7455–7460. doi: 10.1073/pnas.1001006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ramachandran S, Pan CQ, Zimmermann SC, et al. Structural basis for exploring the allosteric inhibition of human kidney type glutaminase. Oncotarget. 2016;7(36):57943–57954. doi: 10.18632/oncotarget.10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McDermott LA, Iyer P, Vernetti L, et al. Design and evaluation of novel glutaminase inhibitors. Bioorganic Med. Chem. 2016;24(8):1819–1839. doi: 10.1016/j.bmc.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; • McDermott et al. designed heterocyclic derivatives of bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide (BPTES) with superior pharmacological properties relative to CB-839. This is the only current nonpatent publication describing a large number of such compounds.
- 65.Thangavelu K, Chong QY, Low BC, Sivaraman J. Structural basis for the active site inhibition mechanism of human kidney-type glutaminase (KGA) Sci. Rep. 2014;4:3827. doi: 10.1038/srep03827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ferreira AP, Cassago A, Goncalves K de A, et al. Active glutaminase C self-assembles into a supratetrameric oligomer that can be disrupted by an allosteric inhibitor. J. Biol. Chem. 2013;288(39):28009–28020. doi: 10.1074/jbc.M113.501346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thangavelu K, Pan CQ, Karlberg T, et al. Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism. Proc. Natl Acad. Sci. USA. 2012;109(20):7705–7710. doi: 10.1073/pnas.1116573109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.DeLaBarre B, Gross S, Fang C, et al. Full-length human glutaminase in complex with an allosteric inhibitor. Biochemistry. 2011;50(50):10764–10770. doi: 10.1021/bi201613d. [DOI] [PubMed] [Google Scholar]
- 69.Patel M, McGivan JD. Partial purification and properties of rat liver glutaminase. Biochem. J. 1984;220(2):583–590. doi: 10.1042/bj2200583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Møller M, Nielsen SS, Ramachandran S, et al. Small angle x-ray scattering studies of mitochondrial glutaminase C reveal extended flexible regions, and link oligomeric state with enzyme activity. PLoS ONE. 2013;8(9):e74783. doi: 10.1371/journal.pone.0074783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Katt WP, Cerione RA. Glutaminase regulation in cancer cells: a druggable chain of events. Drug Discov. Today. 2014;19(4):450–457. doi: 10.1016/j.drudis.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lukey MJ, Wilson KF, Cerione RA. Therapeutic strategies impacting cancer cell glutamine metabolism. Future Med. Chem. 2013;5(14):1685–1700. doi: 10.4155/fmc.13.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.DeBerardinis RJ, Cheng T. Q's next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29(3):313–324. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tennant DA, Duran RV, Gottlieb E. Targeting metabolic transformation for cancer therapy. Nat. Rev. Cancer. 2010;10(4):267–277. doi: 10.1038/nrc2817. [DOI] [PubMed] [Google Scholar]
- 75.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat. Rev. Cancer. 2011;11(2):85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 76.Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J. Clin. Invest. 2013;123(9):3678–3684. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Erickson JW, Cerione RA. Glutaminase: a hot spot for regulation of cancer cell metabolism? Oncotarget. 2010;1(8):734–740. doi: 10.18632/oncotarget.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cline MS, Craft B, Swatloski T, et al. Exploring TCGA pan-cancer data at the UCSC Cancer Genomics Browser. Sci. Rep. 2013;3:2652. doi: 10.1038/srep02652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cancer Genome Atlas Research Network. Weinstein JN, Collisson EA, et al. The cancer genome atlas pan-cancer analysis project. Nat. Genet. 2013;45(10):1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Seltzer MJ, Bennett BD, Joshi AD, et al. Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res. 2010;70(22):8981–8987. doi: 10.1158/0008-5472.CAN-10-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Emadi A, Jun SA, Tsukamoto T, Fathi AT, Minden MD, Dang CV. Inhibition of glutaminase selectively suppresses the growth of primary acute myeloid leukemia cells with IDH mutations. Exp. Hematol. 2014;42(4):247–251. doi: 10.1016/j.exphem.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 82.Willems L, Jacque N, Jacquel A, et al. Inhibiting glutamine uptake represents an attractive new strategy for treating acute myeloid leukemia. Blood. 2013;122(20):3521–3532. doi: 10.1182/blood-2013-03-493163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Katt WP, Ramachandran S, Erickson JW, Cerione RA. Dibenzophenanthridines as inhibitors of glutaminase C and cancer cell proliferation. Mol. Cancer Ther. 2012;11(6):1269–1278. doi: 10.1158/1535-7163.MCT-11-0942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gross MI, Demo SD, Dennison JB, et al. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol. Cancer Ther. 2014;13(4):890–901. doi: 10.1158/1535-7163.MCT-13-0870. [DOI] [PubMed] [Google Scholar]; • Introduces CB-839, a potent BPTES derivative currently considered to be the best-in-class molecule for inhibiting GLS.
