Abstract
Background
Degenerative mitral valve disease is associated with variable and complex defects in valve morphology. Three-dimensional echocardiography (3DE) has shown promise in aiding preoperative planning for patients with this disease but to date has not been as transformative as initially predicted. The clinical usefulness of 3DE has been limited by the laborious methods currently required to extract quantitative data from the images.
Methods
To maximize the utility of 3DE for preoperative valve evaluation, this work describes an automated 3DE image analysis method for generating models of the mitral valve that are well suited for both qualitative and quantitative assessment. The method is unique in that it captures detailed alterations in mitral leaflet and annular morphology and produces image-derived models with locally varying leaflet thickness. The method is evaluated on midsystolic transesophageal 3DE images acquired from 22 subjects with myxomatous degeneration and from 22 subjects with normal mitral valve morphology.
Results
Relative to manual image analysis, the automated method accurately represents both normal and complex leaflet geometries with a mean boundary displacement error on the order of one image voxel. A detailed quantitative analysis of the valves is presented and reveals statistically significant differences between normal and myxomatous valves with respect to numerous aspects of annular and leaflet geometry.
Conclusions
This work demonstrates a successful methodology for the relatively rapid quantitative description of the complex mitral valve distortions associated with myxomatous degeneration. The methodology has the potential to significantly improve surgical planning for patients with complex mitral valve disease.
Degenerative mitral valve disease is the most common cause of mitral regurgitation requiring surgical treatment in developed nations and is frequently associated with mitral valve prolapse [1]. Consideration of surgical intervention in patients with this disease involves evaluation of numerous variables, many of which are determined by echocardiographic examination [2]. Valve repair, which is preferable to valve replacement when the likelihood of successful repair is high, requires specific surgical maneuvers and techniques, the success of which are dependent on a thorough understanding of patient-specific valve defects. In the setting of myxomatous degeneration, these defects range from isolated prolapse of a single leaflet segment to thickening and aneurysmal billowing of all or most of the leaflet tissue.
Although two-dimensional echocardiography (2DE) has long been the preferred imaging modality to assess mitral valve pathology, the commercial development of three-dimensional echocardiography (3DE) imaging over the past decade has led to the awareness that subtle, yet significant, defects can be detected preoperatively with 3DE that may otherwise be discovered intraoperatively [3, 4]. Nevertheless, 3DE has not had the transformative impact on mitral valve repair that trans-esophageal 2DE had in the 1980s. Maximizing the potential of real-time 3DE imaging technology requires advanced image analysis techniques that enable the surgeon to interactively visualize the valve in 3D from any perspective and to derive quantitative measures of valve morphology that are meaningful to the surgical intervention. Commercial software packages, such as Mitral Valve Quantification (Philips Medical Systems, Andover, MA), are steps toward this goal and have demonstrated potential for morphological distinction of mitral valves with degenerative pathology [5]. With a desire for increased automation, several advanced image analysis methods for 3DE assessment of the mitral valve have been proposed [6–8], but few have been demonstrated to capture the complex valve distortions associated with mitral valve prolapse secondary to myxomatous degeneration. This stems from the inherent challenges in automatically generating models of the myxomatous mitral valve from 3DE images: the image intensity characteristics of the leaflets are similar to other structures in the image, which can make the leaflets difficult to localize, distinguish, and extract without extensive user interaction; the 3DE images may contain signal dropouts or shadowing artifacts; and the pathologic distortions in myxomatous valve morphology are diverse, which makes valve modeling more difficult. For these reasons, the existing valve modeling techniques require intensive user interaction, are restricted to simpler pathologies, or may lack patient-specific detail.
