Abstract
Background:
Pancreatic ductal adenocarcinoma (PDA) is a lethal cancer with complex genomes and dense fibrotic stroma. This study was designed to identify clinically relevant somatic aberrations in pancreatic cancer genomes of patients with primary and metastatic disease enrolled and treated in two clinical trials.
Methods:
Tumour nuclei were flow sorted prior to whole genome copy number variant (CNV) analysis. Targeted or whole exome sequencing was performed on most samples. We profiled biopsies from 68 patients enrolled in two Stand Up to Cancer (SU2C)-sponsored clinical trials. These included 38 resected chemoradiation naïve tumours (SU2C 20206-003) and metastases from 30 patients who progressed on prior therapies (SU2C 20206-001). Patient outcomes including progression-free survival (PFS) and overall survival (OS) were observed.
Results:
We defined: (a) CDKN2A homozygous deletions that included the adjacent MTAP gene, only its’ 3′ region, or excluded MTAP; (b) SMAD4 homozygous deletions that included ME2; (c) a pancreas-specific MYC super-enhancer region; (d) DNA repair-deficient genomes; and (e) copy number aberrations present in PDA patients with long-term (⩾ 40 months) and short-term (⩽ 12 months) survival after surgical resection.
Conclusions:
We provide a clinically relevant framework for genomic drivers of PDA and for advancing novel treatments.
Keywords: pancreatic cancer, flow sorting, CNVs, homozygous deletions, focal amplicons, KRAS mutations
Pancreatic ductal adenocarcinoma (PDA) is projected to be the second leading cause of cancer deaths by 2020 and over the following decade (Rahib et al, 2014). Surgical resection remains the only modality capable of delivering curative outcomes for PDA patients. However, only ∼20% of PDA patients are candidates for surgical resection, and of these ∼80% are unable to achieve disease-free survival of >4 years. Most recurrences in surgically resected patients arise within 2 years and are almost universally fatal, typically within 1–2 years of recurrent disease (Neoptolemos et al, 2010; Oettle et al, 2013). Most patients are diagnosed with advanced metastatic disease adding to the dismal prognosis. Recent clinical trials have reported improved responses for subsets of patients highlighting the potential for discovery of somatic events in the variable landscape of PDA genomes that can be exploited for improved patient outcomes (Conroy et al, 2011; Von Hoff et al, 2013). However, the identification of drivers of clinical responses is challenged by the complexity and variability of PDA genomes and the heterogeneous cellularity and dense stroma present in tissue samples, as cancer cells represent on average only 25% of the cells within the tumour (Seymour et al, 1994; Evers et al, 2014). Furthermore, biopsies from patients who have received one or more prior therapies often contain high levels of necrosis and cellular debris.
The diverse mutational landscape of PDA genomes has been catalogued (Waddell et al, 2015; Bailey et al, 2016). Integrated analyses of genomic data with associated expression patterns have identified subtypes that correlate with histopathological features and may drive clinical outcomes (Collisson et al, 2011; Biankin and Maitra, 2015). These genomic-based research studies of PDA typically rely on optimally collected resection specimens with relatively high (>40–50%) levels of tumour cells. Various approaches have been used to extend genomic studies to bio specimens with low tumour content and high levels of necrosis (Jones et al, 2008; Song et al, 2012; Moffitt et al, 2015). These include patient-derived cell lines and xenografts, in silico tumour purity determination based on sequencing reads for KRAS mutations, and algorithms designed to deconvolute single nucleotide polymorphism (SNPs)-based signatures prior to sequencing. However additional processing steps and reliance on specialised algorithms for biospecimen evaluation and data interpretation limits the application of these methods for PDA biopsies collected during clinical trials.
To address the challenges associated with routinely collected clinical biopsies, we have validated DNA content-based flow sorting of diploid, tetraploid and aneuploid tumour populations in PDA and other solid tumours (Ruiz et al, 2011; Holley et al, 2012). Our methods yield purified (>95%) tumour samples for whole genome analyses from a variety of clinical samples. These include fresh frozen and formalin fixed paraffin embedded tissues with low tumour content (<10–20%) and high amounts (>90%) of necrosis and debris. In this current study we applied our flow sorting-based methods to PDA biopsies collected from 68 patients enrolled in two Stand Up to Cancer (SU2C)-sponsored clinical trials. These included 38 surgically resected chemoradiation naïve tumours (SU2C 20206-003) and metastases, primarily from liver, from 30 patients with advanced disease who had progressed on prior therapies (SU2C 20206-001). Patients were followed prospectively and clinical outcomes including progression-free survival (PFS) and overall survival (OS) were observed. Here we show that by combining prospectively collected clinical data, clonal genomic analyses and employing novel information analytical tools for data integration, we can provide insights into the genomic landscapes and clinical phenotypes of primary and metastatic PDA. This integrated approach enables linkage of data to external sources and databases. Our initial results include the detection of homozygous deletions targeting known and novel tumour suppressor genes, the mapping of a PDA MYC amplicon cassette containing a cluster of pancreas-specific super enhancers, the identification of DNA repair defects including a BRCA-like copy number aberration signature, and the identification of genomic landscapes associated with improved and limited survival after resection.
Methods
Clinical samples
Fresh frozen PDA samples were obtained under a WIRB protocol (20040832) for AACR/SU2C-sponsored clinical trials (20206-001 and 20206-003). All patients gave informed consent for collection and use of all the samples which were collected in liquid nitrogen and stored at −80 °C. All tumour samples were histopathologically evaluated prior to genomic analysis. All research conformed to the Helsinki Declaration (https://www.wma.net/policies-post/wma-declaration-of-helsinki-ethical-principles-for-medical-research-involving-human-subjects/).
Metastatic patients 20206-001
The SU2C 20206-001 trial was a phase II study of therapy selected by molecular profiling in patients with previously treated metastatic disease. Eligible patients (⩾18 years of age) had a diagnosis of metastatic PDA and had received ⩾1 prior therapy for the treatment of metastatic disease. One or more metastatic tumours had to be measurable by CT scan and be accessible for a tumour biopsy. Other pertinent eligibility criteria were acceptable bone marrow, kidney and liver function, prior therapy must have been completed ⩾3 weeks before starting study and side effects of prior therapy must have resolved to ⩽grade 1. A Karnofsky performance status (KPS)⩾70 was required. All patients signed an institutional review board approved consent form before participating in any study-related activities. Patients who met eligibility criteria underwent a percutaneous CT or US guided biopsy of an accessible metastatic lesion using an 18 gauge needle with at least three passes. Treatment decision was determined by immunohistochemistry (IHC). Commercially available treatment regimens prescribed included FOLFIRI, FOLFOX, irinotecan and doxorubicin. The response (RECIST) was 9%. The median survival was 5.6 months (94% CI 3.8–7.2) and the 1-year survival was 20% (95% CI, 7–33%). A tissue from each metastatic core biopsy was pooled, then flash frozen before flow sorting and array-based comparative genomic hybridisation (aCGH).
Surgical resection patients 20206-003
The SU2C 20206-003 trial was a tissue acquisition protocol to support metabolomic and genetic profiling for predicting patient outcomes in early-stage resectable pancreatic cancer. Eligible patients (⩾18 years of age) had a diagnosis of primary pancreatic cancer and did not receive any prior therapies. Informed consent for research use was obtained from all patients at the enrolling institution (University of Pennsylvania) before tissue banking and study approval was obtained. PDA specimens and matched germline specimens (from peripheral blood) from patients were acquired for genomic analyses. Primary tumour samples for genomic analyses were selected from patients with resectable stage I, II, or III disease, and reviewed by histopathological assessment.
Flow cytometry and PDA biopsies
Biopsies were minced in the presence of NST buffer and DAPI according to published protocols (Rabinovitch et al, 2001; Holley et al, 2012). Nuclei were disaggregated then filtered through a 40 μm mesh before flow sorting with an Influx cytometer (Becton-Dickinson, San Jose, CA, USA) with ultraviolet excitation and DAPI emission collected at >450 nm. DNA content and cell cycle were analysed using the software program MultiCycle (Phoenix Flow Systems, San Diego, CA, USA).