- 85.Huang F, Zhang Q, Ma H, Lv Q, Zhang T. Expression of glutaminase is upregulated in colorectal cancer and of clinical significance. Int. J. Clin. Exp. Pathol. 2014;7(3):1093–1100. [PMC free article] [PubMed] [Google Scholar]
- 86.Gameiro PA, Yang J, Metelo AM, et al. In vivo HIF-mediated reductive carboxylation is regulated by citrate levels and sensitizes VHL-deficient cells to glutamine deprivation. Cell Metab. 2013;17(3):372–385. doi: 10.1016/j.cmet.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shroff EH, Eberlin LS, Dang VM, et al. MYC oncogene overexpression drives renal cell carcinoma in a mouse model through glutamine metabolism. Proc. Natl Acad. Sci. USA. 2015;112(21):6539–6544. doi: 10.1073/pnas.1507228112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.van den Heuvel AP, Jing J, Wooster RF, Bachman KE. Analysis of glutamine dependency in non-small cell lung cancer. Cancer Biol. Ther. 2012;13(12):1185–1194. doi: 10.4161/cbt.21348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hernandez-Davies JE, Tran TQ, Reid MA, et al. Vemurafenib resistance reprograms melanoma cells towards glutamine dependence. J. Transl. Med. 2015;13(1):1–11. doi: 10.1186/s12967-015-0581-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Roy S, Maity P. Modulation of metastatic potential of B16F10 melanoma cells by acivicin: synergistic action of glutaminase and potentiation of cisplatin cytotoxicity. Asian Pacific J. Cancer Prev. 2007;8(2):301–306. [PubMed] [Google Scholar]
- 91.Son J, Lyssiotis CA, Ying H, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496(7443):101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cheng T, Sudderth J, Yang C, et al. Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proc. Natl Acad. Sci. USA. 2011;108(21):8674–8679. doi: 10.1073/pnas.1016627108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lu W, Pelicano H, Huang P. Cancer metabolism: is glutamine sweeter than glucose? Cancer Cell. 2010;18(3):199–200. doi: 10.1016/j.ccr.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yudkoff M, Pleasure D, Cregar L, et al. Glutathione turnover in cultured astrocytes: studies with [15N]glutamate. J. Neurochem. 1990;55(1):137–145. doi: 10.1111/j.1471-4159.1990.tb08831.x. [DOI] [PubMed] [Google Scholar]
- 95.Shanware NP, Mullen AR, DeBerardinis RJ, Abraham RT. Glutamine: pleiotropic roles in tumor growth and stress resistance. J. Mol. Med. 2011;89(3):229–236. doi: 10.1007/s00109-011-0731-9. [DOI] [PubMed] [Google Scholar]
- 96.Gao P, Tchernyshyov I, Chang TC, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458(7239):762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Martín-Rufián M, Nascimento-Gomes R, Higuero A, et al. Both GLS silencing and GLS2 overexpression synergize with oxidative stress against proliferation of glioma cells. J. Mol. Med. 2014;92(3):277–290. doi: 10.1007/s00109-013-1105-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Antonyak MA, Wilson KF, Cerione RA. R(h)oads to microvesicles. Small GTPases. 2013;3(4):219–224. doi: 10.4161/sgtp.20755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Santana SM, Antonyak MA, Cerione RA, Kirby BJ. Cancerous epithelial cell lines shed extracellular vesicles with a bimodal size distribution that is sensitive to glutamine inhibition. Phys. Biol. 2014;11(6):65001. doi: 10.1088/1478-3975/11/6/065001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gaglio D, Metallo CM, Gameiro PA, et al. Oncogenic K-Ras decouples glucose and glutamine metabolism to support cancer cell growth. Mol. Sys. Biol. 2011;7(1):1–15. doi: 10.1038/msb.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Weinberg F, Hamanaka R, Wheaton WW, et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc. Natl Acad. Sci. USA. 2010;107(19):87889–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Davidson SM, Papagiannakopoulos T, Olenchock BA, et al. Environment impacts the metabolic dependencies of Ras-driven non-small cell lung cancer. Cell Metab. 2016;23(3):517–528. doi: 10.1016/j.cmet.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hensley CT, Faubert B, Yuan Q, et al. Metabolic heterogeneity in human lung tumors. Cell. 2016;164(4):681–694. doi: 10.1016/j.cell.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Laterza OF, Curthoys NP. Effect of acidosis on the properties of the glutaminase mRNA pH-response element binding protein. J. Am. Soc. Nephrol. 2000;11(9):1583–1588. doi: 10.1681/ASN.V1191583. [DOI] [PubMed] [Google Scholar]
