Abstract
RNase II is the most active exoribonuclease in Escherichia coli cell extracts. Yet, its removal appears to have no deleterious effect on growing cells. Here, we show that RNase II is required for cell survival during prolonged stationary phase and upon starvation. The absence of RNase II leads to greatly increased rRNA degradation and to the accumulation of rRNA fragments, both of which lead to a decline in cell survival. The deleterious effects of RNase II removal can be completely reversed by the simultaneous absence of a second exoribonuclease, RNase PH, an enzyme known to be required to initiate ribosome degradation in starving cells. We have now found that the role of RNase II in this process is to regulate the amount of RNase PH present in starving cells, and it does so at the level of RNase PH stability. RNase PH normally decreases as much as 90% during starvation because the protein is unstable under these conditions; however, in the absence of RNase II the amount of RNase PH remains relatively unchanged. Based on these observations, we propose that in the presence of RNase II, nutrient deprivation leads to a dramatic reduction in the amount of RNase PH, thereby limiting the extent of rRNA degradation and ensuring cell survival during this stress. In the absence of RNase II, RNase PH levels remain high, leading to excessive ribosome loss and ultimately to cell death. These findings provide another example of RNase regulation in response to environmental stress.
Keywords: exoribonucleases, ribosome, rRNA, Escherichia coli
INTRODUCTION
Bacteria need to continually sense and respond to environmental stress, and this requires gene regulation at both the transcriptional and post-transcriptional levels. Ribonucleases (RNases) play an important role in these responses inasmuch as they participate in all aspects of RNA metabolism and consequently are crucial for determining the intracellular levels of RNA species (Bandyra and Luisi 2013; Hui et al. 2014; Deutscher 2015b; Durand et al. 2015). Currently, close to 20 ribonucleases have been characterized in Escherichia coli, among which are eight exoribonucleases, including six that are hydrolytic and two that are phosphorolytic (Li and Deutscher 2004).
RNase II, a 3′–5′ processive exoribonuclease, is the major hydrolytic exoribonuclease, accounting for ∼90% of the total degradative activity in a cell extract (Nossal and Singer 1968; Cheng and Deutscher 2002). RNase II is not essential for normal growth in E. coli, but mutant cells lacking both polynucleotide phosphorylase (PNPase) and RNase II are inviable (Donovan and Kushner 1986), indicating that these two RNases have significant functional overlap. It is known that these enzymes, as well as the exoribonucleases RNase R and RNase PH, are particularly important for ribosomal RNA (rRNA) metabolism as they participate in both rRNA maturation and degradation (Basturea et al. 2011; Sulthana and Deutscher 2013; Sulthana et al. 2016). Despite their importance, overproduction of these RNases, or RNases in general, often can be deleterious to a cell as it may lead to degradation of functionally important RNAs, which either are not protected or which become exposed under certain stress conditions (Lehnik-Habrink et al. 2012; Arraiano et al. 2013; Deutscher 2015a). Likewise, too little of a required RNase can lead to poor adaptation to a particular stress. Based on these considerations, it is becoming clear that RNases need to be regulated (Deutscher 2015a) and that regulation of RNases is particularly important in various stress situations.
Ribosomes are the protein-synthesizing machines of the cell, and as such, they play a pivotal role in determining the overall gene expression profile of the cell. Although generally stable in growing E. coli cells, ribosomes can undergo degradation under conditions of stress due to reduced translation that increases the free subunit population, rendering them susceptible to ribonucleases (Okamura et al. 1973; Cohen and Kaplan 1977; Zundel et al. 2009). During starvation, rRNA degradation is RNase PH dependent as this enzyme initiates removal of nucleotides from the 3′ end of 16S rRNA that ultimately leads to endonucleolytic cleavages by RNase E and complete breakdown of rRNA (Basturea et al. 2011; Sulthana et al. 2016). In addition, rRNA degradation is greatly affected by RNase II as its removal leads to as much as a threefold increase in rRNA breakdown during starvation by a heretofore unknown mechanism (Basturea et al. 2011).
RNase PH is a 3′–5′ phosphorolytic exoribonuclease that participates in the maturation of tRNA and degradation of structured RNAs (Deutscher 1990; Jain 2012). In addition, the enzyme was found to be beneficial for acquired stress resistance in E. coli during laboratory selection (Herring et al. 2006; Conrad et al. 2009; Dragosits et al. 2013). In fact, deletion of the rph gene was the most frequently observed mutation during the laboratory evolution of E. coli cells over many generations in minimal medium and under stress conditions. However, why the absence of RNase PH would be beneficial to cells under these conditions is still unknown. RNase PH is also known to be absent in certain widely used E. coli K-12 strains, such as MG1655 (Jensen 1993), further emphasizing that the enzyme is readily lost.
