Abstract
The respiratory tract is heavily populated with innate immune cells, but the mechanisms that control such cells are poorly defined. Here we found that the E3 ubiquitin ligase TRIM29 was a selective regulator of the activation of alveolar macrophages, the expression of type I interferons and the production of proinflammatory cytokines in the lungs. We found that deletion of TRIM29 enhanced macrophage production of type I interferons and protected mice from infection with influenza virus, while challenge of Trim29−/− mice with Haemophilus influenzae resulted in lethal lung inflammation due to massive production of proinflammatory cytokines by macrophages. Mechanistically, we demonstrated that TRIM29 inhibited interferon-regulatory factors and signaling via the transcription factor NF-κB by degrading the adaptor NEMO and that TRIM29 directly bound NEMO and subsequently induced its ubiquitination and proteolytic degradation. These data identify TRIM29 as a key negative regulator of alveolar macrophages and might have important clinical implications for local immunity and immunopathology.
Acute viral and bacterial infections in the respiratory tract remain major causes of morbidity and mortality, with an estimated 3.5 million annual deaths worldwide1. The innate immune system constitutes the first line of host defense against pathogens2. Among the innate immune cells, macrophages are essential for host defense in the lungs through their ability to survey the exposed airways and regulate innate and adaptive immune responses3. In the lungs, macrophages are the most abundant immune cells under homoeostatic conditions4. Alveolar macrophages (AMs) are found in the airspace of the alveoli and form 90–95% of the cellular content within the alveoli under normal conditions, which makes them the natural ‘gatekeepers’ of the respiratory tract5,6. Indeed, AMs are the main effector cells in innate host defense in the lungs and also have a crucial role in lung development, surfactant homeostasis and immunosurveillance6,7. AMs interact with alveolar epithelial cells, dendritic cells and T cells via a network of cell-surface receptors, chemokines and cytokines to ‘fine tune’ the immune responses to viral, bacterial and fungal infections in the lungs6,7. They exert these effects via phagocytic activities and the secretion of antimicrobial factors, nitric oxide, tumornecrosis factor (TNF) and type I and type II interferons3. Because of their potent phagocytic ability, AMs are essential in clearance of bacteria and fungi, including Streptococcus pneumonia, Mycobacteria tuberculosis, Pseudomonas aeruginosa and Pneumocystis carinii8–11. Among viral respiratory infections, the best-studied example is infection with influenza virus, which is the leading cause of death from infection worldwide12. Some evidence suggests that AMs are critical for protection from respiratory failure and associated morbidity after infection with influenza virus. In animal models, a lack of AMs in mice leads to impaired clearance of virus and increased mortality in response to infection with low doses of influenza virus13–16, while overexpression of the cytokine GM-CSF prevents AM apoptosis and provides resistance to a lethal dose of influenza virus14. AMs are less-efficient producers of infectious viral particles than are alveolar epithelial cells15, but they potently upregulate the production of type I interferons13,16 and thereby interfere with viral spreading17. This ameliorates morbidity through the induction of interferon-inducible antiviral proteins18,19, which inhibit the recruitment of neutrophils as well as the production of proinflammatory cytokines17. Although activation of AMs is critical for killing pathogens, this activation must be tightly regulated for the host to effectively control and eliminate invading microorganisms while limiting unwanted tissue damages. Inappropriate activation and excessive production of cytokines have been linked to severe immunological disorders, such as systemic lupus erythematosus20, asthma21, tissue inflammation and lung damage22. Much effort has been devoted to the development of strategies to control excessive activation of the immune system and tissue damage23. Among those, the ubiquitination and degradation of critical signaling molecules have emerged as a pivotal mechanism in the control of innate immunity24. Here we report that AMs specifically expressed the E3 ubiquitin ligase TRIM29, a member of the ‘tripartite interaction motif ’ (TRIM) family, and that TRIM29 had a crucial role in the regulation of macrophage activation in response to viral and bacterial infections in the respiratory tract.
RESULTS
TRIM29 inhibits cytokine production in AMs
Because of the importance of AMs and the role of ubiquitination in regulating innate immune cells, we screened all 70 members of the TRIM family of E3 ubiquitin ligases in the mouse AM cell line MH-S by a small-interfering-RNA approach. We found that knocking down TRIM29 in MH-S cells led to markedly enhanced (over threefold) production of interferon-β(IFN-β) in response to 5′-triphosphorylated RNA (5′pppRNA), the ligand of RIG-I-like receptors (RLRs)), when delivered via Lipofectamine (data not shown). We confirmed those results by stable knockdown of TRIM29 via short hairpin RNA (shRNA) in this cell line and found that two distinct TRIM29-targeting shRNAs produced efficient knockdown of TRIM29 (Fig. 1a). We then stimulated these cells with 5′pppRNA and measured the production of IFN-β and the cytokine IL-6 by the stimulated cells by ELISA. Consistent with published data25–27, knockdown of the signaling adaptor MAVS via shRNA abrogated the production of both IFN-β and IL-6 in macrophages stimulated by cytosolic 5′pppRNA (Fig. 1b,c). In contrast, knockdown of TRIM29 markedly enhanced the production of IFN-β and IL-6 by macrophages by up to fourfold relative to their production by cells treated with control (non-targeting) shRNA (Fig. 1b,c). These data suggested that TRIM29 negatively regulated the production of IFN-β and IL-6 in AMs in response to 5′pppRNA.
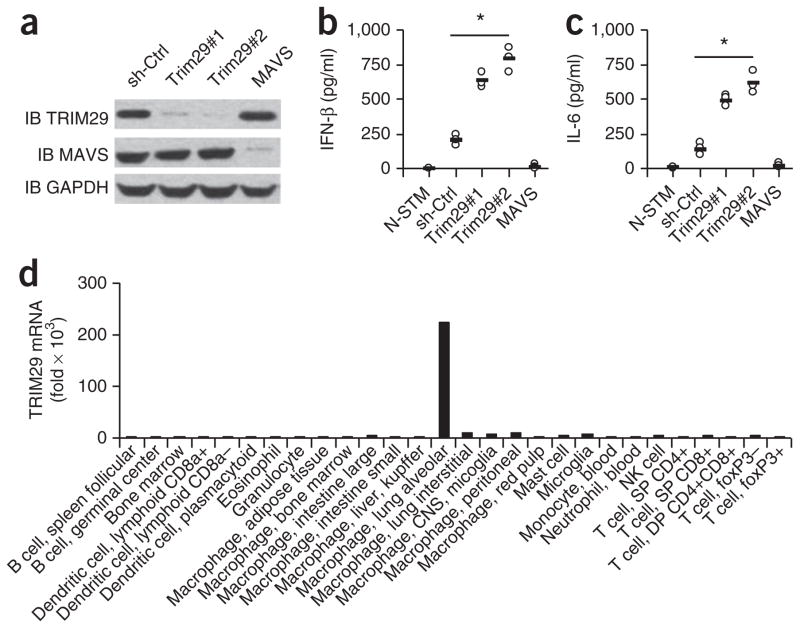
Figure 1.
TRIM29 inhibits cytokine production by AMs in response to 5′ppp RNA. (a) Immunoblot analysis (IB) of TRIM29, MAVS and GAPDH (loading control throughout) in MH-S cells treated with shRNA with a scrambled sequence (as a control (sh-Ctrl)) or shRNA to knock down expression of TRIM29 (Trim29#1 and Trim29#2)) or MAVS (above lanes). (b,c) ELISA of IFN-β (b) and IL-6 (c) in supernatants of MH-S cells left unstimulated and treated with the control shRNA (N-STM) or stimulated for 16 h with 5′pppRNA (1.0 μg/ml) and then treated with shRNA as in a (horizontal axes). Each symbol represents an individual independent experiment; small horizontal lines indicate the average of biological triplicates. (d) Real-time PCR analysis of TRIM29 mRNA in primary cells (horizontal axis) isolated from C57BL/6 mice; results are resented relative to those of GAPDH. *P < 0.0005 (unpaired t-test). Data are representative of three independent experiments with similar results (ac) or two experiments (d).
Sensing of 5′pppRNA, poly(I:C) and RNA viruses by TRIM29
To determine the role of TRIM29 in primary cells, we first analyzed the expression of TRIM29 in primary mouse cells by real-time PCR. We found high expression of TRIM29 in AMs but not in other cell types (Fig. 1d). To further determine the function of TRIM29 in innate immunity, we obtained TRIM29-deficient (Trim29−/−) mice and confirmed loss of TRIM29 in these mice by PCR genotyping and immunoblot analysis (Supplementary Fig. 1a,b). We first isolated primary AMs from wild-type and Trim29−/− mice, then stimulated the cells with 5′pppRNA and the high- molecular-weight synthetic RNA duplex poly(I:C) (poly(I:C) (HMW), the ligand for the RNA helicase and cytosolic receptor MDA5. ELISA showed that Trim29−/− AMs produced three- to fivefold more type I interferons (IFN-α and IFN-β) and proinflammatory cytokines (TNF and IL-6) than wild-type AMs did in response to 5′pppRNA or poly(I:C) at 6 h (Fig. 2a) or 12 h (Fig. 2b,c) after stimulation, indicative of a negative role for TRIM29 in AMs in response to ligands of RLRs.