This work demonstrates an automated method for generating image-derived models of mitral valves with defects associated with myxomatous degeneration. The models are unique from others presented in the valve modeling literature in that they provide locally resolved leaflet thickness measurements, are amenable to statistical shape analysis, and are able to capture patient-specific variations in leaflet pathology. With minimal user interaction, the method generates these models by using a library of previously traced 3DE images of myxomatous mitral valves to generate valve models in new unseen 3DE images. Secondly, the image analysis pipeline incorporates a deformable modeling step that can correct for errors associated with minor to moderate signal dropouts. In this study, we leverage this image analysis method to compare quantitative image-derived measurements in a population of subjects with myxomatous mitral valve pathology and with normal mitral valve morphology. We demonstrate that these valve morphologies are accurately captured in 3DE image data, can be statistically analyzed, and are distinguished both qualitatively and quantitatively. Such methods have potential to enhance image-based surgical planning for mitral valve disease by enhancing diagnostic precision and by providing information that guides the intervention and supports the development of minimally invasive strategies to mitral valve repair.
Material and Methods
Study Population and Image Acquisition
Twenty-two patients with a posterior mitral leaflet defect due to myxomatous degeneration were retrospectively selected from an archived database of 3DE images acquired before mitral valve repair. In all cases, the posterior leaflet defect was predominantly in the P2 segment. A control group of 22 patients with normal mitral valve morphology and function, undergoing cardiac surgery unrelated to the mitral valve, was also selected. All imaging studies were performed after induction of general anesthesia and before initiation of cardiopulmonary bypass. The iE-33 platform (Philips Medical Systems, Andover, MA) was used to acquire volumetric images of the mitral valve through a mid-esophageal view with a 2- to 7-MHz transesophageal matrix-array transducer. A 3DE image of the mitral valve in its entirety at midsystole was acquired and exported in Cartesian format with an approximate isotropic resolution of 0.5 to 0.8 mm. The protocol was approved by the University of Pennsylvania Institutional Review Board.
Automated 3DE Image Analysis
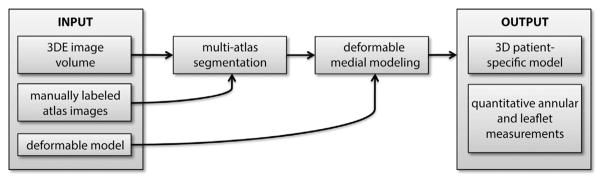
Automated image-based modeling of the mitral leaflets was performed with a method similar to that proposed in Pouch and associates [9], a schematic of which is presented in Figure 1. Given a target 3DE image to analyze, the mitral leaflets are labeled (or segmented) in the target image using a multi-atlas label fusion technique [10]. Then, a model (or template) of the valve is deformed to capture the geometry of the segmentation result. The deformed model is a patient-specific 3D geometry of the mitral leaflets from which quantitative measurements of annular and leaflet morphology can be derived. Each step of the analysis is described below.
Fig 1.
Schematic of the automated three-dimensional echocardiography (3DE) image segmentation and mitral valve modeling method.
MULTI-ATLAS LABEL FUSION
Multi-atlas label fusion is an image segmentation method that uses a set of reference atlases to label or identify an anatomical structure in a target image. An atlas is defined here as a 3DE image in which the mitral leaflets have been manually traced. In the segmentation process, each atlas is warped to the target 3DE image, thereby generating a “candidate” segmentation of that image. Because the individual candidate segmentations may not be very accurate on their own, information from the candidates is combined to produce a more accurate “consensus” segmentation of the leaflets.
To create the set of atlases required for label fusion, an expert manually traced the mitral leaflets in six of 22 images in both the normal and diseased data sets using the interactive segmentation software ITK-SNAP [11]. The anterior and posterior leaflets were individually traced in their entirety, and five landmarks were identified in each image: the anterolateral and posteromedial commissures, the midpoint on the posterior annulus, the anterior aortic peak of the annulus, and a leaflet coaptation point at the center of the valve.
Each of the 22 images in the normal and diseased categories was automatically segmented using five reference atlases from the same category, ensuring that none of the reference atlases used was the same as the target image. The manually identified landmarks were used to automatically initialize multi-atlas segmentation so that the valve was properly localized in the target 3DE image.