In this paper, we show that RNase PH is normally down-regulated during stationary phase, and that this regulation is important for maintaining the proper amounts of ribosomes in stationary phase and upon starvation. Most interestingly, we identify for the first time an essential function for RNase II. Thus we find that the reduction in RNase PH levels is regulated by RNase II. In its absence, the amount of RNase PH remains high, leading to excessive ribosome degradation and ultimately to cell death. In addition to identifying a novel regulatory pathway, this work also may explain why deletion of the rph gene occurs at such high frequency in laboratory evolution experiments and why the absence of RNase PH is beneficial under stress conditions.
RESULTS
Although ribosomes are stable in growing E. coli cells, they may become substrates for degradation under conditions of starvation when the need for functional ribosomes decreases. In earlier experiments exploring the role of various RNases in the degradative process (Basturea et al. 2011), we observed that the absence of RNase II, a processive exoribonuclease, led to a dramatic increase in acid-soluble material derived from rRNA. Although that finding was not explored further at the time, it implied that the presence of RNase II stabilized rRNA, a counterintuitive suggestion. Here, we examine in detail the basis for this unusual observation and explore how RNase II affects rRNA degradation.
RNase II is essential during stationary phase
To begin examination of the role of RNase II, we first determined the effect of its removal on the plating efficiency of wild-type cells (MG1655*I−) and their RNase II− derivative (MG1655*I−II−) during growth in M9/glucose medium and over several days in stationary phase at 31°C and 42°C. Under these conditions, cells naturally depleted nutrients, ultimately leading to a starvation condition. Plating efficiency was determined at periodic intervals by plating on LB medium plates at 37°C. As can be seen in Figure 1, which represents a typical experiment, plating efficiency of the wild-type strain was little changed over a period of 96 h at either 31°C or 42°C, whereas cells lacking RNase II displayed decreased plating efficiency such that by 96 h their plating efficiency was substantially reduced. Although there was some variation in the degree of reduction in plating efficiency of the RNase II− cells, varying from 65% (one experiment at 31°C) to >95% (two experiments at both temperatures) in six independent experiments, in most cases, the decrease was in the range of 80%–90%.
FIGURE 1.
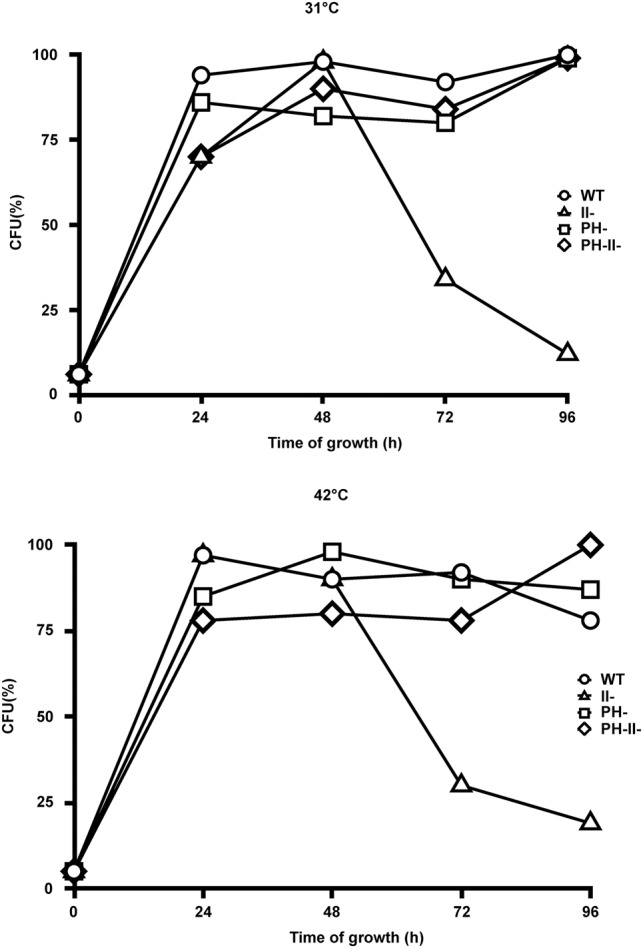
Survival of wild-type and mutant cells during stationary phase. WT, RNase II−, RNase PH−, and RNase II− RNase PH− strains were continually grown in 50 mL M9/glucose media at either 31°C or 42°C. At regular time intervals, 2 mL of cultures were collected, two appropriate sequential dilutions of cells in duplicate were spread on LB plates, and viable cells (CFU) were determined after overnight incubation at 37°C. The data from one of the sequential dilutions at each time point are presented as a percentage in which the time point with the highest number of colonies was set at 100%. A representative experiment from those repeated six times is shown.