Copy number analysis of flow sorted PDA biopsies
DNAs were extracted using Qiagen micro kits (Qiagen, Valencia, CA, USA). For each hybridisation 100 ng of genomic DNA from each sample and of pooled commercial 46XX reference (Promega, Madison, WI, USA) were amplified using the GenomiPhi amplification kit (GE Healthcare, Piscataway, NJ, USA). Subsequently 1 μg of amplified sample and 1μg of amplified reference template were digested with DNaseI then labelled with Cy-5 dUTP and Cy-3 dUTP respectively, using a BioPrime labelling kit (Invitrogen, Carlsbad, CA, USA). All labelling reactions were assessed using a Nanodrop assay (Nanodrop, Wilmington, DE, USA) prior to mixing and hybridisation to CGH arrays with 400 000 or 1 000 000 oligonucleotide features (Agilent Technologies, Santa Clara, CA, USA).
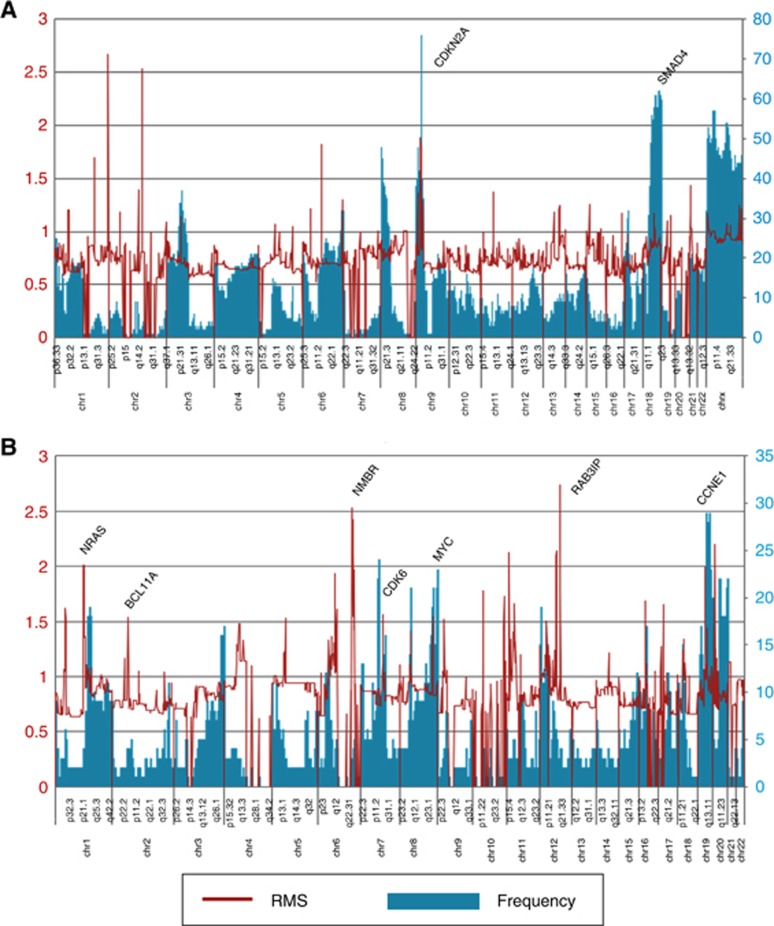
All microarray slides were scanned using an Agilent 2565C DNA scanner and the images were analysed with Agilent Feature Extraction (FE) version 11.0 using default settings according to our published data (Barrett et al, 2015). The aCGH data were assessed with a series of FE QC metrics then analysed using an aberration detection algorithm (ADM2; Lipson et al, 2006). The latter identifies all aberrant intervals in a given sample with consistently high or low log ratios based on the statistical score derived from the average normalised log ratios of all probes in the genomic interval multiplied by the square root of the number of these probes. This score represents the deviation of the average of the normalised log ratios from its expected value of zero and is proportional to the height h (absolute average log ratio) of the genomic interval, and to the square root of the number of probes in the interval. The values for each sample were then imported into ISSAC’s ILLUMINATIVE Analytics platform. For deletions a threshold of log2ratio<−0.5 was initially applied to identify regions of recurrent loss (Figure 1A). We then used a window of 10 kb and a threshold of log2ratio<−2.0 for ADM2 defined intervals with<−3.0 for individual probes to identify homozygous deletions. We used the root mean square (RMS) of the log2ratio genomic interval values with a sliding window of 0.5 Mb and a threshold of log2ratio >0.6 to identify high level and recurring amplicons (Figure 1B). These data were integrated with KRAS (30 metastases) and cancer panel (5 primaries) targeted resequencing, and with whole exome sequencing (25 primaries).
Figure 1.
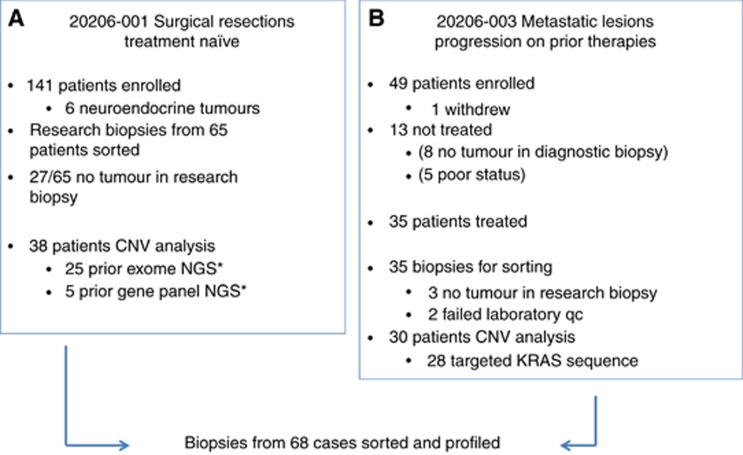
Summary of patients enrolled and samples profiled in SU2C clinical trials. (A) Copy number variant (CNV) analysis was performed on flow sorted primary pancreas cancer samples from 38 patients enrolled in SU2C 20206-003. Next generation sequencing (NGS) data were available from 25 of the 38 samples. (B) CNV analysis was obtained from 30 patients with metastatic pancreatic cancer. Targeted KRAS mutation analysis was done on 28 of these metastatic samples. *Sausen et al., Nat Commun 6: 7686.
Sequencing
From the needle biopsies in the 20206-001 trial, mutation analysis of selected regions for KRAS was performed using Sanger sequencing with M13-linked polymerase chain reaction primers. polymerase chain reaction products were bidirectionally sequenced using the BigDye Terminator v1.1 chemistry (Applied Biosystems, Grand Island, NY, USA), and analysed using the 3730 DNA Analyser (Applied Biosystems). Sequence traces were analyzed using Mutation Surveyor software v3.25 (Soft Genetics, San Francisco, CA, USA). Samples from the 20206-003 trial were processed for either whole exome (n=25) or targeted resequencing with a 111 gene cancer panel (n=5) according to our published methods (Sausen et al, 2015).
Super-enhancer analysis
Genomic intervals from the ADM2 derived CGH analyses were used to interrogate pancreas-specific features within the Super-Enhancer Archive (SEA) http://www.bio-bigdata.com/SEA/index.html (Wei et al, 2016). The response incorporated detailed (epi)genetic information, incorporating cell type specificity, nearby genes, transcriptional factor binding sites, CRISPR/Cas9 target sites, evolutionary conservation, SNPs, H3K27ac, DNA methylation, gene expression and transcription factor ChIP-sequence data.
Data integration and analysis
Genomic results, treatment reports and outcome data from trials 20206-001 and 20206-003 were fused and analysed using The Health Analytic (THA) powered by ILLUMINATIVE Analytics (ISSAC Corp). To perform intra- and inter-trial analysis, data were ingested, fused, analysed and reported-on, iteratively. Data were ingested via loose-heuristic parsing into a novel graph structure supported by SQL and fused and related across multiple dimensions via exact value matching (cardinal and categorical types), similarity assessment (real numbered values) and concept resolution (string types) (Talburt, 2011).
