Abstract
Introduction
Tyrosine kinase inhibitors (TKIs) have been developed to treat patients with epidermal growth factor receptor (EGFR)-mutant lung cancers. However, the therapeutic efficacy of TKIs in patients with uncommon EGFR mutations remains unclear.
Methods
Next-generation sequencing was performed on a patient’s lung adenocarcinoma tumor sample, revealing rare combined in cis (on the same allele) EGFR mutations. Stable Ba/F3 and NIH-3T3 cell lines harboring the mutations were established to investigate the effect of first, second, and third generation EGFR TKIs on cell proliferation by MTS assay and EGFR phosphorylation by Western blotting.
Results
EGFR L858M/L861Q mutations in cis were detected in a non-small cell lung cancer patient’s tumor. The patient demonstrated primary resistance to erlotinib and was subsequently treated with afatinib, which caused tumor regression. In in vitro studies, first and third generation TKIs exhibited a decreased capacity to prevent EGFR phosphorylation and inhibit cell proliferation in EGFR L858M/L861Q cells compared to cells harboring the common EGFR L858R point mutation. In contrast, afatinib treatment reduced proliferation and inhibited EGFR phosphorylation in L858M/L861Q and L858R mutant cells at similar concentrations.
Conclusions
Afatinib may be a beneficial therapeutic option for a subset of lung cancer patients with rare EGFR mutations in their tumors. Understanding how uncommon mutations affect protein structure and TKI binding will be important for identifying effective targeted therapies for these patients.
Keywords: NSCLC, EGFR mutation, afatinib, targeted therapy
INTRODUCTION
Epidermal growth factor receptor (EGFR) mutations are reported in approximately 10–30% of non-small cell lung cancers (NSCLCs) worldwide.1–3 These mutations occur in the tyrosine kinase domain and result in ligand-independent activation of the receptor and signaling through downstream pro-survival and anti-apoptotic pathways. The most common EGFR mutations are in-frame deletions in exon 19 and the L858R point mutation in exon 21.3 Most patients whose tumors harbor these classic mutations have significant radiographic and clinical responses to the first generation reversible ATP-competitive EGFR tyrosine kinase inhibitors (TKIs) erlotinib and gefitinib and the second generation irreversible TKI afatinib.1,4,5
However, less common mutations are found in approximately 10–15% of EGFR-mutant lung cancers,3,6 and the relevance of these mutations as clinical biomarkers for targeted therapy has not been clearly established. Here, we describe the identification of novel EGFR L858M/L861Q mutations in cis (on the same allele) in a NSCLC patient that confer decreased sensitivity to first and third generation EGFR TKIs while retaining responsiveness to afatinib. Our findings suggest that afatinib may be a beneficial therapeutic option for patients with rare EGFR mutations with primary resistance to first-generation EGFR inhibitors.
MATERIALS AND METHODS
Patient information
The patient, a 62-year-old Caucasian female with < 1 pack year smoking history, presented with symptoms of shortness of breath that were initially felt to be panic attacks. Two months later, her exertional shortness of breath worsened. A chest X-ray was obtained and identified a diffuse infiltrative process predominately in the upper lobes. A chest CT identified irregular interlobular septal thickening predominately in the upper lobes along with nodularity in a subpleural distribution. Given the diffuse nature of the process, a video assisted thoracoscopic surgical wedge resection from the right upper and middle lobes was performed. Pathologic evaluation demonstrated extensive involvement by a poorly differentiated adenocarcinoma. Immunohistochemistry demonstrated the tumor cells to be positive for cytokeratin 7, CEA, TTF1 with focal cells also staining for mucin. The patient’s tumor specimen was sent for targeted next generation sequencing (NGS) to help guide treatment.
DNA extraction, NGS, and data analysis
Targeted sequencing was performed using a next generation sequencing assay (“OncoPanel”) to detect single nucleotide variants, small insertion-deletion mutations, copy number variations, and structural variants in tumor DNA extracted from fresh, frozen or formalin-fixed paraffin-embedded samples containing at least 20% tumor cells.7–9 The assay covers the complete exonic sequences of 300 cancer genes and 113 introns across 35 genes for genomic rearrangements.