Importantly, there was no loss in the A600 of the RNase II− cells during the course of growth, suggesting that they were not lysing upon prolonged incubation; this conclusion was supported by the fact that there was no loss of RNA from the cells even though they were pelleted prior to RNA isolation (see below). Also, incubation of the LB plates for as long as 4 d did not lead to the appearance of any additional colonies, indicating that the reduced plating efficiency of the RNase II− cells was not due to a delay in their ability to form colonies. Based on these findings, we believe that the reduced plating efficiency of the RNase II− cells is due to a loss of viability, and therefore, that RNase II is an essential enzyme for E. coli survival during prolonged stationary phase.
Previously, we found that another exoribonuclease, RNase PH, was required to initiate rRNA breakdown during starvation (Basturea et al. 2011). It therefore was of interest to determine whether this enzyme also affected survival during stationary phase. As can be seen in Figure 1, the absence of RNase PH, by itself, did not affect the viability of cells at any temperature. However, removal of RNase PH, in addition to RNase II, completely reversed the loss of viability caused by the absence of RNase II. These findings suggest a close functional relationship between the two exoribonucleases that becomes evident as cells approach starvation.
RNase II is also required during starvation at elevated temperature
Inasmuch as natural depletion of nutrients is time consuming and impractical for routine experiments, we instead used a starvation protocol that has been examined extensively in our previous studies (Zundel et al. 2009; Basturea et al. 2011; Sulthana et al. 2016) for most of the remaining experiments. Cells were initially grown in M9/glucose to early exponential phase at 31°C and then rapidly starved by centrifugation and resuspension in medium lacking glucose. Survival was determined by measurement of viable cells at various times after the initiation of starvation at either 31°C or 42°C (Fig. 2). At 31°C, both wild-type and RNase II− cells remained viable over a period of at least 20 h. In contrast, at 42°C, while wild-type cells remained viable, cells lacking RNase II rapidly lost viability such that only ∼30% remained after 4 h and <10% were viable after 6 h of starvation. This result indicates that, as in the natural depletion of nutrients described above, the presence of RNase II is also required for cell survival during starvation at higher temperature.
FIGURE 2.
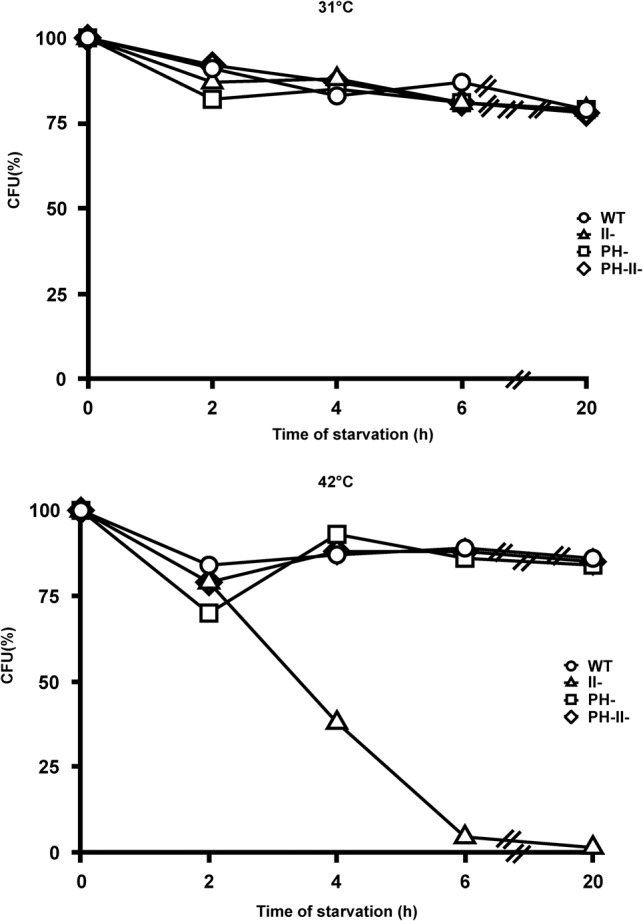
Survival of wild-type and mutant cells during starvation. WT, RNase II−, RNase PH−, and RNase II− RNase PH− strains were first grown in M9/glucose media at 31°C to an A600 of ∼0.2, and then subjected to starvation in M9 salts at either 31°C or 42°C as described in Materials and Methods. At regular time intervals, appropriate sequential dilutions of cells in duplicate were spread on LB plates and viable cells (CFU) were determined after overnight incubation at 37°C. Data are presented as a percentage in which the zero time of starvation was set at 100%. A representative experiment from those repeated two or three times is shown.