DEFORMABLE MEDIAL MODELING
Although multi-atlas segmentation effectively labels the mitral leaflets in a 3DE image, it does not provide a straightforward means to make clinically relevant quantitative measurements. To accomplish that goal, a model of the valve is deformed to capture the geometry of the multi-atlas segmentation result. Quantitative measurements can then be automatically computed from that model. In this work, the deformable model used to represent mitral leaflet geometry is a continuous medial representation (cm-rep), a shape descriptor that defines the mitral leaflets in terms of their medial axis [12, 13], which can be thought of as a surface that passes between the atrial and ventricular leaflet surfaces. Each point on the medial surface is associated with a thickness value, which is the distance between that point on the medial surface and the closest points on the atrial and ventricular surfaces. Stemming from this definition is the unique benefit that locally varying thickness measurements can easily be derived from the deformed model, an advantage that has not elsewhere been described for image-based mitral valve modeling.
MITRAL ANNULAR AND LEAFLET QUANTIFICATION
Once a patient-specific cm-rep of the mitral valve is obtained, quantitative measurements of the mitral annulus and leaflets are automatically computed. The mitral annular contour is defined as points on the outer edge of the deformable model. The following measurements are obtained: annular circumference, septolateral diameter, intercommissural width, annular height, and the regional and maximum annular height to commissural width ratios (AHCWR and AHmaxCWR). The AHmaxCWR is the maximum annular height divided by the intercommissural width, and the AHCWR is the localized annular height divided by the intercommissural width, plotted as a function of rotational position on the mitral annulus. In addition to annular quantification, the atrial surface area of each mitral leaflet is computed. A least squares plane is fitted through the posterior annulus points, and the surface area of the posterior leaflet above that plane is computed. In addition, the maximum height of the posterior leaflet above the posterior annular plane is calculated.
Results