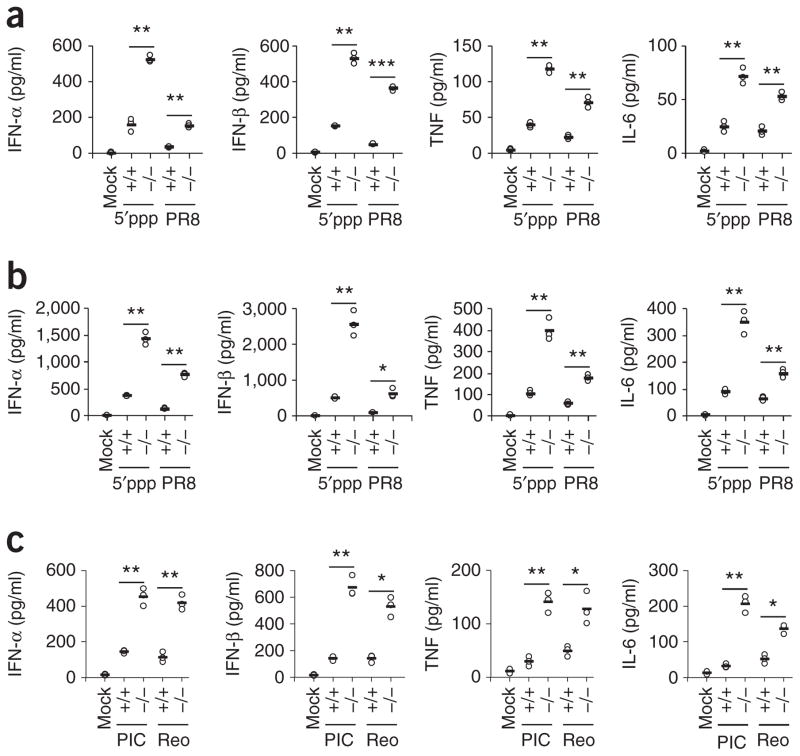
Figure 2.
TRIM29 inhibits the innate immune response to 5′pppRNA, poly(I:C) and RNA viruses in primary AMs. (a,b) ELISA of type I interferons, TNF and IL-6 in supernatants of wild-type AMs without stimulation or infection (Mock) or Trim29+/+ (wild-type) (+/+) and Trim29−/− (−/−) primary AMs stimulated for 6 h (a) or 12 h (b) with 5′pppRNA (5′ppp) (1 μg/ml) or infected for those times with influenza virus at a multiplicity of infection (MOI) of 5 (PR8). (c) ELISA of type I interferons, TNF and IL-6 in supernatants of Trim29+/+ and Trim29−/− primary AMs mock treated (Mock), stimulated for 12 h with high-molecular-weight poly(I:C) (10 μg/ml) (PIC) or infected with reovirus at an MOI of 5 (Reo). Each symbol represents an individual independent experiment; small horizontal lines indicate the average of biological triplicates. *P < 0.005, **P < 0.0005 and ***P < 0.0001 (unpaired t-test). Data are representative of three independent experiments with similar results.
Infection with influenza virus produces typical 5′pppRNA and/or double-stranded RNA, while infection with reovirus produces mainly double-stranded RNA28. We therefore used influenza A virus (PR8 strain) and reovirus (type 3, Dearing strain) to investigate the role of TRIM29 in AMs. We isolated AMs from wild-type and Trim29−/− mice and infected the cells with influenza virus or reovirus. Trim29-deficient AMs produced three- to sevenfold more type I interferons and proinflammatory cytokines at 6 h (Fig. 2a) or 12h (Fig. 2b,c) after infection than did wild-type cells. Similar results were obtained at 20 h after infection by influenza virus (Supplementary Fig. 2a–d). The primary AMs from wild-type and Trim29−/− mice were similar in term of the expression of differentiation markers (Supplementary Fig. 2e). To determine whether TRIM29 has a role in regulating other cytokines and chemokines, we isolated primary AMs from wild-type and Trim29−/− mice and stimulated the cells with 5′pppRNA or influenza virus. Trim29-deficient AMs produced two- to fivefold more chemokines, including MIP-1α (Supplementary Fig. 3a), CCL-2, CCL-5 and CXCL-2 (Supplementary Fig. 3b), than did wild-type cells. In contrast, knockout of TRIM29 had no effect on the production of IL-10 (Supplementary Fig. 3c), IL-4, IL-5 or IL-13 (Supplementary Fig. 3d). To determine whether TRIM29 negatively regulates cytokine production in other cell types, we isolated peritoneal macrophages, splenic macrophages and CD11c+ splenic dendritic cells from wild-type mice and Trim29−/− mice and stimulated the cells with 5′pppRNA or influenza virus. We found that TRIM29 deficiency had no effect on IL-6 production in response to 5′pppRNA in peritoneal macrophages (Supplementary Fig. 4a) or in splenic macrophages (Supplementary Fig. 4b). In contrast, TRIM29-deficient CD11c+ splenic dendritic cells produced twofold more type I interferons and proinflammatory cytokines in response to 5′pppRNA or infection with influenza virus than did wild-type cells (Supplementary Fig. 5a–d), which suggested an inhibitory role for TRIM29 in CD11c+ splenic dendritic cells in response to 5′pppRNA or an RNA virus. Together these data demonstrated a negative role for TRIM29 in primary AMs in response to not only the intracellular RLR ligands 5′pppRNA and poly(I:C) but also RNA viruses.
TRIM29 modulates responses against RNA viruses
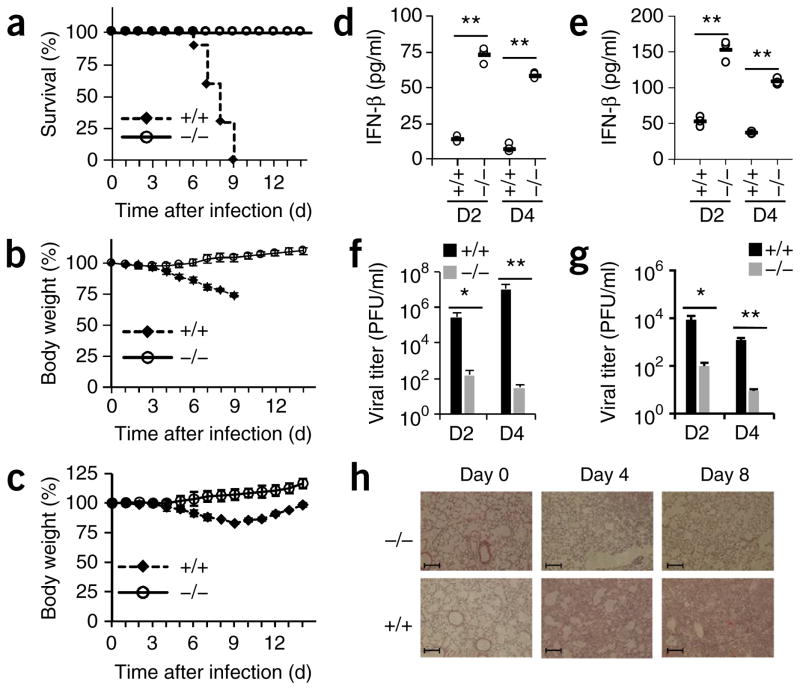
We next evaluated the importance of TRIM29 in vivo in mediating host defense against viral infection. We first infected wild-type and Trim29−/− mice intranasally with influenza virus, which is transmitted by an oral or nasal route in nature, and monitored both survival and body-weight changes over time. The wild-type mice were all dead between day 6 and day 9 after infection with high dose of virus (1 × 105 plaque-forming units (PFU) per mouse). In contrast, none of the Trim29−/− mice succumbed to this infection (Fig. 3a). In addition, the Trim29−/− mice started to regain body weight at 4 or 5 d after infection with a high dose of virus, while the wild-type mice continued to lose body weight until death (Fig. 3b). When infected with a low dose of influenza virus (1 × 102 PFU per mouse), both wild-type mice and Trim29−/− mice survived, but the difference in body-weight changes was striking: wild-type mice continued to lose body weight until day 9 after infection, while Trim29−/− mice gained body weight during this time period (Fig. 3c). In addition, Trim29−/− mice had three- to fivefold more type I interferons in bronchoalveolar lavage fluid (BALF) than did wild-type mice at day 2 or day 4 after infection with either a high dose or a low dose (Fig. 3d,e). Also, there was two- to threefold more IL-6 in BALF from Trim29−/− mice than in that from wild-type mice at day 2 or day 4 after infection (Supplementary Fig. 5e,f). We also measured titers of influenza virus in lung homogenates by plaque-forming assay after infection and found that the viral load was minimal in Trim29−/− mice, in contrast to that in wild-type mice (1 × 102 PFU (Trim29−/−) versus 1 × 107 PFU (wild-type) at day 4 after infection with 1 × 105 PFU per mouse infection, and 9 PFU (Trim29−/−) versus 1 × 103 PFU (wild-type) at day 4 after infection with 1 × 102 PFU per mouse; Fig. 3f,g). Furthermore, histopathology revealed that the edema, alveolar hemorrhaging, alveolar wall thickness and neutrophil infiltration in Trim29−/− lungs was much lower than that of wild-type lungs after viral infection (Fig. 3h). These data indicated that TRIM29 inhibited activation of the innate immune system, especially the production of type I interferons by AMs, and thus allowed the virus to escape attack by the host immune system29.