Another similarity between the natural depletion of nutrients and starvation is that while the absence of RNase PH, by itself, did not affect the viability of cells during starvation at either 31°C or 42°C (Fig. 2), removal of RNase PH, in addition to RNase II, completely reversed the loss of viability caused by the absence of RNase II at 42°C. These findings confirm that the relationship between RNase II and RNase PH also extends to the starvation protocol at elevated temperature. At present, we do not understand why removal of RNase II does not lead to inviability during starvation at 31°C, but we assume it is because certain structural alterations in the ribosome that occur during prolonged stationary phase or starvation at 42°C do not occur during the rapid starvation protocol at the lower temperature. Nevertheless, these findings indicate that the starvation protocol at elevated temperature may serve as a useful alternative to the natural depletion of nutrients.
RNase II affects rRNA degradation during starvation and stationary phase
In earlier experiments (Basturea et al. 2011), we found that the absence of the processive exoribonucleases RNase II and RNase R or RNase II, RNase R and PNPase, during starvation led to the accumulation of fragments derived from 16S and 23S rRNAs, indicating that the RNases are required for removal of these degradation intermediates. We also made the unexpected observation (Basturea et al. 2011) that during starvation the absence of RNase II substantially increased the total amount of acid-soluble material derived from rRNA, suggesting that RNase II somehow affected overall rRNA degradation. In view of the findings presented above that removal of RNase II has profound effects on the survival of cells during starvation and stationary phase, it was of interest to reexamine rRNA metabolism under these conditions in cells lacking only RNase II.
Wild-type and RNase II− cells were grown at 31°C to early exponential phase and then either starved (−glucose) or allowed to grow (+glucose) for 4 h at 42°C. Total RNA was isolated and analyzed by gel electrophoresis (Fig. 3A). In wild type, only 16S and 23S rRNAs were observed, both in growing cells (lane 1) and in those undergoing starvation (lane 2). In the RNase II− strain, growing cells contained only full-length rRNAs (lane 3); however, during starvation, a 16S* band, slightly shorter than 16S rRNA, and fragments between 16S and 23S RNAs, also were observed (lane 4 and Fig. 3B, lane 2). These findings suggest that RNase II, by itself, is a major contributor to the removal of rRNA fragments during starvation, and that in its absence such intermediates accumulate.
FIGURE 3.
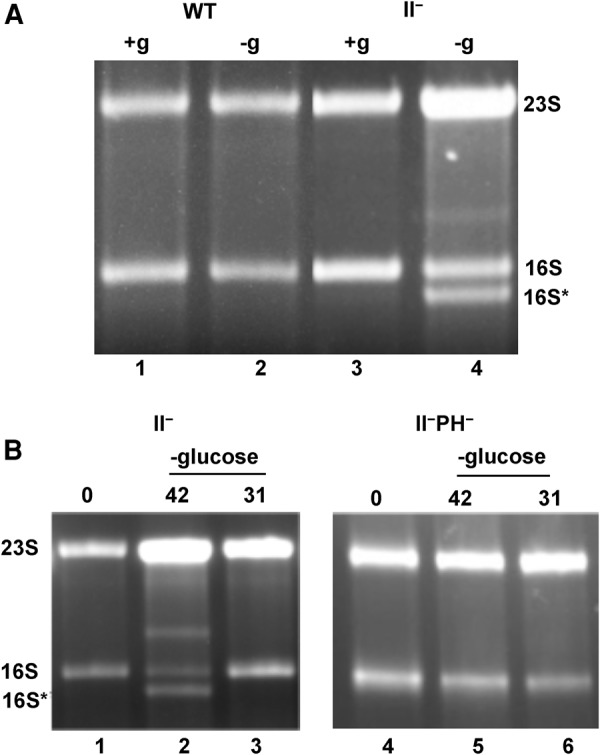
Degradation of rRNA during starvation. (A) WT and RNase II− cells were first grown in M9/glucose at 31°C to an A600 of ∼0.2 and then either starved of glucose for 4 h (−g) or grown in the presence of glucose for 4 h (+g) at 42°C, as described in Materials and Methods. Cells were collected by centrifugation, and total RNA was extracted and resolved by Synergel/agarose gel electrophoresis, as described in Materials and Methods. The gel was stained with ethidium bromide and visualized by UV irradiation. (B) RNase II− or RNase II− RNase PH− cells were first grown in M9/glucose at 31°C to early exponential phase (zero time) and then starved of glucose for an additional 4 h (−glucose) at either 42°C or 31°C. Total RNA was extracted and resolved by Synergel/agarose gel electrophoresis followed by ethidium bromide staining.