Cell lines and reagents
Ba/F3 cells were maintained in RPMI-1640 (Gibco) supplemented with 10% FBS and 1% penicillin/streptomycin (Gibco), and NIH-3T3 cells were maintained in DMEM (Gibco) supplemented with 10% fetal calf serum and 1% penicillin/streptomycin. Generation of EGFR L858R Ba/F3 and EGFR L858R NIH-3T3 cells was previously described.10,11 Gefitinib, afatinib, and osimertinib were purchased from Selleck Chemicals, and stock solutions of drugs were prepared in DMSO.
EGFR L858M/L861Q mutant construct and retroviral infection
The L858M and L861Q point mutations were introduced into human EGFR cDNA by site-directed mutagenesis using the Quick Change Site-Directed Mutagenesis kit (Stratagene) according to the manufacturer’s protocol with the following primers: 5′-ttccgcacccagctgtttggccatcccaaaatctgtg-3′ and 5′-cacagattttgggatggccaaacagctgggtgcggaa-3′. Successful generation of the construct was confirmed by DNA sequencing. The construct was shuttled into the retroviral vector JP1540 using the Agilent Cre Recombinase kit (Agilent). Ba/F3 and NIH-3T3 cells were infected with retrovirus according to standard protocols.12 Stably-transfected cell lines were obtained by puromycin selection (2 μg/mL; Sigma-Aldrich) for NIH-3T3 cells and by puromycin selection and IL-3 withdrawal for Ba/F3 cells. EGFR expression was verified by Western blotting, and pooled stable cells were used for experiments.
Cell proliferation assays
Cell growth/growth inhibition was assessed by MTS assay. 3000 cells were seeded per well in 96 well plates and exposed to drug (3.3 nM to 10 μM) for 72 hours All experimental points included 6 to 12 wells, and data were averaged from three independent experiments. Data were displayed graphically using GraphPad Prism (GraphPad Software, Inc.), and the curves were fitted using a nonlinear regression model with sigmoidal dose response.
Western blotting analysis
EGFR L858R and EGFR L858M/L861Q NIH-3T3 cells were plated at 1×106 cells per 10-cm plate and treated with the indicated concentrations of inhibitors for 6 hours the following day. After 6 hours of treatment, cells were washed with PBS and lysed with NP40 buffer (Calbiochem) supplemented with Complete Mini protease inhibitor and PhosSTOP phosphatase inhibitors (Roche). Lysates were separated by SDS-PAGE gel, transferred to nitrocellulose membranes, and probed the following antibodies: phospho-EGFR(Y1068) (3777), total EGFR (2232) (Cell Signaling), and tubulin (T9026) (Sigma Aldrich). Densitometry was measured using ImageJ. Samples were normalized to tubulin before determining the ratio of phospho-EGFR in treated versus untreated samples.
RESULTS
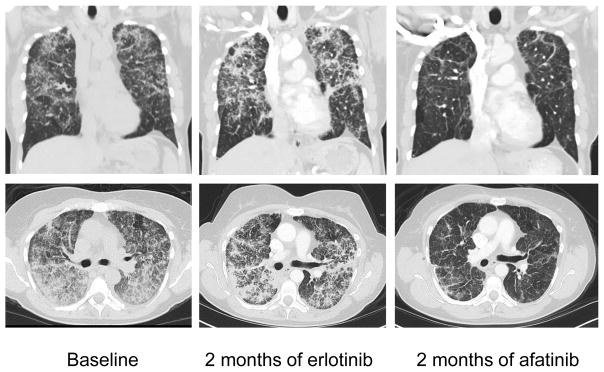
Targeted NGS of the patient’s tumor revealed two EGFR mutations, L858M (in 30% of 190 reads) and L861Q (in 28% of 175 reads). These two mutations were in cis with one another (Fig. 1). Based on these findings, the patient was initiated on erlotinib therapy at 150 mg once daily 4 months after initial presentation (Fig. 2). At the time of erlotinib initiation, the patient was using supplemental oxygen (2–3 liters/minute). After 1 month of treatment, her erlotinib dose had to be reduced to 100 mg once daily due to skin toxicity. Repeat staging 2 months after initiation of erlotinib demonstrated stable disease (Fig. 2). However, after 4 months of erlotinib, her shortness of breath and oxygen requirement (3–4 liters/minute) had worsened. She was started on combination chemotherapy with carboplatin and pemetrexed and received 4 cycles of treatment. Repeat imaging demonstrated a minor response. The patient still required supplemental oxygen (3 liters/minute) and had substantial shortness of breath. After 4 months of chemotherapy, and following the development of worsening shortness of breath, she was switched to afatinib 40 mg once daily. The patient noted improvement in her breathing with just a few days of treatment. Following 2 weeks of afatinib therapy, she no longer required supplemental oxygen and her shortness of breath had markedly improved. Repeated imaging after 2 months of afatinib demonstrated a dramatic radiographic response (Fig. 2). She has remained on afatinib, with grade 1 diarrhea as her only side effect, for 10 months and continues treatment as of the writing of this report.