To determine how accurately the deformable model captured patient-specific valve geometry, the mean boundary displacement between each deformed model and its manual atlas segmentation was computed. This leave-one-out cross-validation was performed for each of the six atlases in the normal and diseased categories. (Note that five reference atlases were used to segment each target image so that the target and atlas images were never the same.) The mean boundary displacement error was 0.4 ± 0.1 mm for the normal valves and 0.6 ± 0.1 mm for the diseased valves. This error is on the order of one voxel and is comparable to the accuracy achieved in previous studies with less complex valve morphologies [9, 14].
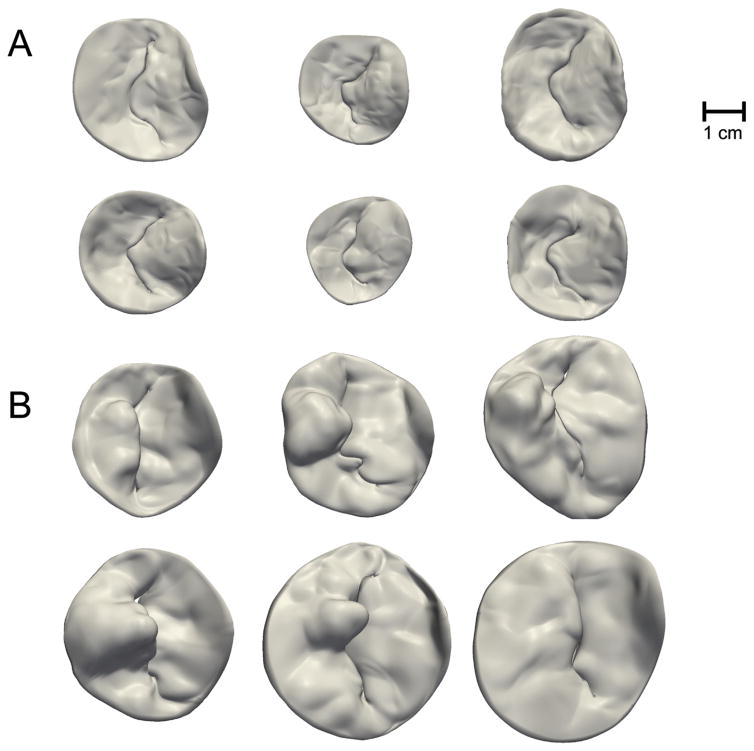
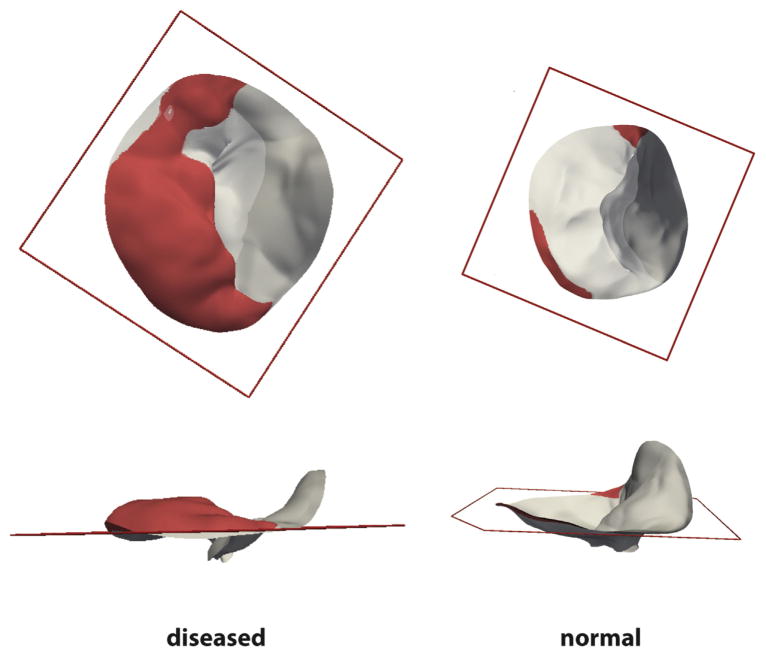
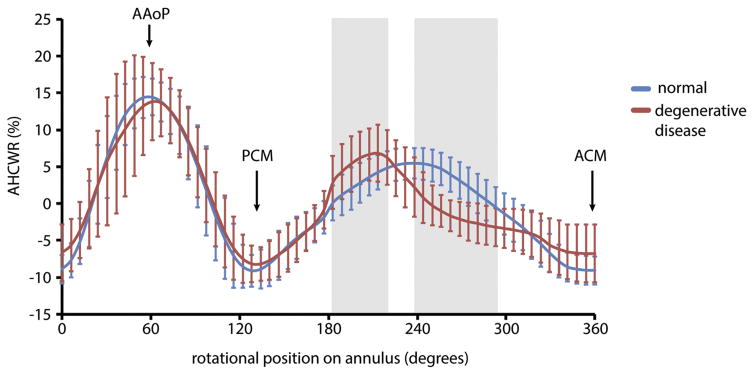
Figure 2 illustrates representative patient-specific models of normal and diseased mitral leaflets generated from the automated 3DE image analysis approach described in Methods. The valves are shown to scale and visually demonstrate the enlarged size and abnormal leaflet morphology of valves with myxomatous degeneration. Quantitative image-derived measurements of mitral annular and leaflet morphology are given in Table 1. Each of the measurements except for AHmaxCWR was statistically significant (p < 0.01) in differentiating the myxomatous and normal valves based on an independent Student’s t test. In the diseased cases, most of the posterior leaflet surface protruded above the posterior annular plane into the left atrium. In the control cases, most of the posterior leaflet surface was below the posterior annular plane, as shown in Figure 3. Although there was no statistically significant difference between AHmaxCWR measurements in the normal and diseased groups (Table 1), there were statistically significant differences in localized AHCWR measurements along the posterior annulus, as shown by the gray regions in Figure 4. Statistically significant differences in localized AHCWR were determined by computing independent Student’s t tests on the AHCWR measurements at incremental positions on the annular contour, with false discovery rate adjustment to account for multiple comparisons. The well-described saddle shape of the mitral annulus can be appreciated in the mean AHCWR curve of the normal subjects, but prominent distortions are apparent in the posterior annulus of valves with myxomatous degeneration. A similar observation is noted in Jassar and associates [15], where it was found that AHmaxCWR does not differ significantly between patients with normal and degenerative mitral valves, but there are significant localized distortions in the posterior annulus of myxomatous valves. This finding underscores the importance of assessing regional annular geometry, in addition to computing global annular measures such as diameters and maximum height.