- 105.Katt WP, Antonyak MA, Cerione RA. Simultaneously targeting tissue transglutaminase and kidney type glutaminase sensitizes cancer cells to acid toxicity and offers new opportunities for therapeutic intervention. Mol. Pharm. 2015;12(1):46–55. doi: 10.1021/mp500405h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Huang W, Choi W, Chen Y, et al. A proposed role for glutamine in cancer cell growth through acid resistance. Cell Res. 2013;23(5):724–727. doi: 10.1038/cr.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wise DR, DeBerardinis RJ, Mancuso A, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl Acad. Sci. USA. 2008;105(48):18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rathore MG, Saumet A, Rossi JF, et al. The NF-kappa B member p65 controls glutamine metabolism through miR-23a. Int. J. Biochem. Cell Biol. 2012;44(9):1448–1456. doi: 10.1016/j.biocel.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 109.Reynolds MR, Lane AN, Robertson B, et al. Control of glutamine metabolism by the tumor suppressor Rb. Oncogene. 2014;33(5):556–566. doi: 10.1038/onc.2012.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.McGuirk S, Gravel SP, Deblois G, et al. PGC-1α supports glutamine metabolism in breast cancer. Cancer Metab. 2013;1(1):1–11. doi: 10.1186/2049-3002-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lukey MJ, Greene KS, Erickson JW, Wilson KF, Cerione RA. The oncogenic transcription factor c-Jun regulates glutaminase expression and sensitizes cells to glutaminase-targeted therapy. Nat. Commun. 2016;7:11321. doi: 10.1038/ncomms11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sebastián C, Zwaans BM, Silberman DM, et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell. 2012;151(6):1185–1199. doi: 10.1016/j.cell.2012.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Polletta L, Vernucci E, Carnevale I, et al. SIRT5 regulation of ammonia-induced autophagy and mitophagy. Autophagy. 2015;11(2):253–270. doi: 10.1080/15548627.2015.1009778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Redis RS, Vela LE, Lu W, et al. Allele-specific reprogramming of cancer metabolism by the long non-coding RNA CCAT2. Mol. Cell. 2016;61(4):520–534. doi: 10.1016/j.molcel.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Szeliga M, Obara-Michlewska M, Matyja E, et al. Transfection with liver-type glutaminase cDNA alters gene expression and reduces survival, migration and proliferation of T98G glioma cells. Glia. 2009;57(9):1014–1023. doi: 10.1002/glia.20825. [DOI] [PubMed] [Google Scholar]
- 116.Szeliga M, Zgrzywa A, Obara-Michlewska M, Albrecht J. Transfection of a human glioblastoma cell line with liver-type glutaminase (LGA) down-regulates the expression of DNA-repair gene MGMT and sensitizes the cells to alkylating agents. J. Neurochem. 2012;123(3):428–436. doi: 10.1111/j.1471-4159.2012.07917.x. [DOI] [PubMed] [Google Scholar]
- 117.Xiang L, Xie G, Liu C, et al. Knock-down of glutaminase 2 expression decreases glutathione, NADH, and sensitizes cervical cancer to ionizing radiation. Biochim. Biophys. Acta. 2013;1833(12):2996–3005. doi: 10.1016/j.bbamcr.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 118.Suzuki S, Tanaka T, Poyurovsky MV, et al. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc. Natl Acad. Sci. USA. 2010;107(16):7461–7466. doi: 10.1073/pnas.1002459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lamonte G, Tang X, Chen JL, et al. Acidosis induces reprogramming of cellular metabolism to mitigate oxidative stress. Cancer Metab. 2013;1(1):23. doi: 10.1186/2049-3002-1-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Xiao D, Ren P, Su H, et al. Myc promotes glutaminolysis in human neuroblastoma through direct activation of glutaminase 2. Oncotarget. 2015;6(38):40655–40666. doi: 10.18632/oncotarget.5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang R, Dillon CP, Shi LZ, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35(6):871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Thomas AG, O'Driscoll CM, Bressler J, Kaufmann W, Rojas CJ, Slusher BS. Small molecule glutaminase inhibitors block glutamate release from stimulated microglia. Biochem. Biophys. Res. Commun. 2014;443(1):32–36. doi: 10.1016/j.bbrc.2013.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pinkus LM. Glutamine binding sites. Methods Enzymol. 1977;46:414–427. doi: 10.1016/s0076-6879(77)46049-x. [DOI] [PubMed] [Google Scholar]
- 124.Robinson MM, McBryant SJ, Tsukamoto T, et al. Novel mechanism of inhibition of rat kidney-type glutaminase by bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide (BPTES) Biochem. J. 2007;406(3):407–414. doi: 10.1042/BJ20070039. [DOI] [PMC free article] [PubMed] [Google Scholar]; • The discovery by Robinson and coworkers of the GLS inhibitor BPTES, which has subsequently been used in numerous preclinical studies.