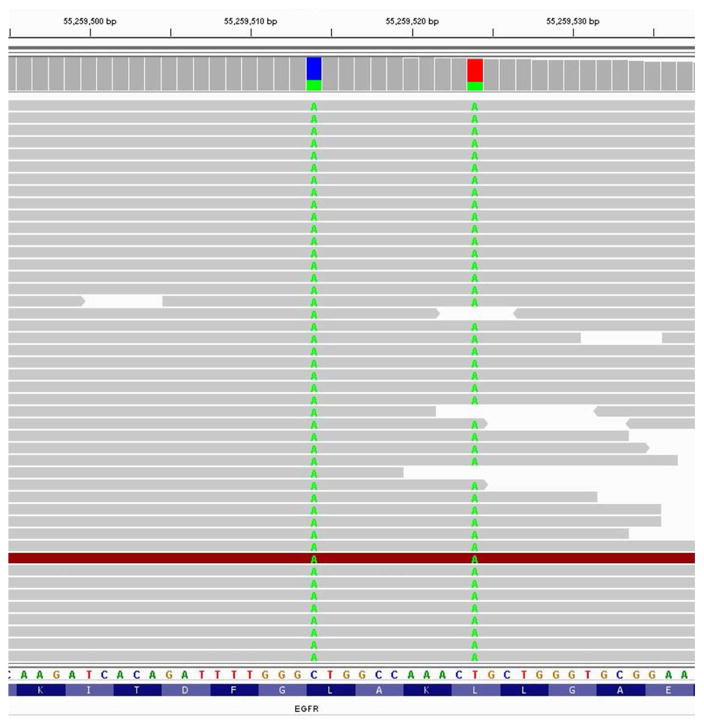
Figure 1. Identification of EGFR L858M and L861Q mutations in cis from lung cancer tumor biopsy.
Alignment of NGS sequencing reads from the patient’s tumor reveals the 2572C>A and 2582T>A nucleotide substitutions resulting in L858M and L861Q missense mutations in exon 21 of EGFR. These substitutions co-occurred in the same reads, indicating that the mutations are in cis.
Figure 2. Computed tomography (CT) scans of a NSCLC patient exhibiting primary resistance to erlotinib and subsequent response to afatinib.
Coronal and transverse CT scan images at baseline before treatment, after two months of erlotinib treatment, and after two months of afatinib treatment.
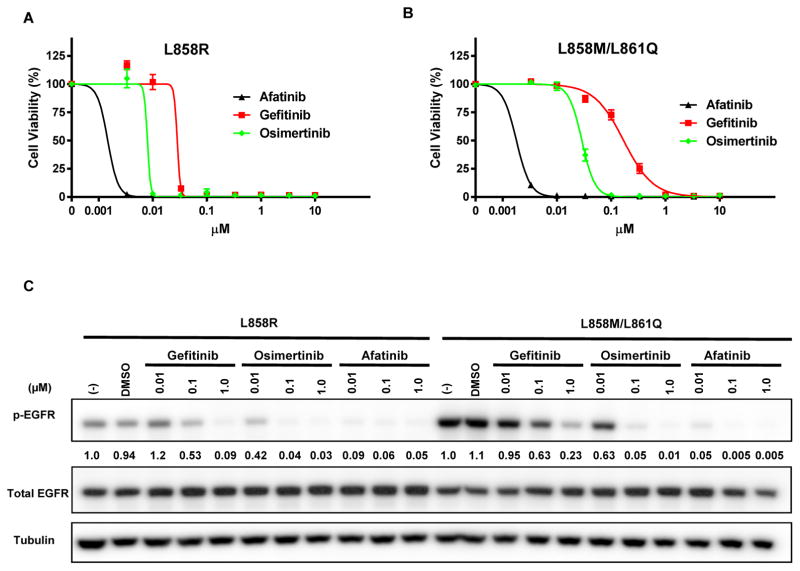
To understand whether the patient’s resistance to erlotinib and sensitivity to afatinib could be explained by the unique EGFR L858M/L861Q mutations, the effect of the L858M/L861Q mutations on drug sensitivity was examined in vitro. Stable EGFR L858M/L861Q Ba/F3 cells were generated, and cell viability/proliferation after treatment with EGFR TKIs was measured by MTS assay. Compared to EGFR L858R mutant cells, L858M/L861Q cells demonstrated reduced sensitivity to gefitinib treatment (L858R IC50 28.3 nM vs. L858M/L861Q IC50 169.1 nM) (Fig. 3A, B), but the response to afatinib was similar for both cell lines (L858R IC50 1.5 nM vs. L858M/L861Q IC50 1.8 nM). In addition, L858M/L861Q cells exhibited decreased sensitivity to osimertinib, a third generation irreversible and mutant-specific EGFR inhibitor (L858R IC50 8.0 nM vs. L858M/L861Q IC50 28.8 nM) (Fig. 3A, B). Using stable EGFR L858M/L861Q NIH-3T3 cells, we examined the capacity of gefitinib, afatinib, and osimertinib to prevent EGFR phosphorylation by Western blotting. Gefitinib and osimertinib inhibited EGFR L858M/L861Q phosphorylation less potently than EGFR L858R (Fig. 3C), while afatinib treatment significantly reduced EGFR L858M/L861Q phosphorylation even at low concentrations.
Figure 3. In vitro studies demonstrate reduced sensitivity of EGFR L858M/L861Q to gefitinib and osimertinib compared to EGFR L858R.