Results
Tumour content of pancreas biopsies
There were 141 patients enrolled in SU2C 20206-003 trial. Biopsies from 65 resections were available for genomic analysis and processed for flow cytometry (Figure 1). Based on combined flow cytometry and copy number analysis we detected ⩾1–5% tumour in the research biopsy in 38/65 (58.5%) of these optimally collected resected cases including two pancreatic neuroendocrine tumours (PNETs). Given the superior outcomes for patients with neuroendocrine tumours and the absence of KRAS mutations the PNET samples were analysed separately from the PDA cases (Jiao et al, 2011). Forty-nine patients were accrued for the advanced previously treated SU2C 20206-001 trial. One patient withdrew consent prior to a biopsy, and 48 were scheduled and underwent a biopsy without major complications. Thirteen patients did not start protocol therapy either due to insufficient tumour for analysis on biopsy (n=8) or due to worsening cancer-related symptoms after biopsy which precluded further treatment (n=5). A total of 35 metastatic PDA biopsies from treated patients were processed with flow cytometry and aCGH. In three of these cases there was no tumour detected whereas two additional cases failed quality control metrics in the aCGH analysis. A final total of 68 prospectively collected tumour biopsies (38 resected and 30 metastases) were included in this study. Tumour content in the biopsies varied extensively (<5% to >50%). However, the use of flow sorted samples enabled objective thresholds for the identification of genomic lesions and for the discrimination of homozygous deletions and focal amplicons in each tumour.
Selected genomic lesions
Mutations
KRAS codon 12 mutations were detected by targeted resequencing in 27 of 28 (96%) metastatic PDA samples. Strikingly we detected a novel homozygous deletion affecting RASA1; a negative regulator of wild-type KRAS, in the one confirmed case that was KRAS wild type (Supplementary Figure 1). Thirty resected cases were sequenced including 25 by whole exome as previously described (Sausen et al, 2015). Twenty-seven (90%) of these had a detectable KRAS mutation. In addition, mutations in TP53 (77% patients) and in multiple chromatin regulation and modification genes (MLL1, MLL2, MLL3 and ARID1A) were detected in the resected patient samples (Sausen et al, 2015).
Copy number deletions
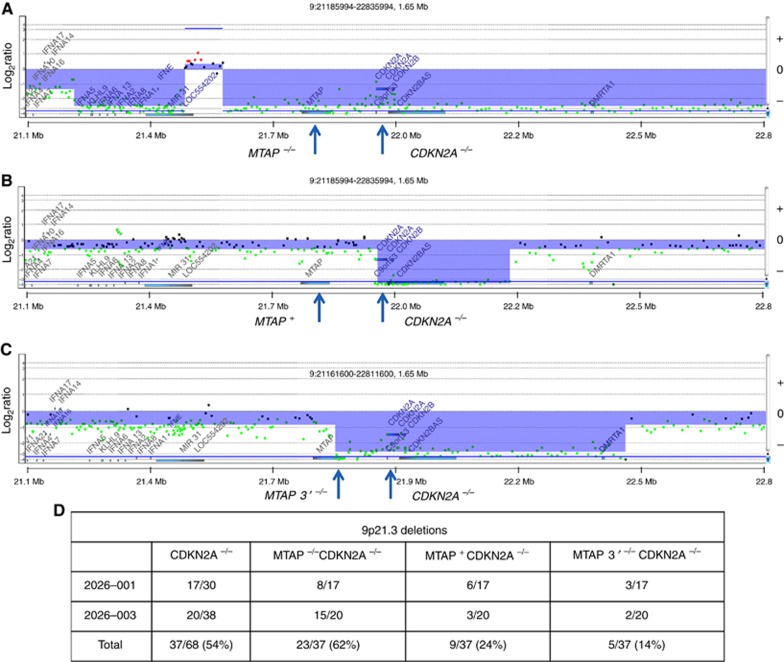
Two of the most frequent copy number aberrations were losses of 9p21 in 53/68 patients (78%) and 18q21 in 58/68 patients (85%) (Figure 2A). These included homozygous deletions targeting CDKN2A (9p21.3) in 37/68 cases (54%) and SMAD4 (18q21.1) in 3/68 cases (4.0%), tumour suppressor genes with known roles in pancreatic cancer. The homozygous 9p21.3 deletions targeting CDKN2A varied in length from 76 kb to 7.6 mb and included methylthioadenosine phosphorylase (MTAP) which maps approximately 100 kb proximal to CDKN2A in 23/37 of these cases (62%) while 3/37 cases (8.0%) had homozygous deletions that extended only to the 3′ region of MTAP (Figure 3). The remaining 11/37 cases (30%) had at least one intact copy of MTAP. The three cases with SMAD4 homozygous deletions also included the adjacent malic enzyme 2 (ME2) locus, a context that may be exploited through a synthetic lethal targeting of ME3 (Dey et al, 2017). Homozygous deletions were also present in genes within common fragile sites including FHIT, WWOX, and in multiple tumour suppressor genes including unique deletions in RB1, MAP2K4 and MSH6 (Supplementary Figure 2).
Figure 2.
Genome-wide view of recurring copy number aberrations in PDA genomes. (A) Deletions: The root mean square (RMS) of the log2ratio genomic interval values with a sliding window of 0.5 Mb and a threshold of log2ratio<−0.5 was initially applied to identify regions of recurrent loss. A window of 10 kb and a threshold of log2ratio<−2.0 for ADM2 defined intervals with<−3.0 for individual probes was used to identify homozygous deletions. (B) Amplicons: The root mean square (RMS) of the log2ratio genomic interval values with a sliding window of 0.5 Mb and a threshold of log2ratio >0.6 was used to identify high level and recurring amplicons.
Figure 3.
Chromosome 9p24.1 homozygous deletions. Homozygous deletions were detected and mapped that include (A) CDKN2A and MTAP in patient 20206-003 041, (B) CDKN2A in patient 20206-003 039, and (C) CDKN2A and the 3′ region of MTAP in patient 20206-001 015. Blue shade areas denote ADM2 defined genomic intervals. (D) Summary of homozygous 9p21.3 deletions in the SU2C trials.
The resected tumour with the MSH6 homozygous deletion which was not associated with a germ line variant had over 400 somatic nucleotide variants (SNVs) in the exome. This level of SNVs is consistent with microsatellite instability (MSI). MSI represents a clinical phenotype that has been exploited therapeutically with immune checkpoint blockade in a variety of indications (Le et al, 2015). However, MSI PDAs are very rare and their clinical significance has not been validated (Laghi et al, 2012). Other novel homozygous deletions detected in our cohort included those within a series of chromatin and transcriptional regulators including SMYD3, L3MBTL4, PRDM15, KLF13, ASXL2 and MLL3. The latter two genes also had mutations in one and four cases respectively highlighting their role and that of deregulated transcriptional programming in PDA. Additional homozygous deletions were detected in genes with a variety of known and putative biological roles including chromosome stability (STAG1, FRY), cell signalling (RGS7), autophagy (TRIM13) and cell migration (SLITRK6, SLITRK5).
Copy number amplifications
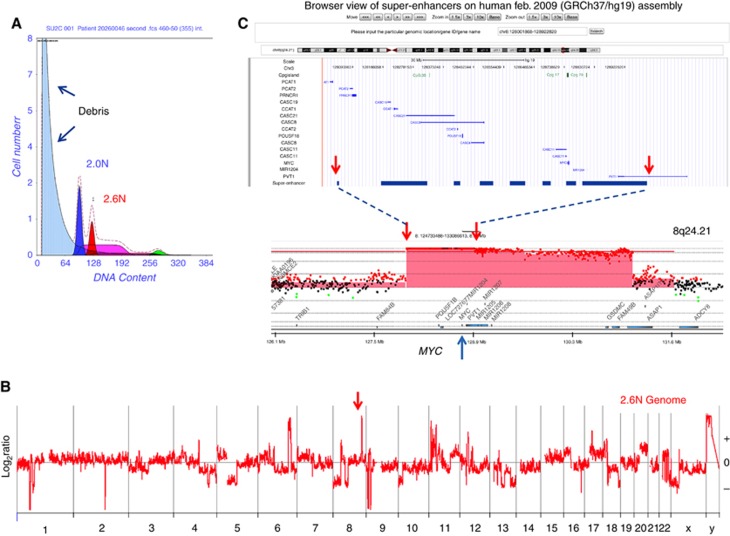
The most frequent amplicons included chromosomes 8q24.3 in 21/68 (31%) patients and 19q12 in 29/68 (43%) patients (Figure 2B). These amplicons included focal events (RMS>1.5) whose shortest region of overlap (SRO) targeted c-MYC and CCNE1 respectively. We used the coordinates of the amplicon SROs to interrogate the SEA database and identified a pancreas-specific super-enhancer region that is co-amplified with the c-MYC driver oncogene (Figure 4). In contrast the SRO of the CCNE1 amplicon did not contain any known pancreas-specific super-enhancers. Additional recurring high-level amplicons targeted chromosomes 12q15 in 4/68 (6.0%) of patients and 7q21 in 10/68 (15%) patients. The 7q21 amplicon included CDK6 and was present in a genome with a co-occurring CDKN2A homozygous deletion suggesting these two selected oncogenic copy number events arose independently of each other and have non-overlapping cancer phenotypes (Supplementary Figure 3). Strikingly the two most focal 12q15 amplicons included MDM2 but were present in cases with mutations in TP53, including the R175H gain of function, suggesting that a gene other than MDM2 was the target of this selected event (Muller and Vousden, 2014). Notably the guanine exchange factor (GEF) RAB3IP, which can activate RAS superfamily members RAB8A and RAB8B, maps to the SRO of this amplicon.