Figure 3.
TRIM29 has an important role in host defense against infection with RNA viruses in vivo. (a–c) Survival (a) and body weight (b,c) of age- and sex-matched 6-week-old Trim29+/+ and Trim29−/− mice (n = 10 per strain) after intranasal infection with a high dose (1 × 105 PFU per mouse) (a,b) or low dose (1 × 102 PFU per mouse) (c) of influenza virus PR8. (d–g) ELISA of IFN-β in BALF (d,e) and pulmonary viral titers (f,g) of 6-week-old Trim29+/+ and Trim29−/− mice (n = 3 per strain) at day 2 (D2) or day 4 (D4) after intranasal infection with a high dose (d,f) or low dose (e,g) (as in a–c) of influenza virus PR8. Each symbol (d,e) represents an individual independent experiment; small horizontal lines indicate the average of biological triplicates. (h) Hematoxylin-and-eosin staining of lung sections from Trim29+/+ and Trim29−/− mice on day 0, day 4 or day 8 after infection with influenza virus PR8 (1 × 105 PFU per mouse). Scale bars, 200 μm. *P < 0.0005 and **P < 0.0001 (unpaired t-test). Data are representative of two independent experiments with similar results (mean + s.d. in f,g).
Role of TRIM29 in bacteria-induced acute lung inflammation
As TRIM29 has high expression specifically in lung primary AMs, we reasoned that the TRIM29 might also protect lungs from injury by acute bacterial infection. Lipopolysaccharide (LPS) is the main component of the outer membrane of Gram-negative bacteria and often induces strong immune responses in animals. We therefore stimulated wild-type and TRIM29-deficent primary AMs with LPS and compared their production of proinflammatory cytokines. Notably, challenge of wild-type AMs with LPS induced tenfold greater production of TNF and IL-6 than that of unstimulated wild-type AMs but induced minimal production of type I interferon (Fig. 4a and Supplementary Fig. 6a). TRIM29-deficient AMs produced up to sevenfold more proinflammatory cytokines than did wild-type cells in response to LPS challenge (Fig. 4a). We next used an LPS-induced septic-shock model to further investigate the in vivo function of TRIM29 in acute lung inflammation. In this acute model, death occurs within hours, mainly due to the production of TNF30,31. Age-matched wild-type and Trim29−/− littermates were inoculated intranasally with LPS. LPS caused the death of most Trim29−/− mice within 24 h (Fig. 4b). In contrast, only 25% of wild-type mice showed septic shock within 36 h, a time at which all Trim29−/− mice were dead (Fig. 4b).
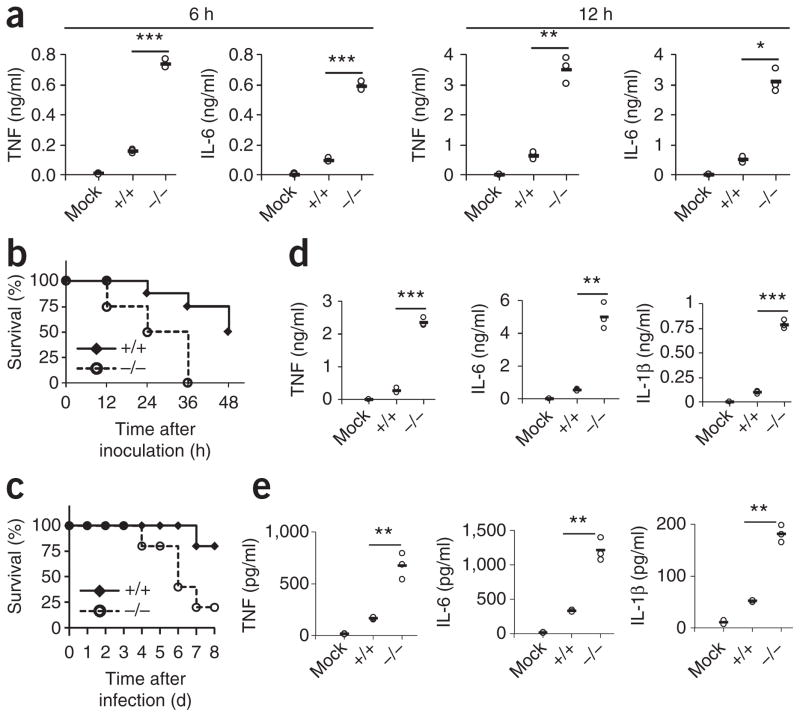
Figure 4.
TRIM29 prevents septic shock induced by LPS and infection with H. influenzae in the lungs. (a) ELISA of TNF and IL-6 in supernatants of Trim29+/+ primary AMs left unstimulated (Mock) or Trim29+/+ and Trim29−/− primary AMs stimulated for 6 h or 12 h (above plots) with LPS (10 ng/ml). (b) Survival of age- and sex-matched 6-week-old Trim29+/+ and Trim29−/− mice (n = 8 per strain) inoculated intranasally with LPS (80 mg per kg body weight) and monitored every hour for 48 h. (c) Survival of age- and sex-matched 6-week-old Trim29+/+ mice left uninfected (Mock) or Trim29+/+ and Trim29−/− mice (n = 5 per strain) inoculated intratracheally with H. influenzae (1 × 107 CFU per mouse) and monitored daily for 8 d. (d,e) ELISA of TNF, IL-6 and IL-1β in BALF of Trim29+/+ mice left untreated or uninfected (Mock) or Trim29+/+ and Trim29−/− mice at 12 h after intranasal inoculation with LPS (80 mg per kg body weight) (d) or at day 1 after intratracheal inoculation with H. influenzae (1 × 107 CFU per mouse) (e). Each symbol (a,d,e) represents an individual independent experiment; small horizontal lines indicate the average of biological triplicates. *P < 0.005, **P < 0.0005 and ***P < 0.0001 (unpaired t-test). Data are representative of two independent experiments with similar results.
We then used the respiratory bacteria H. influenzae to further investigate the role of TRIM29 in acute lung inflammation. We delivered H. influenzae intratracheally into wild type and Trim29−/− mice and monitored the survival of these mice. We found that mortality in Trim29−/− mice was 80% at 8 d after infection, while the mortality of wild-type mice was only 20% (Fig. 4c). We also collected BALF from LPS-challenged mice or H. influenzae–infected mice and measured the concentrations of proinflammatory cytokines. Consistent with the survival results, concentrations of the proinflammatory cytokines TNF, IL-6 and IL-1β (Fig. 4d,e) as well as that of MIP-1α (Supplementary Fig. 6b) were more than tenfold higher (LPS) or fourfold higher (H. influenzae) in Trim29−/− mice than in wild-type mice. The production of type I interferons did not show much difference in wild type mice relative to that in Trim29−/− mice (Supplementary Fig. 6c,d). These results suggested a critical role for TRIM29 in limiting LPS- or H. influenza–induced acute lung injury.