Fig 2.
(A) Atrial views of six normal mitral valves. (B) Atrial views of six myxomatous mitral valves with posterior leaflet defects.
Table 1.
Mitral Annular and Leaflet Measurements Computed From Automated Three-Dimensional Echocardiography Image Analysis
| Measurement | Normal | Diseased | p Value |
|---|---|---|---|
| Annulus | |||
| Annular circumference | 114.2 ± 11.9 mm | 143.4 ± 16.2 mm | <0.01 |
| Septolateral diameter | 31.0 ± 3.1 mm | 39.8 ± 5.6 mm | <0.01 |
| Commissural width | 33.1 ± 4.1 mm | 41.4 ± 5.5 mm | <0.01 |
| Annular height | 8.7 ± 1.6 mm | 10.9 ± 2.4 mm | <0.01 |
| AHCWR | 26.4 ± 3.8% | 26.6 ± 5.9% | 0.88 |
| Leaflets | |||
| AL surface area | 5.09 ± 1.14 cm2 | 8.35 ± 2.08 cm2 | <0.01 |
| PL surface area | 6.40 ± 1.18 cm2 | 11.00 ± 2.72 cm2 | <0.01 |
| PL surface area above PAP | 20.9 ± 14.2% | 74.2 ± 17.3% | <0.01 |
| PL maximum height above PAP | 1.9 ± 0.9 mm | 8.1 ± 3.4 mm | <0.01 |
| Mean AL thickness | 1.6 ± 0.2 mm | 2.0 ± 0.2 mm | <0.01 |
| Maximum AL thickness | 2.8 ± 0.5 mm | 3.7 ± 0.7 mm | <0.01 |
| Mean PL thickness | 1.6 ± 0.2 mm | 1.9 ± 0.2 mm | <0.01 |
| Maximum PL thickness | 2.7 ± 0.3 mm | 4.6 ± 0.9 mm | <0.01 |
AHCWR = annular height to commissural width ratio; AL = anterior leaflet; PAP = posterior annular plane; PL = posterior leaflet.
Fig 3.
Mitral valve with posterior leaflet prolapse (left) and normal morphology (right) are shown from (top row) atrial and (bottom row) medial viewpoints. The portion of the posterior leaflet above the posterior annular plane, whose bounds are indicated by the red square, is shown in red.
Fig 4.
Mean ± SD of the annular height to commissural width ratio (AHCWR) plotted as a function of rotation position along the mitral annulus. The AHCWR curve for the myxomatous valves is shown in red; the curve for the normal valves is shown in blue. Statistically significant differences in regional AHCWR are indicated in gray. (AAoP = anterior aortic peak of annulus; ACM = anterior commissure; PCM = posterior commissure; SD = standard deviation.)
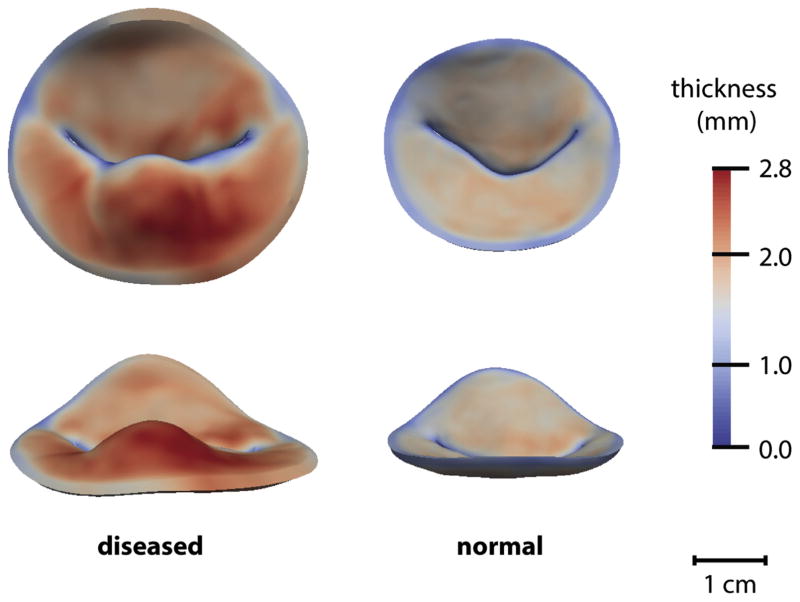
Generalized Procrustes analysis [16] was used to compute mean models of the degenerate and normal valves. Figure 5 illustrates mean models of myxomatous and normal mitral valves from two viewpoints, with thickness measurements displayed in color. The mean diseased model shows increased leaflet thickness in the prolapsed segment of the posterior leaflet.