Figure 4.
Shortest region of overlap (SRO) of 8q24.1 MYC amplicon and superhancer loci. (A) Flow sort histogram of biopsy from patient 2026-001 046. (B) Whole genome CNV plot of sorted 2.6N PDA population. (C) Pancreas-specific superhancer loci and SRO of MYC amplicon mapped using genome browser and genome build HG19. Red shaded areas denote ADM2 defined amplicon in 20206-001 046.
Clinical outcomes for chemoradiation naïve surgical resection patients (20206-003)
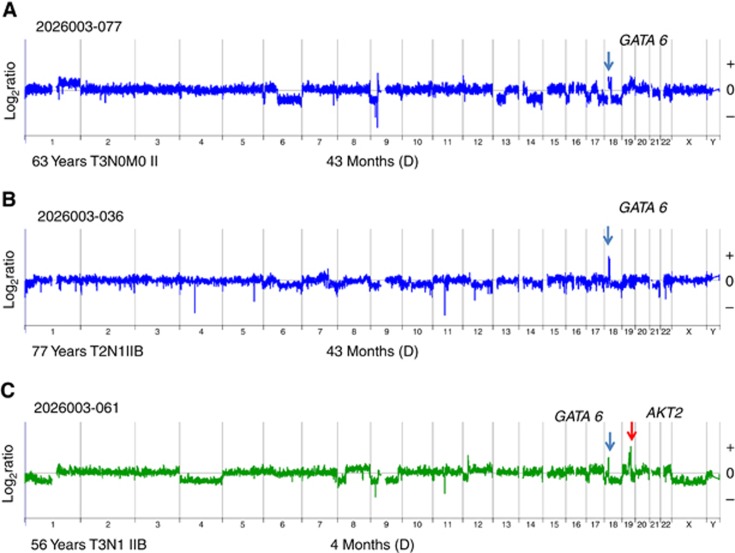
Fifty-three per cent of the evaluable patients were males with an age range of 47–85 (median 66 years). None of the individual recurring copy number aberrations was significantly associated with clinical outcomes in this trial. Furthermore there was no association with increased number of copy number aberrations and OS after surgical resection. However to investigate potential correlates of survivorship we examined those cases with the most positive and the most negative outcomes post surgery. There were six PDA patients who underwent surgical resection who survived at least 40 months after surgery followed by adjuvant chemotherapy (Table 1A). The genomic profiles for these six patients included the mismatch DNA repair phenotype associated with a MSH6 homozygous deletion, two separate tumours with co-occurring high-level amplicons (log2ratio >3.0) targeting CCNE1 and CTPS, and two patients with a focal GATA6 amplicon as the most significant copy gain in their tumour (Supplementary Figures 2, 4) (Figure 5A and B). These genomic profiles were unique to this subset of resected PDAs.
Table 1. Survival SU2C 20206-003.
| Patient | Stage | Months | D/Aa | Genomics | Age/Sex | Adjuvant |
|---|---|---|---|---|---|---|
|
(A) Long term >40 months | ||||||
| 077 | IIa T3N0 | 43 | D | 18q11 GATA6+++b | 63 XX | Y |
| 034 | IIb T3N1 | 41 | D | MSH6-/-c MSI+ | 76 XX | Y |
| 062 | IIb T3N1 | 48 | A | 19q11 CCNE+++ | 70 XY | Y |
| 038 | IIa T3N0 | 41 | D | 1p34.2 CTPS+++ | 72 XX | Y |
| 036 | IIb T2N1 | 43 | D | 18q11 GATA6+++ | 77 XX | Y |
| 035 | IIa T3N0 | 51 | A | BRCA2969Q>X | 72 XX | Y |
|
(B) Short term ⩽12 months | ||||||
| 115 | IIb | 10 | D | 18q-/+d | 63 XX | ? |
| 039 | IIa T3N0 | 12 | D | SMAD4-/- | 65 XY | Y |
| 090 | IIb T3N1 | 8 | D | 18q-/+ | 59 XY | Y |
| 087 | T3N1 | 12 | D | 18q-/+ | 69 XY | Y |
| 061 | IIb T3N1 | 4 | D | AKT2+++ SMAD4 insertion | 56 XX | Y |
| 021 | IIa T3N0 | 9 | D | SMAD4361R>H | 82 XX | N |
| 042 | IIb T3N1 | 8 | D | 18q-/+ | 58 XY | Y |
D, dead; A, alive.
+++ Amplified.
−/− Homozygous deletion.
−/+ Partial loss.
Figure 5.
GATA6 amplification and long-term 20206-003 survivors. Patients (A) 077 and (B) 036 from 20206-003 trial had ⩾40-month survival post surgery. Each had a GATA6 amplicon as the most significant copy number aberration in the genomes of their PDA. (C) In contrast patient the PDA genome of patient 061, 4-month survival after surgery, had additional AKT2 amplicon that exceeded the height of the co-occurring GATA6 amplicon.
Seven PDA patients survived less than 12 months despite surgical resection and adjuvant treatment (Table 1B). The genomic profiles of these seven short-term survivors shared heterozygous deletions of 18q spanning the SMAD4 locus. These included two somatic mutations and a homozygous deletion consistent with previous studies associating inactivation of SMAD4 with the development of a more aggressive cancer phenotype in the pancreas (Supplementary Figure 5) (Iacobuzio-Donahue et al, 2009). However each of the six long-term survivors also had heterozygous loss of 18q and 2/4 cases that were sequenced had somatic mutations in SMAD4 suggesting that additional genomic and/or epigenetic events contribute to the aggressiveness of disease.
There were two cases (5%) of resected PDA that had HER2 amplicons as their most significant copy number aberration. One of these cases subsequently developed multiple low-attenuation masses in the residual body and tail of pancreas with a large mass in the tail that spread around the spleen into the subdiaphragmatic region 11 months after surgery. The prevalence and clinical history is similar to previous reports of HER2 positive PDA (Chou et al, 2013).
The outcomes for the six PNET patients were uniformly superior when compared to the PDAs with each surviving beyond 46 months after surgery. Five of the six patients with PNETs had no evidence of disease even without receiving any adjuvant therapy. The fifth patient relapsed at 13 months post surgery but achieved a complete response to chemotherapy and was reported alive after more than 60 months from time of surgery. The biopsies from two of these cases contained tumour cells that were sorted and profiled for copy number (Supplementary Figure 6). The two neuroendocrine genomes each had whole chromosome loss of chromosome 11 and gain of chromosome 5, two distinct features of this histologic subtype (Zhao et al, 2001). In addition we did not detect KRAS mutations in either of the two PNETs that were sequenced. Strikingly one of these sequenced tumours had a homozygous deletion of SMYD3 (SET and MYND domain containing 3) histone methyltransferase.