TRIM29 induces NEMO degradation in AMs
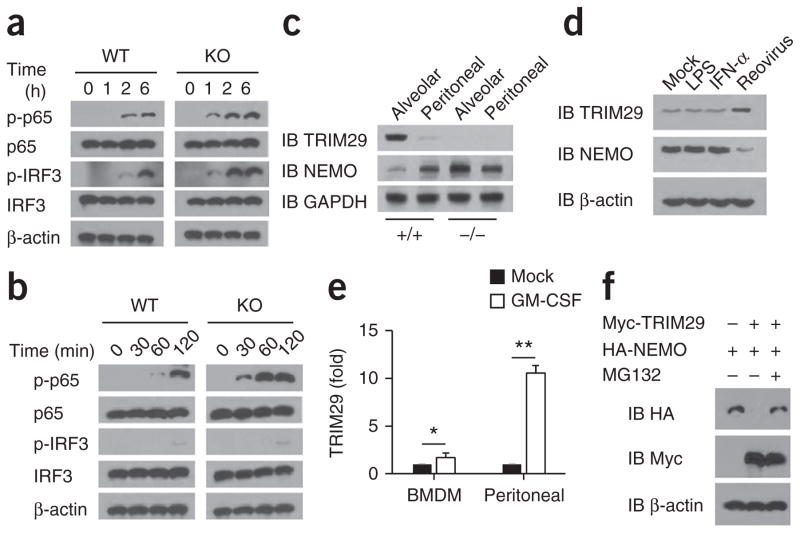
To investigate the mechanisms by which the enhanced production of type I interferons and expression inflammatory cytokines were achieved in AMs in Trim29−/− mice, we isolated AMs from wild-type and Trim29−/− mice and infected the cells with influenza virus or treated them with LPS for various times, then assessed activation of the transcription factors IRF3 and NF-κB by immunoblot analysis. We found that phosphorylation of IRF3 and the NF-κB subunit p65 in TRIM29-deficient AMs was enhanced relative to that in wild-type AMs after viral infection (Fig. 5a). Additionally, treatment with LPS induced rapid and robust upregulation of the expression and phosphorylation of p65, but not the phosphorylation of IRF3, in TRIM29-deficient AMs relative to that in wild-type AMs (Fig. 5b). To further investigate how TRIM29 regulates such signaling molecules, we used immunoprecipitation with an antibody specific to TRIM29 to identify TRIM29-interacting proteins in lysates of AMs, followed by protein sequencing by liquid chromatography–mass spectrometry. Among a group of TRIM29-interacting proteins, we identified NEMO, the regulatory subunit of the NF-κB complex (Supplementary Table 1). Because TRIM29 is expressed specifically in resting AMs, we isolated both primary AMs and peritoneal macrophages to measure endogenous TRIM29 and NEMO. The abundance of TRIM29 was much greater in wild-type AMs than in peritoneal macrophages; in contrast, the abundance of NEMO was much lower in AMs than in peritoneal macrophages (Fig. 5c). Notably, the amount of NEMO in AMs and peritoneal macrophages from Trim29−/− mice was similar to that in their counterparts from wild-type mice (Fig. 5c). These data indicated that TRIM29 expression in AMs might limit NEMO expression. To investigate that possibility, we isolated macrophages and bone-marrow-derived dendritic cells from wild-type mice and treated them for 6 h with LPS, IFN-α or infection with reovirus. We found that TRIM29 expression was induced in dendritic cells by infection with reovirus but not by treatment with LPS or IFN-α (Fig. 5d) and could not be further induced in macrophages (Supplementary Fig. 7). The amount of NEMO was much lower in bone-marrow-derived dendritic cells infected with reovirus than those treated with LPS or IFN-α (Fig. 5d). AMs have high expression of TRIM29, and GM-CSF is known to be a critical regulator of protein expression in the airspaces1. We next determined whether GM-CSF was able induce TRIM29 expression in macrophages. TRIM29 expression was substantially induced in peritoneal macrophages by treatment with GM-CSF but was marginally induced in bone-marrow-derived macrophages by GM-CSF (Fig. 5e).
Figure 5.
TRIM29 inhibits the activation of IRF3 and p65 in AMs after viral infection or LPS treatment by degrading NEMO. (a,b) Immunoblot analysis of total and phosphorylated (p−) p65 and IRF3, as well as β-actin (loading control throughout), in lysates of Trim29+/+ (WT) and Trim29−/− (KO) AMs infected for various times (above lanes) with influenza virus PR8 at an MOI of 5 (a) or stimulated for various time (above lanes) with LPS (20 ng/ml) (b). (c) Immunoblot analysis of TRIM29 and NEMO in primary AMs (Alveolar) or peritoneal macrophages (Peritoneal) from Trim29+/+ and Trim29−/− mice. (d) Immunoblot analysis of TRIM29 and NEMO in bone-marrow-derived dendritic cells given mock treatment or treated for 6 h with LPS (20 ng/ml), IFN-μ (25 ng/ml) or reovirus. (e) Real-time PCR analysis of TRIM29 mRNA in wild-type bone-marrow-derived macrophages (BMDM) and peritoneal macrophages l treated for 6 h with GM-CSF (50 ng/ml); results are presented relative to those of untreated cells (Mock), set as 1. (f) Immunoblot analysis HEK293T cells transfected to express various combinations (above lanes) of Myc-tagged TRIM29 and HA-tagged NEMO and treated with MG132 (+) or not (−). *P < 0.05 and **P < 0.001 (unpaired t-test). Data are representative of three independent experiments with similar results (mean + s.d. in e).
Because TRIM29 is an E3 ubiquitin ligase, we next determined whether NEMO is the target of TRIM29 for ubiquitination and degradation. Coexpression of NEMO with TRIM29 showed that TRIM29 induced complete degradation of NEMO, and treatment with the proteasome inhibitor MG132 rescued it from this degradation (Fig. 5f), which suggested that TRIM29 was able to induce proteasome-dependent degradation of NEMO. Together these data indicated that TRIM29 inhibited the activation of IRF3 and p65 in AMs by degrading NEMO in response to viral infection or LPS treatment.
TRIM29 binds to and colocalizes with NEMO in lysosome
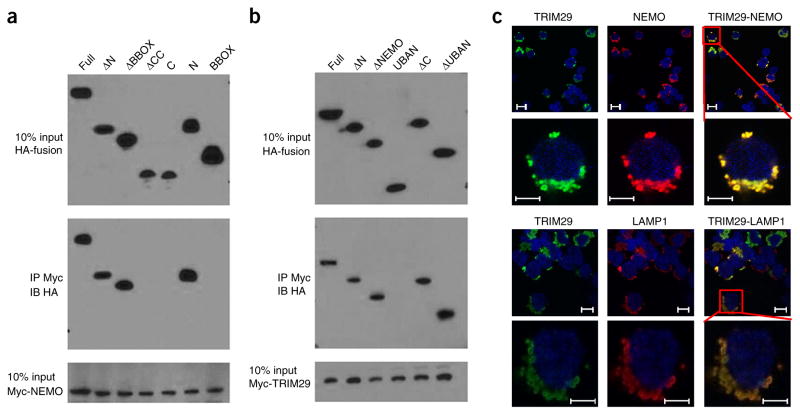
To ascertain the physical interactions between TRIM29 and NEMO, we investigated the interactions between Myc-tagged recombinant NEMO and hemagglutinin (HA)-tagged full-length TRIM29, as well as truncation mutants of TRIM29 (Supplementary Fig. 8a). Both full-length TRIM29 and the OmpH (‘outer membrane protein H’) domain of TRIM29 bound NEMO (Fig. 6a). Additionally, the mapping results for Myc-tagged recombinant TRIM29 and HA-tagged full-length NEMO and their truncation mutants (Supplementary Fig. 8b) showed that the OmpH domain of NEMO bound TRIM29 (Fig. 6b); this suggested that the interaction between TRIM29 and NEMO was mediated by the OmpH-OmpH domains. Next, we expressed Myc-tagged NEMO together with HA-tagged full-length TRIM29 or mutant TRIM29 lacking the NEMO-binding site (TRIM29 BBOX) in human embryonic kidney cells (HEK293T cells) to determine their subcellular localization. Immunofluorescence analysis showed that full-length TRIM29 colocalized with NEMO in the cytosol (Fig. 6c). In contrast, TRIM29 BBOX did not colocalize with NEMO (Supplementary Fig. 8c). Furthermore, subcellular colocalization analysis showed that TRIM29 colocalized with the lysosome marker LAMP1 (Fig. 6c) but not with a mitochondrial marker, the endosomal marker TfR or the autophagosome marker LC3A (Supplementary Fig. 8d). Additionally, viral infection had no effect on the colocalization of TRIM29 with LAMP1 (Supplementary Fig. 8e). These data suggested that TRIM29 bound to NEMO via the OmpH-OmpH domains and that it colocalized with NEMO in the cytosolic lysosome.
Figure 6.
TRIM29 binds to and colocalizes with NEMO in the lysosome. (a) Immunoblot analysis of purified Myc-tagged NEMO with anti-Myc (bottom), and immunoblot analysis (with anti-HA) of purified HA-tagged full-length TRIM29 (Full) or TRIM29 with deletion of the N terminus (ΔN), the B-box zinc-finger domain (ΔBBOX) or the coiled-coil domain (ΔCC) or containing only the carboxyl terminus (C), the N terminus (N) or the BBOX domain (above lanes), alone (top) or after incubation with Myc-tagged NEMO and immunoprecipitation (IP) with anti-Myc (middle). (b) Immunoblot analysis of purified Myc-tagged TRIM29 with anti-Myc (bottom), and immunoblot analysis (with anti-HA) of purified HA-tagged full-length NEMO (Full) and NEMO with deletion of the N terminus (ΔN), a fragment of the NEMO domain (ΔNEMO), the carboxyl terminus (ΔC) or the UBAN domain (ΔUBAN) or containing only the UBAN (UBAN), alone (top) or after incubation with Myc-tagged TRIM29 and immunoprecipitation with anti-Myc (middle). (c) Confocal microscopy of HEK293T cells cotransfected with expression plasmids for HA-tagged TRIM29 and Myc-tagged NEMO and stained with Alexa Fluor 488–conjugated antibody to HA (green) and Alexa Fluor 555–conjugated antibody to Myc (red); LAMP1 serves as a marker of lysosome (red), and the DNA-binding dye DAPI serves as a marker of nuclei (blue). Bottom rows are an enlargement of the areas outlined at top right. Scale bars, 10μm (top rows) or 5 μm (bottom rows). Data are representative of three independent experiments with similar results.