Fig 5.
Mean models of a myxomatous mitral valve (left) and normal mitral valve (right) shown to scale (top row, atrial viewpoint; and bottom row, along the septolateral direction). Leaflet thickness is displayed in color.
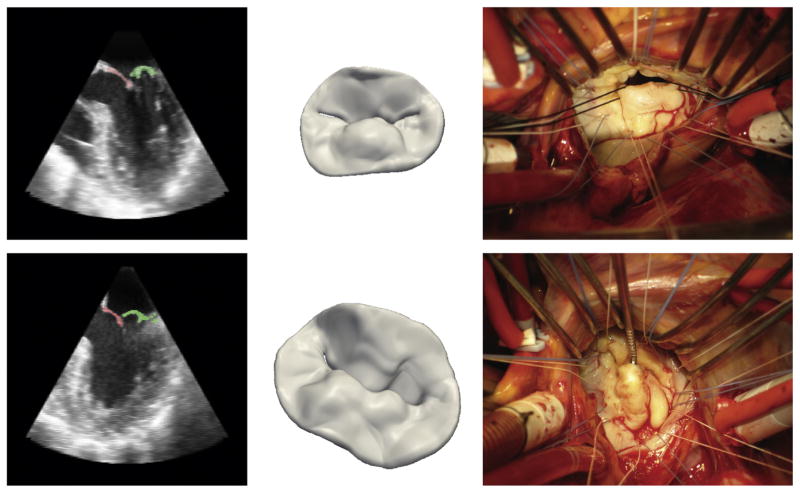
Figure 6 shows intraoperative views of two mitral valves with prolapse of the P2 segment of the posterior leaflet. A 2D cross-section of the preoperative 3DE image and the 3D model derived from automated image analysis are displayed. It is evident that the physiologic state of the mitral valve at midsystole cannot be fully appreciated once the heart is arrested and the valve is exposed; the flaccid leaflets are retracted into the left ventricle and must be manipulated to visualize the posterior leaflet defect, thereby reinforcing the value of preoperative 3DE imaging in assessing diseased valve morphology.
Fig 6.
(Left) Septolateral cross-sections of the three-dimensional image volume showing segmentation of the posterior leaflet in green and anterior leaflet in red. (Center) Three-dimensional image-derived models of the mitral valve. (Right) Intraoperative atrial views of two valves with posterior leaflet prolapse. The anterolateral and posteromedial commissures are on the left and right, respectively. The top and bottom rows are examples of two different patients’ valves.
Comment
The image analysis method described in this work is a step toward maximizing the potential of 3DE imaging in treating mitral valve disease associated with complex distortions in valve morphology. It demonstrates that complex valve morphologies associated with myxomatous degeneration can be accurately described and can be distinguished from normal valve morphologies in 3DE images. There are four potential contributions automated 3DE image analysis can make to surgical intervention. Firstly, detailed image-derived geometric models of the valve can increase diagnostic precision, enabling an accurate interpretation of the pathophysiology of MR. The technology provides the surgeon a direct quantitative analysis of in vivo valve morphology that can help to precisely describe patient-specific lesions before the valve is exposed and viewed in a nonphysiological state, as shown in Figure 6. Most important, Figure 2 demonstrates that the image analysis method described in this work has the ability to describe not only normal valve morphology, but also a spectrum of complex lesions that are observed in patients with myxomatous degeneration.