Clinical outcomes for advanced previously treated patients (20206-001)
The demographics of evaluable patients (n=35) were males (55%), the majority had KPS of 70–80 and age range was 34–81 (median 63 years). Time from first diagnosis of metastatic PDA to biopsy ranged from 5.8 to 26.7 months (median 16.1 months). All patients had prior gemcitabine-based therapy with a median of two prior regimens (range 1–6). The time from patient signing consent to performing the biopsy was a median of 4 days (range 1–9 days) and from biopsy to availability of the IHC report was a median of 13 days (range 4–16 days). Time on study ranged from 0 to 8.6 months (median 1.3 months, 95% CI 1–1.6 months). All patients have died, with survival ranging from 0.4 months to 26.8 months. The median PFS was 2.4 months (95% CI 2.1–3.9 months) and median survival was 5.6 months (94% CI 3.8–7.2 months). The 1-year survival was 20% (95% CI, 7–33%) for all 35 patients. In the study population (n=35) nine patients had stable disease and there were three partial responses (9%); these patients received FOLFOX (20206–001 023), FOLFIRI (20206-001 029) and nab-paclitaxel/capecitabine (20206-001 033) for a disease control rate of 35% (95% CI, 19–54%). The 30 metastatic tumour samples that were successfully sorted and profiled included 24 patients who received treatment. Tumour biopsies from two of the three patients who achieved partial responses were evaluated. In both cases the 1p34.2 amplicon (CTPS) and the 18q11 amplicon (GATA6) detected in primary tumours of long-term survivors were present as co-occurring aberrations in the metastatic PDA genomes (Supplementary Figure 7).
Discussion
The genomic landscapes of primary and metastatic human pancreatic tumours have been surveyed in detail (Waddell et al, 2015; Bailey et al, 2016). However most of these studies do not include clinical trial biopsies and prospective outcomes. The two SU2C studies provide an opportunity to profile both newly diagnosed surgically resected patients (SU2C 20026-003) and those with advanced metastatic disease who had progressed on prior therapies (SU2C 20206-001). Our data provide a framework for developing new hypotheses for the drivers of clinical outcomes in PDA and the development of effective therapeutic strategies.
CDKN2A homozygous deletions
Our detailed mapping of homozygous deletions targeting CDKN2A in flow sorted clinical samples (54% of cases) reveals that these events have been under reported in conventional bulk tumour analyses (Waddell et al, 2015; Bailey et al, 2016). Recent reports suggest that co-deletion of MTAP with CDKN2A disrupts tumour methionine metabolism and creates a synthetic lethal condition that can be exploited clinically (Kryukov et al, 2016; Mavrakis et al, 2016). Given the high prevalence of CDKN2A homozygous deletions in both primary and metastatic lesions, PDA represents a favourable clinical model for future studies to test this therapeutic concept. Notably, homozygous deletions of CDKN2A may extend through MTAP (62%), include only the 3′ region (8.0%), or exclude MTAP altogether (30%) (Figure 3).This level of precision in genomic studies is needed to develop effective personalised therapies for patients with pancreatic cancer.
Genomic lesions targeting G1/S regulators
The loss of G1/S regulation is a hallmark of most solid tumours including PDA. This can arise as a consequence of multiple genomic and epigenomic lesions including deletions of CDK inhibitors, amplification of CDKs and disruption of cell cycle checkpoints. Accordingly, in addition to highly prevalent CDKN2A homozygous deletions we observed events such as homozygous deletions targeting RB1 and amplification of CDK6 in our cohort. The development of small molecule inhibitors of CDKs has shown promise in clinical trials (Asghar et al, 2015; Finn et al, 2015). Inhibitors with dual specificity for CDK4/6 are being advanced to target those tumours with deregulated G1/S control. Elevated CDK6 activity is associated with different solid tumours (Mendrzyk et al, 2005; Ismail et al, 2011; Sherr et al, 2016). The role of CDK6 in regulating G1/S is believed to be synonymous with CDK4. Therefore high level CDK6 amplification and homozygous deletion of CDKN2A should be mutually exclusive. Given the high prevalence of CDKN2A homozygous deletions PDAs represent a highly favourable clinical model for advancing CDK inhibitors. However the co-occurrence of these two highly selected lesions suggests that CDK6 amplification can drive additional tumour pathways beyond the cell cycle (Supplementary Figure 3). Notably, CDK6 has also been shown to have a transcriptional role in tumour angiogenesis (Kollmann et al, 2013).
The high definition detection and mapping of homozygous deletions identified other novel contexts that can be used to inform new therapeutic strategies. For example mouse studies have implicated SMYD3 as an activator of MEK signalling suggesting that SMYD3 represents a highly favourable clinical target for RAS-driven tumours including those arising in the pancreas (Mazur et al, 2014). Furthermore, the complete loss of SMYD3 function has no visible phenotype in mice. However the presence of a focal SMYD3 homozygous deletion in a PNET suggests a tumour suppressor role for this histone lysine methyltransferase in KRAS wild-type tumours that needs to be addressed in future clinical studies.
MYC amplification
Mapping of recurring MYC amplicons identified an SRO for this frequently amplified driver oncogene that targets a pancreas-specific super-enhancer region. Super-enhancer specific amplicons have been identified from TCGA data sets for multiple cancers (Zhang et al, 2016). Notably an amplified super enhancer that drives MYC expression was mapped distal to the PVT1 locus in lung adenocarcinoma and endometrial carcinoma. We did not detect this super-enhancer-specific amplicon in our flow sorted PDA data. Rather the PDA MYC amplicon consisted of a cassette-like structure that when highly amplified retained distinct core pancreas-specific super-enhancer regions spanning the PVT1 locus and loci proximal to MYC. Copy number profiles of 30 established PDA cell lines also mapped the MYC PDA super-enhancer to this same amplified region in PSN1 cells which retained the most focal and the highest level MYC amplicon.
Recent preclinical studies have demonstrated that inhibitors of transcriptional activation-driven through super-enhancers may have clinical activity in a variety of tumours (Chipumuro et al, 2014; Christensen et al, 2014). Super-enhancers associated with MYC have been shown to be highly sensitive to small molecules that reduce levels of the transcriptional activator BRD4 (Loven et al, 2013). The extensive occupancy and high density of BRD4 binding sites associated with the MYC super-enhancer in tumour cells creates an oncogenic dependency that may be exploited therapeutically. The presence of high level focal MYC amplicons in PDA that include pancreas-specific super-enhancers provides a biomarker and a target for emerging classes of compounds that aim to disrupt the ordered complexes that are required to drive MYC expression in tumour genomes. The extent of MYC amplification and expression in PDA may provide an increased therapeutic index for those patients with this amplicon. Of significant interest will be to follow the occupancy and access of the MYC super-enhancers with CHiP-seq and ATAC-seq assays using sorted tumour and non-tumour fractions from biopsies to be collected in our upcoming trials.
GATA6 amplification
Genomic aberrations, including both gains and losses, targeting 18q11 are frequent in PDA (Waddell et al, 2015; Bailey et al, 2016). Amplification of 18q11 that includes the GATA6 locus has been reported in gastrointestinal malignancies including those arising in the pancreas (Fu et al, 2008; Lin et al, 2012; Sulahian et al, 2014). Functional studies suggest that GATA6 can activate oncogenic signalling during development of pancreatic cancer (Zhong et al, 2011). However, GATA6 amplification has also been associated with a favourable prognosis after resection and adjuvant therapy (Martinelli et al, 2016a). GATA6 was also identified as a suppressor of KRASG12V-driven PDA and as a target in a sleeping beauty screen in KRAS-driven mouse models (Perez-Mancera et al, 2012; Martinelli et al, 2016b). In addition, recent studies suggest that GATA6 overexpression promotes epithelial cell differentiation while suppressing inflammatory pathways (Martinelli et al, 2016a, 2016b). Thus the role of GATA6 in pancreatic cancer and its’ clinical utility remain to be determined. Two of the long-term survivor cases in the 20206-003 trial had GATA6 amplification as the most significant (i.e. ‘driver’) copy number aberration in their tumour genomes (Table 1, Figure 5A and B). GATA6 driver amplicons were also present in two out of three patients with advanced metastatic disease who achieved partial responses after chemotherapy (Supplementary Figure 7). These data suggest that GATA6 driver amplicons may be exploited for improved clinical outcomes.