TRIM29 induces NEMO ubiquitination through K48 linkage
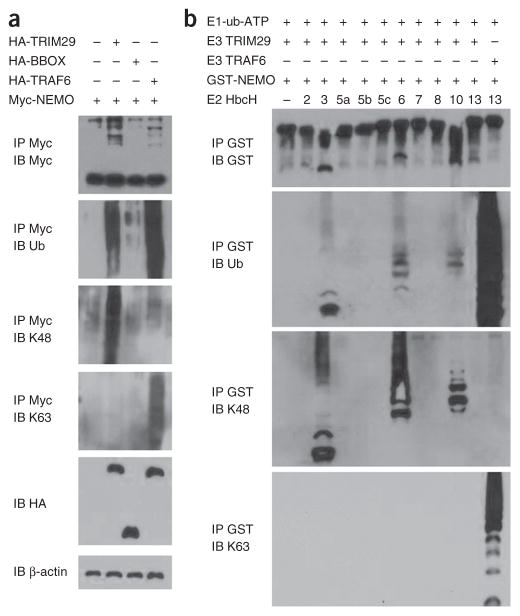
To investigate whether NEMO undergoes ubiquitination and whether such ubiquitination of NEMO is dependent on the binding site at which TRIM29 interacts with NEMO, we transfected HEK293T cells with plasmids expressing Myc-tagged NEMO and HA-tagged full-length TRIM29, the mutant TRIM29 BBOX or full-length ubiquitin ligase TRAF6 to detect the ubiquitination of NEMO. Immunoblot analysis demonstrated that the ubiquitination of NEMO was substantially enhanced by overexpression of TRIM29 but not by the mutant TRIM29 BBOX (Fig. 7a). Immunoblot analysis of Lys48 (K48)-linked or K63-linked ubiquitination further demonstrated that TRIM29 induced the ubiquitination of NEMO by K48-mediated linkage (Fig. 7a), while TRAF6, but not TRIM29, induced the ubiquitination of NEMO by K63-mediated linkage (Fig. 7a).
Figure 7.
TRIM29 induces ubiquitination of NEMO by K48 linkage. (a) Immunoblot analysis with anti-Myc (top blot) or of total ubiquitination (second blot), K48-linked ubiquitination (third blot) or K63-linked ubiquitination (fourth blot) of Myc-tagged NEMO in HEK293T cells transfected with empty vector or expression vector for HA-tagged TRIM29, TRIM29 BBOX or TRAF6 and treated for 3 h with 25 μM MG132, assessed after immunoprecipitation with anti-Myc; and immunoblot analysis of whole-cell lysates with anti-HA (fifth blot) or anti-β-actin (bottom blot). (b) Immunoblot analysis of the degradation and total, K48-mediated or K63-mediated ubiquitination of GST-tagged NEMO, assessed by in vitro ubiquitination assay with various combinations (above lanes) of a mixture of E1 plus ubiquitin plus ATP (E1-ub-ATP), TRIM29 (E3 TRIM29), GST-tagged NEMO (GST-NEMO), GST-tagged TRAF6, and purified E2 enzymes (HbcH; numbers above lanes indicate enzyme), after immunoprecipitation with anti-GST. Data are representative of three independent experiments with similar results.
To determine whether TRIM29 is able to directly modify NEMO, we reconstituted the NEMO-ubiquitination reaction in vitro. We analyzed various combinations of recombinant glutathione S-transferase (GST)-tagged NEMO, recombinant TRIM29 or recombinant TRAF6, purified E2 enzymes (HbcH2, HbcH3, HbcH5a, HbcH5b, HbcH5c, HbcH6, HbcH7, HbcH8, HbcH10 and HbcH13) and a mixture of E1 plus ubiquitin plus ATP, followed by precipitation with GST beads and immunoblot analysis. We observed that K48-linked ubiquitination of NEMO (but not its K63-linked ubiquitination) was induced by TRIM29 plus one of the following E2 enzymes: HbcH3, HbcH6 or HbcH10 (Fig. 7b). In a control assay, the ubiquitination of NEMO by TRAF6 was mediated by K63 linkage (Fig. 7b). These data indicated that TRIM29 induced the ubiquitination and degradation of NEMO by K48-mediated linkage.
TRIM29 induces NEMO ubiquitination at its Lys183 site
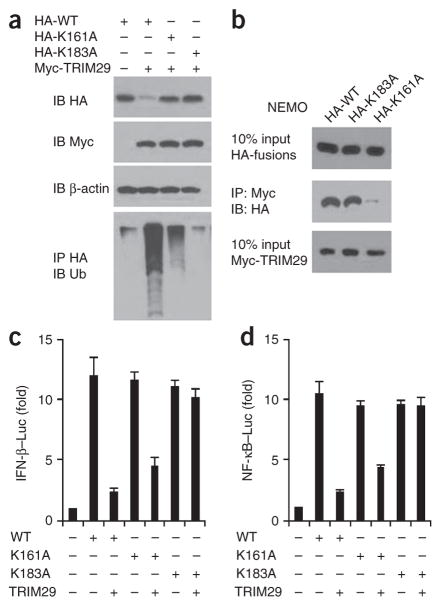
NEMO contains 30 lysine residues, and 22 of these were predicted to be possible ubiquitination sites (Supplementary Table 2). We therefore replaced each of these lysine residues individually with arginine and found that substitution of Lys161 or Lys183 resulted in partial resistance or complete resistance, respectively, to the ubiquitination and degradation induced by TRIM29 (Fig. 8a). All other substitutions had no effect on the degradation of NEMO by TRIM29 (data not shown). We next assessed the interaction between the substitution mutants of NEMO and TRIM29 by co-immunoprecipitation assay. Although NEMO with substitution of Lys183 was still able to bind TRIM29, NEMO with substitution of Lys161 lost most of its binding ability (Fig. 8b). These data suggested that Lys183 is the key ubiquitination site in NEMO. To determine the functional importance of NEMO Lys183, we performed a luciferase reporter assay. Overexpression of TRIM29 inhibited only activation of the promoters of the genes encoding IFN-β or NF-κB induced by wild-type NEMO, not that induced by NEMO with substitution of Lys183 (Fig. 8c,d). These data indicated that Lys183 was a critical site for TRIM29- mediated ubiquitination and regulation of NEMO.
Figure 8.
TRIM29 induces degradation of NEMO after its ubiquitination at Lys183. (a) Immunoblot analysis of HEK293T cells cotransfected an expression vector for HA-tagged wild-type NEMO (HA-WT) or NEMO with substitution of alanine for Lys183 (HA-K183A) or Lys161 (HA-K161A) and with empty vector (−) or an expression vector for Myc-tagged TRIM29 (above lanes). (b) Immunoblot analysis of purified Myc-tagged TRIM29 with anti-Myc (bottom blot), and immunoblot analysis (with anti-HA) of purified HA-tagged NEMO (as in a) alone (top) or after incubation with Myc-tagged TRIM29 and immunoprecipitation with anti-Myc (middle). (c,d) Activation of the promoter of the gene encoding IFN-β (c) or NF-κB (d) in HEK293T cells transfected with luciferase reporters for those genes (Luc), plus expression vector for NEMO (as in a) with or without an expression vector for TRIM29 (below plots); results are presented relative to those of renilla luciferase (cotransfected as an internal control). Data are representative of three independent experiments with similar results (mean + s.d. in c,d).
DISCUSSION
AMs are a major producer of type I interferons and proinflammatory cytokines during respiratory infection, and this effect is initiated by innate sensors and is mediated by several key signaling pathways. Such a response has a key role in local protective immunity as well as in immune-system-mediated pathology32. Studies have shown that many inflammation-associated diseases, including some types of cancers, result from dysregulated innate immunity33. Therefore, understanding of the molecular mechanisms by which innate immunity is dynamically controlled is critical for the development of novel and more-effective treatments for these diseases34. However, the mechanisms that control local innate immunity in the respiratory tract are poorly understood.
In this study, we discovered that AMs specifically expressed the E3 ubiquitin ligase TRIM29 and that TRIM29 served a crucial role in the maintenance of immunological homeostasis in the lungs by regulating the stability of NEMO, a key adaptor in both the IRF-mediated type-I-interferon-production pathway and the NF-κB-mediated proinflammatory signaling pathway. We showed that TRIM29 interacted with NEMO via the OmpH-OmpH domains of NEMO and induced the ubiquitination and degradation of NEMO at Lys183 by K48 linkage. Our data suggest that under homeostatic conditions, TRIM29 expression in AMs limits the abundance of NEMO, so the production of proinflammatory cytokines is inhibited and unwanted lung inflammation is suppressed in response to bacterial infection. However, during viral infection, high levels of TRIM29 also interfere with the production of type I interferons, which hinders the generation of anti-viral immunity and thus renders the mice susceptible to virus-induced death. Indeed, we observed that Trim29−/− mice exhibited significantly reduced morbidity, mortality, lung injury and local inflammation after challenge with influenza virus. On the other hand, Trim29−/− mice were more susceptible and died of uncontrolled inflammation after challenge with LPS or H. influenzae. Clearly, both models highlight the importance of TRIM29 in regulating innate immunity in the lungs.