- 125.Shukla K, Ferraris DV, Thomas AG, et al. Design, synthesis, and pharmacological evaluation of Bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide 3 (BPTES) analogs as glutaminase inhibitors. J. Med. Chem. 2012;55(23):10551–10563. doi: 10.1021/jm301191p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bennett MK, Gross MI, Bromley SD, et al. 2014. WO/2014/089048.
- 127.Zimmermann SC, Wolf EF, Luu A, et al. Allosteric glutaminase inhibitors based on a 1,4-di(5-amino-1,3,4-thiadiazol-2-yl)butane scaffold. ACS Med. Chem. Lett. 2016;7(5):520–524. doi: 10.1021/acsmedchemlett.6b00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Newcomb R, Newcomb M. 2002. US20020115698A1.
- 129.Di Francesco ME, Jones P, Heffernan T, et al. 2016. WO2016004404A2.
- 130.Di Francesco ME, Heffernan T, Soth MJ, et al. 2016. US20160002248A1.
- 131.Di Francesco ME, Jones P, Heffernan T, et al. 2016. WO2016004417A1.
- 132.Lemieux RM, Popovici-Muller J, Salituro FG, Saunders JO, Travins J, Yan S. 2014. p. 54. US20140142146A1.
- 133.Lemieux RM, Popovici-Muller J, Salituro FG, Saunders JO, Travins J, Chen Y. 2014 US20140142081A1. [Google Scholar]
- 134.Bhavar PK, Vakkalanka SKVS, Viswanadha S, Swaroop MG, Babu G. 2015. WO2015101958A2.
- 135.Finlay MRV, Ekwuru CT, Charles MD, Raubo PA, Winter JJG, Nissink JWM. 2015. WO2015181539A1.
- 136.Cianchetta G, Lemieux RM, Cao S, Ding Y, Ye Z. 2015. WO2015143340A1.
- 137.Mackinnon AL, Rodriguez ML. 2016. WO2016014890A1.
- 138.Stalnecker CA, Ulrich SM, Li Y, et al. Mechanism by which a recently discovered allosteric inhibitor blocks glutamine metabolism in transformed cells. Proc. Natl Acad. Sci. USA. 2014;112(2):394–399. doi: 10.1073/pnas.1414056112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gao M, Monian P, Quadri N, Ramasamy R, Jiang X. Glutaminolysis and transferrin regulate ferroptosis. Mol. Cell. 2016;59(2):298–308. doi: 10.1016/j.molcel.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Newell AMB, Yousef GG, Lila MA, Ramírez-Mares MV, Gonzalez de Mejia E. Comparative in vitro bioactivities of tea extracts from six species of Ardisia and their effect on growth inhibition of HepG2 cells. J. Ethnopharmacol. 2010;130(3):536–544. doi: 10.1016/j.jep.2010.05.051. [DOI] [PubMed] [Google Scholar]
- 141.Chang H-S, Lin Y-J, Lee S-J, et al. Cytotoxic alkyl benzoquinones and alkyl phenols from Ardisia virens . Phytochemistry. 2009;70(17–18):2064–2071. doi: 10.1016/j.phytochem.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 142.Yu CC, Wu PJ, Hsu J-L, et al. Ardisianone, a natural benzoquinone, efficiently induces apoptosis in human hormone-refractory prostate cancers through mitochondrial damage stress and survivin downregulation. Prostate. 2013;73(2):133–145. doi: 10.1002/pros.22548. [DOI] [PubMed] [Google Scholar]