Homologous recombination deficient PDA
In addition to specific genomic lesions the overall burden and nature of copy number aberrations represents a potential clinical biomarker for PDA (Bailey et al, 2016). Specifically, the presence of elevated numbers of interstitial aberrations (IAs) throughout a genome is associated with a homologous recombination deficiency (HRD) in BRCA mutant tumours (Lord and Ashworth, 2016). A summary of the patients in the 20206-001 trial showed a range of <10 to >70 in the number of IAs in each sorted tumour population (Supplementary Figure 8). The IAs were defined by the ADM2 step gram algorithm as copy number aberrant intervals with intrachromosomal boundaries (Lipson et al, 2006). These included amplicons, deletions and homozygous losses in the tumour genome. Patient 006 had a pathogenic BRCA2 germ line variant and a PDA genome with 54 IAs that included 20 of 22 autosomes. Notably the highest number of aberrations was observed in the sorted aneuploid population from patient 046. The latter had a stable disease response to an early PARP inhibitor prior to enrolment in the SU2C trial. Strikingly this patient was wild type for BRCA1 and BRCA2. The same HRD signature was observed in a subset of primary PDAs including a patient with a somatic BRCA2Q969X mutation (Supplementary Figure 2B). A fundamental hypothesis from these and other genomic data is that a subset of PDAs, both BRCA mutant and BRCA wild type, have a HRD phenotype and will be responsive to both targeted agents and broad-based chemotherapies that either disrupt DNA repair or increase the burden of DNA double-strand breaks. Our early phase clinical trial of newly diagnosed metastatic PDA combining cisplatin with gemcitabine and nab-paclitaxel has shown over 70% response rate including 30% complete responses (Jameson et al, 2017). These genomic signatures are being combined with strategies targeting other components of PDA tissues, including stroma, stem cells and immune cells, to improve the extent and duration of clinical responses.
Conclusion
We have developed a well-annotated clinical data set using purified flow sorted pancreatic cancer biopsy samples for genomic analyses. These include high resolution mapping of CDKN2A and SMAD4 homozygous deletions and of MYC amplicons, DNA repair deficient tumours and targeted genomic lesions such as GATA6 amplification in patients with positive outcomes in the adjuvant setting. These data provide hypotheses for current models of genomic drivers of this deadly disease as well as the framework for advancing novel treatments for patients.
Acknowledgments
Supported by the Stand Up to Cancer (SU2C-AACR) Dream Team Translational Research Grant, a Program of the Entertainment Industry Foundation (SU2C-AACR-DT0509).
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
The authors declare no conflict of interest.
Supplementary Material
References
- Asghar U, Witkiewicz AK, Turner NC, Knudsen ES (2015) The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat rev Drug discov 14(2): 130–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, Miller DK, Christ AN, Bruxner TJ, Quinn MC, Nourse C, Murtaugh LC, Harliwong I, Idrisoglu S, Manning S, Nourbakhsh E, Wani S, Fink L, Holmes O, Chin V, Anderson MJ, Kazakoff S, Leonard C, Newell F, Waddell N, Wood S, Xu Q, Wilson PJ, Cloonan N, Kassahn KS, Taylor D, Quek K, Robertson A, Pantano L, Mincarelli L, Sanchez LN, Evers L, Wu J, Pinese M, Cowley MJ, Jones MD, Colvin EK, Nagrial AM, Humphrey ES, Chantrill LA, Mawson A, Humphris J, Chou A, Pajic M, Scarlett CJ, Pinho AV, Giry-Laterriere M, Rooman I, Samra JS, Kench JG, Lovell JA, Merrett ND, Toon CW, Epari K, Nguyen NQ, Barbour A, Zeps N, Moran-Jones K, Jamieson NB, Graham JS, Duthie F, Oien K, Hair J, Grutzmann R, Maitra A, Iacobuzio-Donahue CA, Wolfgang CL, Morgan RA, Lawlor RT, Corbo V, Bassi C, Rusev B, Capelli P, Salvia R, Tortora G, Mukhopadhyay D, Petersen GM Australian Pancreatic Cancer Genome IMunzy DM, Fisher WE, Karim SA, Eshleman JR, Hruban RH, Pilarsky C, Morton JP, Sansom OJ, Scarpa A, Musgrove EA, Bailey UM, Hofmann O, Sutherland RL, Wheeler DA, Gill AJ, Gibbs RA, Pearson JV, Waddell N, Biankin AV, Grimmond SM (2016) Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 531(7592): 47–52. [DOI] [PubMed] [Google Scholar]
- Barrett MT, Anderson KS, Lenkiewicz E, Andreozzi M, Cunliffe HE, Klassen CL, Dueck AC, McCullough AE, Reddy SK, Ramanathan RK, Northfelt DW, Pockaj BA (2015) Genomic amplification of 9p24.1 targeting JAK2, PD-L1, and PD-L2 is enriched in high-risk triple negative breast cancer. Oncotarget 6(28): 26483–26493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biankin AV, Maitra A (2015) Subtyping pancreatic cancer. Cancer Cell 28(4): 411–413. [DOI] [PubMed] [Google Scholar]
- Chipumuro E, Marco E, Christensen CL, Kwiatkowski N, Zhang T, Hatheway CM, Abraham BJ, Sharma B, Yeung C, Altabef A, Perez-Atayde A, Wong KK, Yuan GC, Gray NS, Young RA, George RE (2014) CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell 159(5): 1126–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou A, Waddell N, Cowley MJ, Gill AJ, Chang DK, Patch AM, Nones K, Wu J, Pinese M, Johns AL, Miller DK, Kassahn KS, Nagrial AM, Wasan H, Goldstein D, Toon CW, Chin V, Chantrill L, Humphris J, Mead RS, Rooman I, Samra JS, Pajic M, Musgrove EA, Pearson JV, Morey AL, Grimmond SM, Biankin AV (2013) Clinical and molecular characterization of HER2 amplified-pancreatic cancer. Genome Med 5(8): 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen CL, Kwiatkowski N, Abraham BJ, Carretero J, Al-Shahrour F, Zhang T, Chipumuro E, Herter-Sprie GS, Akbay EA, Altabef A, Zhang J, Shimamura T, Capelletti M, Reibel JB, Cavanaugh JD, Gao P, Liu Y, Michaelsen SR, Poulsen HS, Aref AR, Barbie DA, Bradner JE, George RE, Gray NS, Young RA, Wong KK (2014) Targeting transcriptional addictions in small cell lung cancer with a covalent CDK7 inhibitor. Cancer Cell 26(6): 909–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, Cooc J, Weinkle J, Kim GE, Jakkula L, Feiler HS, Ko AH, Olshen AB, Danenberg KL, Tempero MA, Spellman PT, Hanahan D, Gray JW (2011) Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med 17(4): 500–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conroy T, Desseigne F, Ychou M, Bouche O, Guimbaud R, Becouarn Y, Adenis A, Raoul JL, Gourgou-Bourgade S, de la Fouchardiere C, Bennouna J, Bachet JB, Khemissa-Akouz F, Pere-Verge D, Delbaldo C, Assenat E, Chauffert B, Michel P, Montoto-Grillot C, Ducreux M (2011) FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J med 364(19): 1817–1825. [DOI] [PubMed] [Google Scholar]