The innate immune cells sense pathogen-associated molecular patterns such as viral 5′pppRNA and bacterial LPS to produce type I interferons and proinflammatory cytokines. This response is critical in the defense against viral or bacterial infection. Central to the early host defense against viral infection is the production of type I interferons and the synthesis of antiviral interferon-stimulated gene products contain viral dissemination and activate the adaptive immune response35,36. After infection with an RNA virus, the rapid induction of type I interferons and proinflammatory cytokines is regulated by extracellular and intracellular signals to activate both NF-κB and IRFs37,38. The activation of NF-κB is regulated by the inhibitor-of-NF-κB kinase (IKK) complex composed of IKK-α, IKK-β and NEMO. In parallel, the activation of IRF3 and IRF7 is regulated by the IKK-related kinases TBK1 and IKKε39,40. Further studies have demonstrated that NEMO is also essential for virus-induced activation of IRF3 and IRF7 by recruiting TBK1 and IKKε to the RLR-MAVS complex41, which suggests that NEMO is a critical adaptor in both NF-κB signaling and IRF signaling. Published evidence indicates that TRIM29 might have an important role in DNA repair42 and oncogenesis43, which suggests that TRIM29 might have additional functions beyond the regulation of innate immunity. Structurally, TRIM29 belongs to the large TRIM family of E3 ubiquitin ligases, but it lacks a typical RING domain. Instead, TRIM29 has a B-box domain that also has E3 ligase activity. It has been shown that both the RING domain and the B-box domain of TRIM18 have E3 ligase activity, although the B-box domain exhibits weaker E3 ligase activity than that of a typical RING domain29. Furthermore, the E3 ligase Peli1 does not contain a typical RING domain, which suggests that not all E3 ligases use a typical RING domain to catalyze ubiquitination30,31.
Published studies have demonstrated that post-translational modification of NEMO by ubiquitination has a critical role in regulating its function in the IKK complex. It has been shown that the adaptor Bcl10 induces NEMO K63-linked ubiquitination44. Further studies have confirmed that Bcl10 and the adaptor MALT1 form oligomers to induce the oligomerization of TRAF6 for the polyubiquitination of NEMO by the ubiquitin-conjugating enzyme Ubc13 (ref. 45). Another study has indicated that endogenous NEMO undergoes linear ubiquitination after viral infection46. Furthermore, the E3 ubiquitin ligase TRIM23 induces K27-linked ubiquitination of NEMO for positive regulation of TLR3- and RIG-I–MDA5–mediated antiviral responses47. However, it is still unknown whether NEMO undergoes ubiquitination for degradation and thereby negatively regulates the immune responses. Here we have demonstrated that TRIM29 induced the ubiquitination and subsequent degradation of NEMO by K48 linkage to negatively regulate the production of type I interferons as well as proinflammatory cytokines in AMs after infection.
A diverse range of molecules have been identified as important regulators in the signaling pathways of both type I interferon production and inflammatory cytokine production. Our current observations based on the study of Trim29−/− mice have solidified the proposal of a role for TRIM29 as an in vivo checkpoint of type I interferon and inflammatory cytokine responses. We have extended those findings to influenza virus and identified TRIM29 as a negative regulator of the host immune response to pathogens in the respiratory tract. We predict that TRIM29 mutants lacking E3-ubiquitin-ligase activity might serve as biomarkers for the prediction of lung inflammation and susceptibility to influenza virus.
Although the activation of signaling pathways via the sensing of viral nucleic acids in immune cells is critical for the elimination of invading microorganisms48, excessive sensing and overwhelming host cytokine production can lead to tissue damage and autoimmune disease. In this context, the identification of TRIM29 as a key negative regulator of the production of type I interferons as well as proinflammatory cytokines in the lungs might have important implications for the understanding of innate immunity, as well as the study of the pathogenesis of autoimmune diseases.
ONLINE METHODS
Mice
Trim29−/− mice were obtained from the European Mouse Mutant Archive (EMMA). Primary bone marrow cells were collected from wild-type and Trim29−/− mice to prepare bone-marrow-derived dendritic cells and bone-marrow-derived macrophages. All animals were maintained in the specific-pathogen-free facility at Houston Methodist Research Institute in Houston, Texas. Animal use and care were approved by the Houston Methodist Animal Care Committee, in accordance with institutional animal care and use committee guidelines.
Reagents
The 5′pppRNA, high-molecular-weight poly(I:C), and LPS were from Invivogen. Lipofectamine 3000 was from Invitrogen. The proteasome inhibitor MG132 was from Sigma. The following antibodies were used for immunoblot analysis: anti-NEMO (1:1,000) (2685; Cell Signaling), anti-TRIM29 (1:1,000) (sc-33151; H-300; Santa Cruz), anti-MAVS (1:1,000) (AP20074a; Abgent), anti-ubiquitin (1:1,000) (sc-8017; Santa Cruz), K63- specific anti-ubiquitin (1:1,000) (05-1313; Millipore), K48-specific anti- ubiquitin (1:1,000) (05-1307; Millipore), anti-IRF3 (1:1,000) (sc-9082; FL-425; Santa Cruz), antibody to phosphorylated IRF3 (1:1,000) (4947; Cell Signaling), anti-p65 (1:1,000) (4764, Cell Signaling), antibody to phosphorylated p65 (1:1,000) (3033, Cell Signaling), anti-GAPDH (1:10,000) (clone GAPDH-71.1, G9295; Sigma), anti-β-actin (1: 20,000) (Clone AC-15, A3854; Sigma), anti-HA (1: 5,000) (clone HA-7, H6533; Sigma), anti-Myc (1: 5,000) (ab1326; Abcam) and anti-GST (1: 1,000) (ab58626; Abcam). The following antibodies were used for confocal microscopy assay: Alexa Fluor 555–anti-Myc (1:50) (clone 9B11, 3756; Cell Signaling) and Alexa Fluor 488–anti-HA (1:50) (clone 6E2, 2350; Cell Signaling). Anti-HA and anti-Myc beads were from Sigma. Glutathione-Sepharose was from GE Healthcare. Lentiviral vectors for shRNA were from GE Healthcare Dharmacon Inc. (Open Biosystems): TRIM29 (clone TRCN0000241302 (TRIM29-#1) and clone TRCN0000241305 (TRIM29-#2)); MAVS (clone TRCN0000124772). GST-tagged NEMO (ab125589) were from Abcam. TRIM29 (PRO-328) was from ProSpec. Recombinant murine GM-CSF (315-03) was from PeproTech. The Ubiquitin Thioester/Conjugation Initiation kit (K-995) and UbcH Enzyme Set (K-980) were from BostonBiochem. The IFN-β and IFN-α ELISA kits were from PBL InterferonSource. The TNF, IL-6 and MIP-1α ELISA kits were from R&D Systems. The IL-10 ELISA kit was from eBioscience. The Dual-Luciferase Reporter Assay System (E1910) was from Promega. Influenza A virus (PR8 A/Puerto Rico/8/1934(H1N1)) stocks, Reovirus (Reovirus type 3 strain Dearing, T3D), and H. influenzae were from ATCC (ATCC® VR-95™, ATCC® VR-824™ and ATCC® 35056™).
Cell culture, transfection and stimulation
The MH-S cell line was purchased from ATCC (ATCC CRL-2019) and was maintained in RPMI-1640 medium containing 10% heat-inactivated FCS and 1% penicillin-streptomycin (Invitrogen-Gibco). MH-S cells were infected with a pLKO.1 lentiviral vector carrying a target gene sequence or a scrambled shRNA. After 24 h of culture, cells were selected by the addition of puromycin (2 ng/ml) to the medium for 3 d. The knockdown efficiency was detected with immunoblot analysis.
Alveolar macrophages were harvested from the lungs of mice as described49. The lungs were washed four times with PBS. Harvested BALF was then centrifuged and red blood cell lysis buffer (Sigma) was added to remove contaminated red blood cells. The remaining cells (which were >90% alveolar macrophages as judged by differential staining) were harvested by centrifugation, resuspended in RPMI-1640 medium as described17.
Cells were then stimulated with 5′pppRNA (1 μg/ml) delivered by Lipofectamine 3000 or were infected with influenza A virus at a multiplicity of infection (MOI) of 5. The concentration of IFN-α, IFN-β, TNF and IL-6 in culture supernatants was measured by ELISA.
In vivo virus infection
6- to 8-week-old mice were infected intranasally with a high dose (1 × 105 plaque forming units (PFU)) or low dose (1 × 102 PFU) of influenza PR8 virus in 40 μl PBS after anesthesia as described50. The body weight and survival of mice were monitored daily for 14 d. Mice were euthanized at various time points following infection. To obtain bronchoalveolar lavage fluid from mice, tracheas were cannulated after exsanguination and lungs were washed with 1 ml of PBS. BALF samples were centrifuged (800g, 5 min) to isolate cells and supernatants were centrifuged again (13,000g, 1 min) to completely remove remaining cells. The concentration of IFN-β and IL-6 in BALF were measured by ELISA.