Viability of (A) EGFR L858R and (B) EGFR L858M/L861Q Ba/F3 cells after 72 hours of treatment with gefinitib, osimertinib, and afatinib, detected by MTS assay. Error bars represent standard error of the mean. (C) Immunoblotting of lysates from EGFR L858R and EGFR L858M/L861Q NIH-3T3 cells treated with the indicated concentrations of gefitinib, osimertinib, or afatinib. The ratio of phospho-EGFR in treated vs. untreated samples was quantified by densitometry for each cell line.
DISCUSSION
In the era of precision medicine, genotyping of EGFR has become a standard practice for NSCLC, but the identification of a rare mutation does not necessarily indicate a clear course of action for therapy. This is the first report of a patient with EGFR-mutant lung cancer with primary resistance to erlotinib but responsiveness to afatinib, and our in vitro findings indicate that this therapeutic response was the result of rare EGFR L858M/L861Q cis mutations in the patient’s tumor. According to the COSMIC database (v.78), EGFR L858M/L861Q cis mutations have been reported in three other lung cancer patients, although no data is available on the treatment or outcome of these patients.13–15 Previous reports suggest that patients with EGFR L861Q mutations have a response rate of 50–60% to gefitinib and erlotinib, responding less favorably than patients with the more common L858R point mutation or exon 19 deletion.6,16 Similarly, our findings suggest that the potency of first generation inhibitors to prevent EGFR signaling is reduced in the setting of the L858M/L861Q mutation. Additionally, our results indicate that treatment with the mutant-specific inhibitor osimertinib may not sufficiently inhibit EGFR L858M/L861Q signaling, suggesting that afatinib may be the most beneficial first line therapy for a subset of patients with rare EGFR mutations. Together, these data suggest that the various EGFR inhibitors may have distinct capabilities to inhibit signaling of specific EGFR mutants. Additional studies to understand how uncommon mutations affect EGFR protein structure and TKI binding as well as studies to measure the potency of TKIs in the setting of rare mutations will provide valuable therapeutic insight for NSCLC patients with uncommon EGFR mutations in their tumors.
Acknowledgments
Funding: This work was supported by grants from the National Institute of Health (R01 CA135257 (P.A.J.) and R01 CA114465 (P.A.J.) and by the James B. Gillen Thoracic Oncology Research (P.A.J.) and Goldstein Family Research Funds (P.A.J.)