- Dey P, Baddour J, Muller F, Wu CC, Wang H, Liao WT, Lan Z, Chen A, Gutschner T, Kang Y, Fleming J, Satani N, Zhao D, Achreja A, Yang L, Lee J, Chang E, Genovese G, Viale A, Ying H, Draetta G, Maitra A, Wang YA, Nagrath D, DePinho RA (2017) Genomic deletion of malic enzyme 2 confers collateral lethality in pancreatic cancer. Nature 542(7639): 119–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers L, Perez-Mancera PA, Lenkiewicz E, Tang N, Aust D, Knosel T, Rummele P, Holley T, Kassner M, Aziz M, Ramanathan RK, Von Hoff DD, Yin H, Pilarsky C, Barrett MT (2014) STAG2 is a clinically relevant tumor suppressor in pancreatic ductal adenocarcinoma. Genome Med 6(1): 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RS, Crown JP, Lang I, Boer K, Bondarenko IM, Kulyk SO, Ettl J, Patel R, Pinter T, Schmidt M, Shparyk Y, Thummala AR, Voytko NL, Fowst C, Huang X, Kim ST, Randolph S, Slamon DJ (2015) The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): a randomised phase 2 study. lancet oncol 16(1): 25–35. [DOI] [PubMed] [Google Scholar]
- Fu B, Luo M, Lakkur S, Lucito R, Iacobuzio-Donahue CA (2008) Frequent genomic copy number gain and overexpression of GATA-6 in pancreatic carcinoma. Cancer Biol Ther 7(10): 1593–1601. [DOI] [PubMed] [Google Scholar]
- Holley T, Lenkiewicz E, Evers L, Tembe W, Ruiz C, Gsponer JR, Rentsch CA, Bubendorf L, Stapleton M, Amorese D, Legendre C, Cunliffe HE, McCullough AE, Pockaj B, Craig D, Carpten J, Von Hoff D, Iacobuzio-Donahue C, Barrett MT (2012) Deep clonal profiling of formalin fixed paraffin embedded clinical samples. PLoS One 7(11): e50586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobuzio-Donahue CA, Fu B, Yachida S, Luo M, Abe H, Henderson CM, Vilardell F, Wang Z, Keller JW, Banerjee P, Herman JM, Cameron JL, Yeo CJ, Halushka MK, Eshleman JR, Raben M, Klein AP, Hruban RH, Hidalgo M, Laheru D (2009) DPC4 gene status of the primary carcinoma correlates with patterns of failure in patients with pancreatic cancer. J Clin Oncol 27(11): 1806–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail A, Bandla S, Reveiller M, Toia L, Zhou Z, Gooding WE, Kalatskaya I, Stein L, D'Souza M, Litle VR, Peters JH, Pennathur A, Luketich JD, Godfrey TE (2011) Early G(1) cyclin-dependent kinases as prognostic markers and potential therapeutic targets in esophageal adenocarcinoma. Clin Cancer Res 17(13): 4513–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson GS, Borazanci EH, Babiker HM, Poplin E, Niewiarowska AA, Gordon MS, Barrett MT, Ansaldo K, Lebron L, Stoll AC, Rosenthal A, Shemanski LR, Korn RL, Ramanathan RK, Von Hoff DD (2017) A phase Ib/II pilot trial with nab-paclitaxel+gemcitabine+cisplatin in patients (pts) with stage IV pancreatic cancer. In: 2017 Gastrointestinal Cancers Symposium p Abstract 175849 ASCO: San Francisco, CA, USA. [Google Scholar]
- Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, Schulick RD, Tang LH, Wolfgang CL, Choti MA, Velculescu VE, Diaz LA Jr, Vogelstein B, Kinzler KW, Hruban RH, Papadopoulos N (2011) DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science 331(6021): 1199–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, Hong SM, Fu B, Lin MT, Calhoun ES, Kamiyama M, Walter K, Nikolskaya T, Nikolsky Y, Hartigan J, Smith DR, Hidalgo M, Leach SD, Klein AP, Jaffee EM, Goggins M, Maitra A, Iacobuzio-Donahue C, Eshleman JR, Kern SE, Hruban RH, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW (2008) Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321(5897): 1801–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmann K, Heller G, Schneckenleithner C, Warsch W, Scheicher R, Ott RG, Schafer M, Fajmann S, Schlederer M, Schiefer AI, Reichart U, Mayerhofer M, Hoeller C, Zochbauer-Muller S, Kerjaschki D, Bock C, Kenner L, Hoefler G, Freissmuth M, Green AR, Moriggl R, Busslinger M, Malumbres M, Sexl V (2013) A kinase-independent function of CDK6 links the cell cycle to tumor angiogenesis. Cancer Cell 24(2): 167–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryukov GV, Wilson FH, Ruth JR, Paulk J, Tsherniak A, Marlow SE, Vazquez F, Weir BA, Fitzgerald ME, Tanaka M, Bielski CM, Scott JM, Dennis C, Cowley GS, Boehm JS, Root DE, Golub TR, Clish CB, Bradner JE, Hahn WC, Garraway LA (2016) MTAP deletion confers enhanced dependency on the PRMT5 arginine methyltransferase in cancer cells. Science 351(6278): 1214–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laghi L, Beghelli S, Spinelli A, Bianchi P, Basso G, Di Caro G, Brecht A, Celesti G, Turri G, Bersani S, Schumacher G, Rocken C, Grantzdorffer I, Roncalli M, Zerbi A, Neuhaus P, Bassi C, Montorsi M, Scarpa A, Malesci A (2012) Irrelevance of microsatellite instability in the epidemiology of sporadic pancreatic ductal adenocarcinoma. PLoS One 7(9): e46002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, Biedrzycki B, Donehower RC, Zaheer A, Fisher GA, Crocenzi TS, Lee JJ, Duffy SM, Goldberg RM, de la Chapelle A, Koshiji M, Bhaijee F, Huebner T, Hruban RH, Wood LD, Cuka N, Pardoll DM, Papadopoulos N, Kinzler KW, Zhou S, Cornish TC, Taube JM, Anders RA, Eshleman JR, Vogelstein B, Diaz LA Jr (2015) PD-1 blockade in tumors with mismatch-repair deficiency. N Engl j med 372(26): 2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Bass AJ, Lockwood WW, Wang Z, Silvers AL, Thomas DG, Chang AC, Lin J, Orringer MB, Li W, Glover TW, Giordano TJ, Lam WL, Meyerson M, Beer DG (2012) Activation of GATA binding protein 6 (GATA6) sustains oncogenic lineage-survival in esophageal adenocarcinoma. Proc Natl Acad Sci USA 109(11): 4251–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipson D, Aumann Y, Ben-Dor A, Linial N, Yakhini Z (2006) Efficient calculation of interval scores for DNA copy number data analysis. J Comput Biol 13(2): 215–228. [DOI] [PubMed] [Google Scholar]
- Lord CJ, Ashworth A (2016) BRCAness revisited. Nat rev Cancer 16(2): 110–120. [DOI] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA (2013) Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153(2): 320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli P, Carrillo-de Santa Pau E, Cox T, Sainz B Jr, Dusetti N, Greenhalf W, Rinaldi L, Costello E, Ghaneh P, Malats N, Buchler M, Pajic M, Biankin AV, Iovanna J, Neoptolemos J, Real FX (2016. a) GATA6 regulates EMT and tumour dissemination, and is a marker of response to adjuvant chemotherapy in pancreatic cancer. Gut 2016; e-pub ahead of print 20 January 2016; doi:10.1136/gutjnl-2015-311256. [DOI] [PMC free article] [PubMed]
- Martinelli P, Madriles F, Canamero M, Pau EC, Pozo ND, Guerra C, Real FX (2016. b) The acinar regulator Gata6 suppresses KrasG12V-driven pancreatic tumorigenesis in mice. Gut 65(3): 476–486. [DOI] [PubMed] [Google Scholar]
- Mavrakis KJ, McDonald ER 3rd, Schlabach MR, Billy E, Hoffman GR, deWeck A, Ruddy DA, Venkatesan K, Yu J, McAllister G, Stump M, deBeaumont R, Ho S, Yue Y, Liu Y, Yan-Neale Y, Yang G, Lin F, Yin H, Gao H, Kipp DR, Zhao S, McNamara JT, Sprague ER, Zheng B, Lin Y, Cho YS, Gu J, Crawford K, Ciccone D, Vitari AC, Lai A, Capka V, Hurov K, Porter JA, Tallarico J, Mickanin C, Lees E, Pagliarini R, Keen N, Schmelzle T, Hofmann F, Stegmeier F, Sellers WR (2016) Disordered methionine metabolism in MTAP/CDKN2A-deleted cancers leads to dependence on PRMT5. Science 351(6278): 1208–1213. [DOI] [PubMed] [Google Scholar]