Viral plaque titration
After infection of mice with influenza virus PR8, total lung was removed and homogenized to prepare lung extracts in 1 ml of PBS (pH 7.4). Confluent Madin-Darby canine kidney (MDCK) cells were washed with DMEM (Gibco) once and infected with virus for 30 min at room temperature as described17,50. After a wash with PBS, the plate was overlaid with DMEM containing 1% low melting-point agarose and 10 mg/ml of trypsin and incubated at 37 °C for 3 d.
LPS-induced death and cytokine production in mice
Age- and sex-matched Trim29+/+ and Trim29−/− mice were inoculated intranasally with LPS (80 mg/kg). Survival was monitored every hour for 48 h. The lung BALF samples were obtained at 12 h after challenge, and the concentrations of TNF, IL-6, IL-1β, IFN-α and IFN-β in BALF samples were measured by ELISA.
In vivo H. influenzae infection
Age- and sex-matched Trim29+/+ and Trim29−/− mice were anaesthetized and then intratracheally inoculated with H. influenzae at a concentration of 1 × 107 colony-forming units (CFU) per mouse. Survival was monitored daily for 8 d. The lung BALF samples were obtained at day 1 after challenge, and the concentrations of TNF, IL-6, IL-1β, MIP-1α, IFN-α and IFN-β in BALF samples were measured by ELISA.
Histology
Lungs were removed from naive or infected wild-type and Trim29−/− mice and were washed using PBS before being fixed with 4% formaldehyde for 1 h at 4 °C. The tissues were embedded in paraffin and processed by standard techniques. Longitudinal 5-μm sections were stained with hematoxylin and eosin as previously described50.
Luciferase reporter gene assay
HEK293T cells were seeded on 48-well plates (1 × 105 cells per well), then transfected with firefly luciferase reporter vectors (100 ng) for the gene encoding IFN-β or NF-κB and renilla luciferase (1 ng), plus expression vector for wild-type NEMO or NEMO mutants (100 ng) with or without expression vector for TRIM29 (100 ng). Empty control vector was added so that a total of 450 ng of vector DNA was transfected into each well of cells. At 24 h after transfection, cells were stimulated with 5′pppRNA (1 μg/ml) delivered by Lipofectamine 3000. Cells were collected after 6 h of stimulation. Luciferase activity in total cell lysates was detected by Dual-Luciferase Reporter Assay and standardized for comparison.
In vitro precipitation and immunoblot analysis
For the preparation of purified NEMO and TRIM29, HEK293T cells were transfected with expression plasmids encoding full-length or truncated versions of HA- or Myc-tagged NEMO or TRIM29. Lysates were prepared from the transfected cells, followed by incubation with anti-HA or anti-Myc beads. Proteins were eluted from the beads after beads were washed six times with PBS. For precipitation with anti-HA or anti-Myc beads, purified HA-tagged wild-type NEMO or truncations of NEMO were incubated for 2 h with purified Myc-tagged TRIM29 or purified HA-tagged TRIM29 or truncations of TRIM29 were incubated for 2 h with purified Myc-tagged NEMO. Beads were added; after 2 h of incubation, the bound complexes were pelleted by centrifugation. Proteins and beads were analyzed by immunoblot analysis with anti-HA or anti-Myc (identified above).
Ubiquitination
HEK293T cells were transfected with expression plasmids encoding Myc-tagged full-length NEMO and with or without coexpression of HA-tagged TRIM29, TRIM29 BBOX mutant, and TRAF6. At 24 h after transfection, cells were treated with 25 μM MG132 for 3 h and then were collected for analysis. Extracts were analyzed by immunoblot analysis or were immunoprecipitated with anti-Myc beads and then analyzed by immunoblot analysis.
In vitro ubiquitination assay
GST-tagged NEMO (100 ng) and TRIM29 derived from Escherichia coli (100 ng) were incubated with E1 (100 ng), E2 (500 ng) and ubiquitin (2.5 μg) in 50 μl ubiquitination assay buffer (50 mM Tris-HCl, 2.5 mM MgCl2, 0.5 mM DTT and 2 mM ATP). Reactions were incubated for 2 h at 30 °C. Proteins were immunoprecipitated with anti-GST beads and then analyzed by immunoblot analysis.
Confocal microscopy
HEK293T cells were cotransfected with expression plasmids for HA-tagged TRIM29 or TRIM29 BBOX and Myc-tagged NEMO. After 20 h, cells were mock or infected with influenza A PR8 virus for 6 h and then fixed in 4% paraformaldehyde and permeabilized with 0.1% saponin, then blocked for 30 min with 10% goat serum, incubated overnight at 4 °C with Alexa Fluor 555–anti-Myc, Alexa Fluor 488–anti-HA (both identified above), mitochondrial marker MitoTracker Red CMXRos (Cell Signaling), anti-TfR-biotin (555535, BD Bioscience), anti-LAMP1-biotin (555799, BD Bioscience) and anti-LC3A (GTX50635, GeneTex) and then examined with confocal microscopy. Secondary antibodies used were Alexa Fluor 594 goat anti-rabbit IgG (8889S, Cell Signaling) for anti-LC3A, and Alexa Fluor 647 streptavidin (Invitrogen) for anti-TfR-biotin and anti-LAMP1-biotin. Images of ‘zoomed’ single cells were quantified with Leica Confocal Software.
Isolation of CD11c+ splenic dendritic cells
Spleens from age- and sex-matched Trim29+/+ and Trim29−/− mice were harvested and homogenized in collagenase D, and the resulting homogenates were resuspended in PBS, pH 7.2. After incubation with 25 μl of anti-CD11c microbeads (Miltenyi Biotec), 225 μl of cell suspension from one spleen was first sorted for positive selection. The purity of the CD11c+ fraction was >95%.
Quantitative RT-PCR
RNA was isolated using the RNeasy Kit (Qiagen) according to the manufacturer’s instructions. The isolated RNA was used to synthesize cDNA with the iScript cDNA Synthesis Kit (Bio-Rad). iTaq SYBR Green Supermix with ROX (Bio-Rad) was used for quantitative RT-PCR. PCRs were performed in triplicate. The following primers were used: TRIM29 forward, 5′-ACTCCTCCTTCTCCCTGAAA-3′ and reverse, 5′-GACATAGAATGGCCGGTAGTG-3′; CCL-2 forward, 5′-GCTGGGTTCAGTTTCCTTAAGC-3′ and reverse, 5′-CCTAGTCTTTAGCTGTGA GACCTTCTG-3′; CCL-5 forward, 5′-CTGTCATCGCTTGCTCTAGTCCTA-3 ′ and reverse, 5′-CGGATGGAGATGCCGATTT-3′; CXCL-2 forward, 5′-CCACC AACCACCAGGCTACAGGGGC-3′ and reverse, 5′-AGGCTCCTCCTTTCCA GGTCAGTTAGC-3′; IL-4 forward, 5′-AGATGGATGTGCCAAACGTCCTCA-3′ and reverse, 5′-AATATGCGAAGCACCTTGGAAGCC-3′; IL-5 forward, 5′-TCACCGAGCTCTGTTGACAA-3′ and reverse, 5′-CCACACTTCT CTTTTTGGCG-3′; IL-13 forward, 5′-TGAGGAGCTGAGCAACATCACACA-3′ and reverse, 5′-TGCGGTTACAGAGGCCATGCAATA-3′; GAPDH forward, 5′-AACTTTGGCATTGTGGAAGG-3′ and reverse, 5′-ACACATTGGGGGT AGGAACA-3′.
Flow cytometry
Primary AMs were isolated from wild-type mice and Trim29−/− mice. The cells were then fixed and stained with FITC-conjugated anti-mouse CD11c antibody (N418, BioLegend), APC-conjugated anti-mouse F4/80 antibody (BM8, BioLegend) and their isotype-matched control antibodies for assessing their differentiation. Flow cytometry data were acquired on a LSR-II flow cytometer (Beckton Dickinson) and analyzed using FlowJo v10 software (Tree Star).
Statistical analysis
A two-tailed unpaired Student’s t test was used for statistical analysis with Microsoft Excel and GraphPad Prism Software. P values of less than 0.05 and 0.01 were considered significant and very significant, respectively unless specifically indicated otherwise.
Supplementary Material
Acknowledgments
We thank the Wellcome Trust Sanger Institute Mouse Genetics Project (Sanger MGP) and its funders for the mutant mouse line Trim29, and the European Mouse Mutant Archive (http://www.emmanet.org) partner from which the mouse line was received. Funding and associated primary phenotypic information is provided at the Sanger website (http://www.sanger.ac.uk/mouseportal). We also thank L. Minze for operational support. Supported by the US National Institutes of Health (R01AI080779 to X.C.L.).