ABBREVIATIONS
- CT
Computed tomography
- EGFR
Epidermal growth factor receptor
- NGS
Next-generation sequencing
- NSCLC
Non-small cell lung cancer
- TKI
Tyrosine kinase inhibitor
Footnotes
Conflicts of interest:
Jamie A. Saxon: none
Lynette M. Sholl: none
Pasi A. Jänne: Consultant: AstraZeneca, Boehringer Ingelheim, Roche/Genentech, Chugai Pharmaceuticals, Pfizer, Ariad Pharmaceuticals, Merrimack Pharmaceuticals, Ignyta, LOXO Oncology; Sponsored Research: Astellas, AstraZeneca, PUMA, Daiichi Sankyo; Stock ownership: Gatekeeper Pharmaceuticals; Other: post-marketing royalties from DFCI owned intellectual property on EGFR mutations licensed to Lab Corp.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Paez JG, Jänne P, Lee J, et al. EGFR mutations in lung cancer3: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 2.Lynch T, Bell D, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. Science. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 3.Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97(5):339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 4.Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361(10):958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 5.Sequist LV, Yang JC-H, Yamamoto N, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol. 2013;31(27):3327–3334. doi: 10.1200/JCO.2012.44.2806. [DOI] [PubMed] [Google Scholar]
- 6.Wu J-Y, Yu C-J, Chang Y-C, Yang C-H, Shih J-Y, Yang P-C. Effectiveness of tyrosine kinase inhibitors on “uncommon” epidermal growth factor receptor mutations of unknown clinical significance in non-small cell lung cancer. Clin Cancer Res. 2011;17(11):3812–3821. doi: 10.1158/1078-0432.CCR-10-3408. [DOI] [PubMed] [Google Scholar]
- 7.Wagle N, Berger MF, Davis MJ, et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;2(1):82–93. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abo RP, Ducar M, Garcia EP, et al. BreaKmer: detection of structural variation in targeted massively parallel sequencing data using kmers. Nucleic Acids Res. 2015;43(3):e19. doi: 10.1093/nar/gku1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Awad MM, Oxnard GR, Jackman DM, et al. MET exon 14 mutations in non-small-cell lung cancer are associated with advanced age and stage-dependent MET genomic amplification and c-Met overexpression. J Clin Oncol. 2016;34(7):721–730. doi: 10.1200/JCO.2015.63.4600. [DOI] [PubMed] [Google Scholar]
- 10.Greulich H, Chen TH, Feng W, et al. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. PLoS Med. 2005;2(11):1167–1176. doi: 10.1371/journal.pmed.0020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang J, Greulich H, Jänne PA, Sellers WR, Meyerson M, Griffin JD. Epidermal growth factor-independent transformation of Ba/F3 cells with cancer-derived epidermal growth factor receptor mutants induces gefitinib-sensitive cell cycle progression. Cancer Res. 2005;65(19):8968–8974. doi: 10.1158/0008-5472.CAN-05-1829. [DOI] [PubMed] [Google Scholar]
- 12.Engelman JA, Mukohara T, Zejnullahu K, et al. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J Clin Invest. 2006;116(10):2695–2706. doi: 10.1172/JCI28656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forbes SA, Beare D, Gunasekaran P, et al. COSMIC: Exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43(D1):D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tam IYS, Chung LP, Suen WS, et al. Distinct epidermal growth factor receptor and KRAS mutation patterns in non-small cell lung cancer patients with different tobacco exposure and clinicopathologic features. Clin Cancer Res. 2006;12(5):1647–1653. doi: 10.1158/1078-0432.CCR-05-1981. [DOI] [PubMed] [Google Scholar]
- 15.Skronski M, Chorostowska-Wynimko J, Szczepulska E, et al. Reliable detection of rare mutations in EGFR gene codon L858 by PNA-LNA PCR clamp in non-small cell lung cancer. Adv Exp Med Biol. 2013;756:321–331. doi: 10.1007/978-94-007-4549-0_39. [DOI] [PubMed] [Google Scholar]
- 16.Yeh P, Chen H, Andrews J, Naser R, Pao W, Horn L. DNA-mutation Inventory to Refine and Enhance Cancer Treatment (DIRECT): A catalog of clinically relevant cancer mutations to enable genome-directed anticancer therapy. Clin Cancer Res. 2013;19(7):1894–1901. doi: 10.1158/1078-0432.CCR-12-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]