- Mazur PK, Reynoird N, Khatri P, Jansen PW, Wilkinson AW, Liu S, Barbash O, Van Aller GS, Huddleston M, Dhanak D, Tummino PJ, Kruger RG, Garcia BA, Butte AJ, Vermeulen M, Sage J, Gozani O (2014) SMYD3 links lysine methylation of MAP3K2 to Ras-driven cancer. Nature 510(7504): 283–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendrzyk F, Radlwimmer B, Joos S, Kokocinski F, Benner A, Stange DE, Neben K, Fiegler H, Carter NP, Reifenberger G, Korshunov A, Lichter P (2005) Genomic and protein expression profiling identifies CDK6 as novel independent prognostic marker in medulloblastoma. J Clin Oncol 23(34): 8853–8862. [DOI] [PubMed] [Google Scholar]
- Moffitt RA, Marayati R, Flate EL, Volmar KE, Loeza SG, Hoadley KA, Rashid NU, Williams LA, Eaton SC, Chung AH, Smyla JK, Anderson JM, Kim HJ, Bentrem DJ, Talamonti MS, Iacobuzio-Donahue CA, Hollingsworth MA, Yeh JJ (2015) Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet 47(10): 1168–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PA, Vousden KH (2014) Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell 25(3): 304–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neoptolemos JP, Stocken DD, Bassi C, Ghaneh P, Cunningham D, Goldstein D, Padbury R, Moore MJ, Gallinger S, Mariette C, Wente MN, Izbicki JR, Friess H, Lerch MM, Dervenis C, Olah A, Butturini G, Doi R, Lind PA, Smith D, Valle JW, Palmer DH, Buckels JA, Thompson J, McKay CJ, Rawcliffe CL, Buchler MW European Study Group for Pancreatic C (2010) Adjuvant chemotherapy with fluorouracil plus folinic acid vs gemcitabine following pancreatic cancer resection: a randomized controlled trial. Jama 304(10): 1073–1081. [DOI] [PubMed] [Google Scholar]
- Oettle H, Neuhaus P, Hochhaus A, Hartmann JT, Gellert K, Ridwelski K, Niedergethmann M, Zulke C, Fahlke J, Arning MB, Sinn M, Hinke A, Riess H (2013) Adjuvant chemotherapy with gemcitabine and long-term outcomes among patients with resected pancreatic cancer: the CONKO-001 randomized trial. Jama 310(14): 1473–1481. [DOI] [PubMed] [Google Scholar]
- Perez-Mancera PA, Rust AG, van der Weyden L, Kristiansen G, Li A, Sarver AL, Silverstein KA, Grutzmann R, Aust D, Rummele P, Knosel T, Herd C, Stemple DL, Kettleborough R, Brosnan JA, Li A, Morgan R, Knight S, Yu J, Stegeman S, Collier LS, ten Hoeve JJ, de Ridder J, Klein AP, Goggins M, Hruban RH, Chang DK, Biankin AV, Grimmond SM Australian Pancreatic Cancer Genome IWessels LF, Wood SA, Iacobuzio-Donahue CA, Pilarsky C, Largaespada DA, Adams DJ, Tuveson DA (2012) The deubiquitinase USP9X suppresses pancreatic ductal adenocarcinoma. Nature 486(7402): 266–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovitch PS, Longton G, Blount PL, Levine DS, Reid BJ (2001) Predictors of progression in Barrett's esophagus III: baseline flow cytometric variables. Am J Gastroenterol 96(11): 3071–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM (2014) Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 74(11): 2913–2921. [DOI] [PubMed] [Google Scholar]
- Ruiz C, Lenkiewicz E, Evers L, Holley T, Robeson A, Kiefer J, Demeure MJ, Hollingsworth MA, Shen M, Prunkard D, Rabinovitch PS, Zellweger T, Mousses S, Trent JM, Carpten JD, Bubendorf L, Von Hoff D, Barrett MT (2011) Advancing a clinically relevant perspective of the clonal nature of cancer. Proc Natl Acad Sci USA 108(29): 12054–12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sausen M, Phallen J, Adleff V, Jones S, Leary RJ, Barrett MT, Anagnostou V, Parpart-Li S, Murphy D, Kay LiQ, Hruban CA, Scharpf R, White JR, O'Dwyer PJ, Allen PJ, Eshleman JR, Thompson CB, Klimstra DS, Linehan DC, Maitra A, Hruban RH, Diaz LA Jr, Von Hoff DD, Johansen JS, Drebin JA, Velculescu VE (2015) Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat Commun 6: 7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour AB, Hruban RH, Redston M, Caldas C, Powell SM, Kinzler KW, Yeo CJ, Kern SE (1994) Allelotype of pancreatic adenocarcinoma. Cancer Res 54(10): 2761–2764. [PubMed] [Google Scholar]
- Sherr CJ, Beach D, Shapiro GI (2016) Targeting CDK4 and CDK6: From Discovery to Therapy. Cancer Discov 6(4): 353–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Nones K, Miller D, Harliwong I, Kassahn KS, Pinese M, Pajic M, Gill AJ, Johns AL, Anderson M, Holmes O, Leonard C, Taylor D, Wood S, Xu Q, Newell F, Cowley MJ, Wu J, Wilson P, Fink L, Biankin AV, Waddell N, Grimmond SM, Pearson JV (2012) qpure: a tool to estimate tumor cellularity from genome-wide single-nucleotide polymorphism profiles. PLoS One 7(9): e45835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulahian R, Casey F, Shen J, Qian ZR, Shin H, Ogino S, Weir BA, Vazquez F, Liu XS, Hahn WC, Bass AJ, Chan V, Shivdasani RA (2014) An integrative analysis reveals functional targets of GATA6 transcriptional regulation in gastric cancer. Oncogene 33(49): 5637–5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talburt JR (2011) Entity Resolution and Information Quality. Elsevier: Burlington. [Google Scholar]
- Von Hoff DD, Ervin T, Arena FP, Chiorean EG, Infante J, Moore M, Seay T, Tjulandin SA, Ma WW, Saleh MN, Harris M, Reni M, Dowden S, Laheru D, Bahary N, Ramanathan RK, Tabernero J, Hidalgo M, Goldstein D, Van Cutsem E, Wei X, Iglesias J, Renschler MF (2013) Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl j med 369(18): 1691–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, Johns AL, Miller D, Nones K, Quek K, Quinn MC, Robertson AJ, Fadlullah MZ, Bruxner TJ, Christ AN, Harliwong I, Idrisoglu S, Manning S, Nourse C, Nourbakhsh E, Wani S, Wilson PJ, Markham E, Cloonan N, Anderson MJ, Fink JL, Holmes O, Kazakoff SH, Leonard C, Newell F, Poudel B, Song S, Taylor D, Wood S, Xu Q, Wu J, Pinese M, Cowley MJ, Lee HC, Jones MD, Nagrial AM, Humphris J, Chantrill LA, Chin V, Steinmann AM, Mawson A, Humphrey ES, Colvin EK, Chou A, Scarlett CJ, Pinho AV, Giry-Laterriere M, Rooman I, Samra JS, Kench JG, Pettitt JA, Merrett ND, Toon C, Epari K, Nguyen NQ, Barbour A, Zeps N, Jamieson NB, Graham JS, Niclou SP, Bjerkvig R, Grutzmann R, Aust D, Hruban RH, Maitra A, Iacobuzio-Donahue CA, Wolfgang CL, Morgan RA, Lawlor RT, Corbo V, Bassi C, Falconi M, Zamboni G, Tortora G, Tempero MA, Gill AJ, Eshleman JR, Pilarsky C, Scarpa A, Musgrove EA, Pearson JV, Biankin AV, Grimmond SM (2015) Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 518(7540): 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Zhang S, Shang S, Zhang B, Li S, Wang X, Wang F, Su J, Wu Q, Liu H, Zhang Y (2016) SEA: a super-enhancer archive. Nucleic acids res 44(D1): D172–D179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Choi PS, Francis JM, Imielinski M, Watanabe H, Cherniack AD, Meyerson M (2016) Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat genet 48(2): 176–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Moch H, Scheidweiler AF, Baer A, Schaffer AA, Speel EJ, Roth J, Heitz PU, Komminoth P (2001) Genomic imbalances in the progression of endocrine pancreatic tumors. Genes chromosomes cancer 32(4): 364–372. [DOI] [PubMed] [Google Scholar]
- Zhong Y, Wang Z, Fu B, Pan F, Yachida S, Dhara M, Albesiano E, Li L, Naito Y, Vilardell F, Cummings C, Martinelli P, Li A, Yonescu R, Ma Q, Griffin CA, Real FX, Iacobuzio-Donahue CA (2011) GATA6 activates Wnt signaling in pancreatic cancer by negatively regulating the Wnt antagonist Dickkopf-1. PLoS One 6(7): e22129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.