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
J.X. designed and performed most of the experiments; L.W., B.Y., Z.W., L.J., R.J. and H.L. helped with some of the experiments; X.C.L., Y.-J.L. and Z.Z. wrote the manuscript; and Z.Z. supervised the project.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Guilliams M, et al. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J Exp Med. 2013;210:1977–1992. doi: 10.1084/jem.20131199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 3.Vlahos R, Bozinovski S. Role of alveolar macrophages in chronic obstructive pulmonary disease. Front Immunol. 2014;5:435. doi: 10.3389/fimmu.2014.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kopf M, Schneider C, Nobs SP. The development and function of lung-resident macrophages and dendritic cells. Nat Immunol. 2015;16:36–44. doi: 10.1038/ni.3052. [DOI] [PubMed] [Google Scholar]
- 6.Hussell T, Bell TJ. Alveolar macrophages: plasticity in a tissue-specific context. Nat Rev Immunol. 2014;14:81–93. doi: 10.1038/nri3600. [DOI] [PubMed] [Google Scholar]
- 7.Westphalen K, et al. Sessile alveolar macrophages communicate with alveolar epithelium to modulate immunity. Nature. 2014;506:503–506. doi: 10.1038/nature12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ballinger MN, et al. Role of granulocyte macrophage colony-stimulating factor during gram-negative lung infection with Pseudomonas aeruginosa. Am J Respir Cell Mol Biol. 2006;34:766–774. doi: 10.1165/rcmb.2005-0246OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonzalez-Juarrero M, et al. Disruption of granulocyte macrophage-colony stimulating factor production in the lungs severely affects the ability of mice to control Mycobacterium tuberculosis infection. J Leukoc Biol. 2005;77:914–922. doi: 10.1189/jlb.1204723. [DOI] [PubMed] [Google Scholar]
- 10.Paine R, III, et al. Granulocyte-macrophage colony-stimulating factor in the innate immune response to Pneumocystis carinii pneumonia in mice. J Immunol. 2000;164:2602–2609. doi: 10.4049/jimmunol.164.5.2602. [DOI] [PubMed] [Google Scholar]
- 11.LeVine AM, Reed JA, Kurak KE, Cianciolo E, Whitsett JA. GM-CSF-deficient mice are susceptible to pulmonary group B streptococcal infection. J Clin Invest. 1999;103:563–569. doi: 10.1172/JCI5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murphy SL, Xu J, Kochanek KD. Deaths: final data for 2010. Natl Vital Stat Rep. 2013;61:1–117. [PubMed] [Google Scholar]
- 13.Schneider C, et al. Alveolar macrophages are essential for protection from respiratory failure and associated morbidity following influenza virus infection. PLoS Pathog. 2014;10:e1004053. doi: 10.1371/journal.ppat.1004053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang FF, et al. GM-CSF in the lung protects against lethal influenza infection. Am J Respir Crit Care Med. 2011;184:259–268. doi: 10.1164/rccm.201012-2036OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Riel D, et al. Highly pathogenic avian influenza virus H5N1 infects alveolar macrophages without virus production or excessive TNF-α induction. PLoS Pathog. 2011;7:e1002099. doi: 10.1371/journal.ppat.1002099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumagai Y, et al. Alveolar macrophages are the primary interferon-α producer in pulmonary infection with RNA viruses. Immunity. 2007;27:240–252. doi: 10.1016/j.immuni.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 17.Seo SU, et al. Type I interferon signaling regulates Ly6Chi monocytes and neutrophils during acute viral pneumonia in mice. PLoS Pathog. 2011;7:e1001304. doi: 10.1371/journal.ppat.1001304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Everitt AR, et al. IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484:519–523. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bailey CC, Huang IC, Kam C, Farzan M. Ifitm3 limits the severity of acute influenza in mice. PLoS Pathog. 2012;8:e1002909. doi: 10.1371/journal.ppat.1002909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Münz C, Lünemann JD, Getts MT, Miller SD. Antiviral immune responses: triggers of or triggered by autoimmunity? Nat Rev Immunol. 2009;9:246–258. doi: 10.1038/nri2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fitzpatrick AM, Holguin F, Teague WG, Brown LA. Alveolar macrophage phagocytosis is impaired in children with poorly controlled asthma. J Allergy Clin Immunol. 2008;121:1372–1378. 1378. doi: 10.1016/j.jaci.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemarie E, et al. Alveolar macrophage dysfunction in malignant lung tumours. Thorax. 1984;39:448–452. doi: 10.1136/thx.39.6.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liew FY, Xu D, Brint EK, O’Neill LA. Negative regulation of toll-like receptor-mediated immune responses. Nat Rev Immunol. 2005;5:446–458. doi: 10.1038/nri1630. [DOI] [PubMed] [Google Scholar]
- 24.Jiang X, Chen ZJ. The role of ubiquitylation in immune defence and pathogen evasion. Nat Rev Immunol. 2011;12:35–48. doi: 10.1038/nri3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 26.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 27.Xu LG, et al. VISA is an adapter protein required for virus-triggered IFN-β signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 28.Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev. 2009;227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han X, Du H, Massiah MA. Detection and characterization of the in vitro e3 ligase activity of the human MID1 protein. J Mol Biol. 2011;407:505–520. doi: 10.1016/j.jmb.2011.01.048. [DOI] [PubMed] [Google Scholar]
- 30.Jin W, Chang M, Sun SC. Peli: a family of signal-responsive E3 ubiquitin ligases mediating TLR signaling and T-cell tolerance. Cell Mol Immunol. 2012;9:113–122. doi: 10.1038/cmi.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang M, Jin W, Sun SC. Peli1 facilitates TRIF-dependent Toll-like receptor signaling and proinflammatory cytokine production. Nat Immunol. 2009;10:1089–1095. doi: 10.1038/ni.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 33.Ting JP, Kastner DL, Hoffman HM. CATERPILLERs, pyrin and hereditary immunological disorders. Nat Rev Immunol. 2006;6:183–195. doi: 10.1038/nri1788. [DOI] [PubMed] [Google Scholar]
- 34.Wang RF, Miyahara Y, Wang HY. Toll-like receptors and immune regulation: implications for cancer therapy. Oncogene. 2008;27:181–189. doi: 10.1038/sj.onc.1210906. [DOI] [PubMed] [Google Scholar]
- 35.Liu SY, Sanchez DJ, Cheng G. New developments in the induction and antiviral effectors of type I interferon. Curr Opin Immunol. 2011;23:57–64. doi: 10.1016/j.coi.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- 38.Hiscott J, Nguyen TL, Arguello M, Nakhaei P, Paz S. Manipulation of the nuclear factor-κB pathway and the innate immune response by viruses. Oncogene. 2006;25:6844–6867. doi: 10.1038/sj.onc.1209941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharma S, et al. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- 40.Fitzgerald KA, et al. IKKε and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 41.Zhao T, et al. The NEMO adaptor bridges the nuclear factor-κB and interferon regulatory factor signaling pathways. Nat Immunol. 2007;8:592–600. doi: 10.1038/ni1465. [DOI] [PubMed] [Google Scholar]
- 42.Masuda Y, et al. TRIM29 regulates the assembly of DNA repair proteins into damaged chromatin. Nat Commun. 2015;6:7299. doi: 10.1038/ncomms8299. [DOI] [PubMed] [Google Scholar]
- 43.Palmbos PL, et al. ATDC/TRIM29 drives invasive bladder cancer formation through miRNA-mediated and epigenetic mechanisms. Cancer Res. 2015;75:5155–5166. doi: 10.1158/0008-5472.CAN-15-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou H, et al. Bcl10 activates the NF-κB pathway through ubiquitination of NEMO. Nature. 2004;427:167–171. doi: 10.1038/nature02273. [DOI] [PubMed] [Google Scholar]
- 45.Sun L, Deng L, Ea CK, Xia ZP, Chen ZJ. The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol Cell. 2004;14:289–301. doi: 10.1016/s1097-2765(04)00236-9. [DOI] [PubMed] [Google Scholar]
- 46.Belgnaoui SM, et al. Linear ubiquitination of NEMO negatively regulates the interferon antiviral response through disruption of the MAVS-TRAF3 complex. Cell Host Microbe. 2012;12:211–222. doi: 10.1016/j.chom.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 47.Arimoto K, et al. Polyubiquitin conjugation to NEMO by triparite motif protein 23 (TRIM23) is critical in antiviral defense. Proc Natl Acad Sci USA. 2010;107:15856–15861. doi: 10.1073/pnas.1004621107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 49.Zhang X, Goncalves R, Mosser DM. The isolation and characterization of murine macrophages. Current Protocols in Immunology. 2008;Ch. 14(Unit 14):11. doi: 10.1002/0471142735.im1401s83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lupfer C, et al. Receptor interacting protein kinase 2-mediated mitophagy regulates inflammasome activation during virus infection. Nat Immunol. 2013;14:480–488. doi: 10.1038/ni.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.