Secondly, image-based morphologic assessment, in combination with the patient’s known comorbidities, can be a means of risk stratification to determine whether a patient will benefit from valve repair over replacement. With the ability to quantitatively assess and statistically analyze large numbers of 3DE data sets, automated image analysis may help identify a combination of image-derived metrics that predict the likelihood of long-term repair durability. Moreover, an image-based assessment can aid in selecting the specific technique or combination of interventions required to effectively repair a particular lesion. As ongoing work suggests [17, 18], image-derived models of an individual patient’s valve can be used as input to biomechanical simulations that estimate leaflet stresses and predict postoperative outcomes.
Thirdly, by identifying the interventions necessary for a successful repair, advanced imaging can more effectively facilitate a match between the patient-specific lesion and the expertise of the surgeon performing the operation. Adams and colleagues [19] suggest that valve repair is currently underutilized owing to the lack of appropriate referral to surgeons with the experience necessary to operate on valves with complex pathology, commonly associated with Barlow’s disease. With the ability to produce highly detailed geometric models of valve pathology as shown in this work, 3DE image analysis can contribute to an optimized referral system that more effectively meets the needs of patients with complex cases of myxomatous degeneration and avoids unnecessary valve replacement.
Finally, the development of minimally invasive mitral valve repair strategies must occur in tandem with advances in imaging technology. Automated analysis of real-time 3DE imaging provides an interactive visualization that can guide a percutaneous valve procedure, which is essential when direct visualization is not feasible. Moreover, automated analysis of 3DE images is not only useful intraoperatively; the capability to analyze a large quantity of 3DE datasets can also reveal anatomic and functional variations in normal and diseased valves that can lead to the development of novel devices and approaches to valve repair.
Several challenges must still be overcome before automated 3DE image analysis becomes routine in pre-operative surgical planning. The commercialized software packages and image analysis methods proposed in the literature use methods that vary in the time required for analysis, the detail with which patient-specific morphologies are represented, and the quantitative measures that can be computed from the analysis. An optimal tradeoff between these must be sought. In its current form, the image analysis pipeline presented in this work can be completed in 5 to 10 minutes with the use of parallel processing, which is a promising performance time for use in the immediate preoperative period. Roughly 1 minute of this time is required for a user to identify five landmarks on the valve, which is the only step requiring manual interaction. To construct a time-resolved model that can be used for simulation or to analyze valve dynamics, the multiple-step analysis presented here must be performed on a frame-by-frame basis. Efforts are under way to increase computational efficiency, eliminate the need for manual landmarking in 3DE images with varying field of view, and perform a fully four-dimensional segmentation so that measurements of valve dynamics can be incorporated in a more comprehensive disease assessment. With these advances, automated 3DE mitral valve analytics has potential to improve surgical interventions for even the most complex valve diseases.
Acknowledgments
Funding for this work was provided by the National Institutes of Health: HL119010, HL63954, HL73021, HL103723, HL108330, HL113216, and HL119297.
References
- 1.Enriquez-Sarano M, Akins CW, Vahanian A. Mitral regurgitation. Lancet. 2009;373:1382–94. doi: 10.1016/S0140-6736(09)60692-9. [DOI] [PubMed] [Google Scholar]
- 2.Castillo JG, Solis J, Gonzalez-Pinto A, Adams DH. Surgical echocardiography of the mitral valve. Rev Esp Cardiol. 2011;64:1169–81. doi: 10.1016/j.recesp.2011.06.025. [DOI] [PubMed] [Google Scholar]
- 3.Grewal J, Mankad S, Freeman WK, et al. Real-time three-dimensional transesophageal echocardiography in the intraoperative assessment of mitral valve disease. J Am Soc Echocardiogr. 2009;22:34–41. doi: 10.1016/j.echo.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 4.Pepi M, Tamborini G, Maltagliati A, et al. Head-to-head comparison of two- and three-dimensional transthoracic and transesophageal echocardiography in the localization of mitral valve prolapse. J Am Coll Cardiol. 2006;48:2524–30. doi: 10.1016/j.jacc.2006.02.079. [DOI] [PubMed] [Google Scholar]
- 5.Chandra S, Salgo IS, Sugeng L, et al. Characterization of degenerative mitral valve disease using morphologic analysis of real-time three-dimensional echocardiographic images: objective insight into complexity and planning of mitral valve repair. Circ Cardiovasc Imaging. 2011;4:24–32. doi: 10.1161/CIRCIMAGING.109.924332. [DOI] [PubMed] [Google Scholar]
- 6.Ionasec RI, Voigt I, Georgescu B, et al. Patient-specific modeling and quantification of the aortic and mitral valves from 4-D cardiac CT and TEE. IEEE Trans Med Imaging. 2010;29:1636–51. doi: 10.1109/TMI.2010.2048756. [DOI] [PubMed] [Google Scholar]
- 7.Schneider RJ, Tenenholtz NA, Perrin DP, Marx GR, del Nido PJ, Howe RD. Patient-specific mitral leaflet segmentation from 4D ultrasound. Med Image Comput Comput Assist Interv. 2011;14:520–7. doi: 10.1007/978-3-642-23626-6_64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verhey JH, Nathan NS, Rienhoff O, Kikinis R, Rakebrandt F, D’Ambra MN. Finite-element-method (FEM) model generation of time-resolved 3D echocardiographic geometry data for mitral-valve volumetry. Biomed Eng Online. 2006;5:17. doi: 10.1186/1475-925X-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pouch AM, Wang H, Takabe M, et al. Fully automatic segmentation of the mitral leaflets in 3D transesophageal echocardiographic images using multi-atlas joint label fusion and deformable medial modeling. Med Image Anal. 2014;18:118–29. doi: 10.1016/j.media.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang HZ, Suh JW, Das SR, Pluta JB, Craige C, Yushkevich PA. Multi-atlas segmentation with joint label fusion. IEEE Trans Pattern Anal Mach Intell. 2013;35:611–23. doi: 10.1109/TPAMI.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yushkevich PA, Piven J, Hazlett HC, et al. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31:1116–28. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 12.Pizer SM, Fletcher PT, Joshi S, et al. Deformable M-reps for 3D medical image segmentation. Int J Comput Vision. 2003;55:85–106. doi: 10.1023/a:1026313132218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yushkevich PA, Zhang H, Gee JC. Continuous medial representation for anatomical structures. IEEE Trans Med Imaging. 2006;25:1547–64. doi: 10.1109/tmi.2006.884634. [DOI] [PubMed] [Google Scholar]
- 14.Jassar AS, Brinster CJ, Vergnat M, et al. Quantitative mitral valve modeling using real-time three-dimensional echocardiography: technique and repeatability. Ann Thorac Surg. 2011;91:165–71. doi: 10.1016/j.athoracsur.2010.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jassar AS, Vergnat M, Jackson BM, et al. Regional annular geometry in patients with mitral regurgitation: implications for annuloplasty ring selection. Ann Thorac Surg. 2014;97:64–70. doi: 10.1016/j.athoracsur.2013.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dryden IL, Mardia KV. Statistical shape analysis. Chichester, UK: John Wiley & Sons; 1998. [Google Scholar]
- 17.Sprouse C, Mukherjee R, Burlina P. Mitral valve closure prediction with 3D personalized anatomical models and anisotropic hyperelastic tissue assumptions. IEEE Trans Biomed Eng. 2013;60:3238–47. doi: 10.1109/TBME.2013.2272075. [DOI] [PubMed] [Google Scholar]
- 18.Votta E, Le TB, Stevanella M, et al. Toward patient-specific simulations of cardiac valves: state-of-the-art and future directions. J Biomech. 2013;46:217–28. doi: 10.1016/j.jbiomech.2012.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams A, Anyanwu AC. Seeking a higher standard for degenerative mitral valve repair: begin with etiology. J Thorac Cardiovasc Surg. 2008;136:551–6. doi: 10.1016/j.jtcvs.2007.10.060. [DOI] [PubMed] [Google Scholar]