To further examine the role of RNase II, we also determined the effect of starvation temperature and the influence of RNase PH in an RNase II− background. While fragments accumulated in RNase PH+ cells at 42°C (Fig. 3B, lane 2 and as already noted above), few fragments were present at 31°C (Fig. 3B, lane 3). In contrast, there was no degradation in the strain lacking RNase PH, even at 42°C (Fig. 3B, lane 5). These findings agree with our previous data indicating that rRNA degradation during starvation in cells lacking multiple exoribonucleases is dependent on RNase PH (Basturea et al. 2011; Sulthana et al. 2016), but the data presented here extend the requirement for RNase PH to cells lacking just RNase II. Moreover, based on these results, it is clear that the effect of RNase II removal on rRNA degradation closely mirrors what was found for cell viability (Fig. 2), suggesting that the findings are likely to be related.
As noted above, rapid starvation is quite different than the natural depletion of nutrients that occurs as cells enter stationary phase. In the latter process, morphological and physiological changes occur associated with changes in the expression of many stress-related genes. As a consequence, there are alterations in ribosome structure and in enzymes that act on ribosomes that do not occur when cells are rapidly put into starvation mode. Therefore, to examine the effect of RNase II removal on rRNA under these conditions, we isolated RNA at intervals from various strains grown at 31°C and 42°C for up to 96 h.
As shown in Figure 4, rRNA is essentially stable in wild-type and RNase PH– cells during this entire period at both incubation temperatures. On the other hand, there is a significant reduction in rRNA in RNase II− cells by 48 h, and almost all of the rRNA is degraded by 96 h at both 31°C and 42°C. These observations indicate that the absence of RNase II leads to extensive degradation of rRNA during stationary phase of E. coli. Moreover, removal of RNase PH largely reverses this degradation, again confirming that rRNA degradation upon nutrient depletion does not occur in the absence of RNase PH. Taken together, these findings reveal a high degree of similarity between the breakdown of ribosomes in RNase II− cells and their loss of viability during stationary phase. They also show that the rapid starvation protocol is a convenient experimental model for what occurs naturally during the prolonged stationary phase.
FIGURE 4.
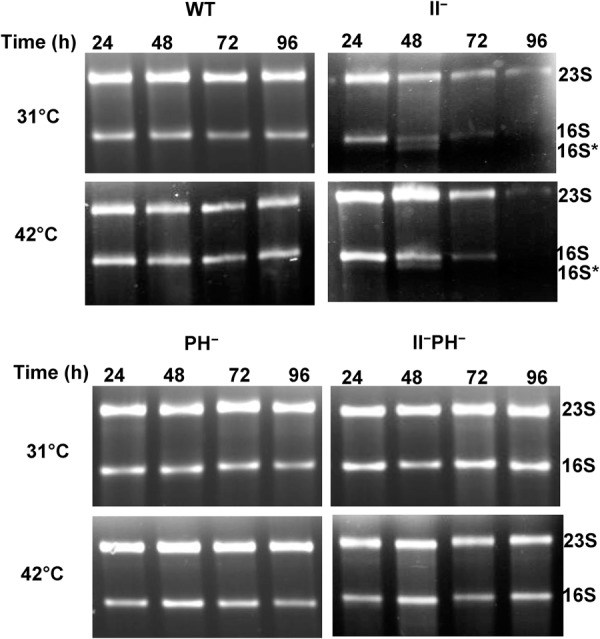
Degradation of rRNA during extended stationary phase. WT, RNase II−, RNase PH−, and RNase II− RNase PH− cells were grown in 50 mL M9/glucose at either 31°C or 42°C, and sample volumes corresponding to an equal number of cells were taken every 24 h. Cells were collected by centrifugation, and total RNA was extracted and resolved by Synergel/agarose gel electrophoresis, as described in Materials and Methods. The gel was stained with ethidium bromide and visualized by UV.
RNase II action does not require membrane association
It has been suggested that RNase II organizes into structures that coil around the E. coli cell periphery, and that RNase II is associated with the cytoplasmic membrane through its amino-terminal amphipathic helix (Lu and Taghbalout 2013). It was also suggested that these higher-order structures and the membrane association are required for the normal functioning of RNase II within the cell. If so, we reasoned that preventing RNase II association with the membrane might be akin to removing RNase II, leading to an accumulation of rRNA fragments and an increase in rRNA degradation during starvation. To test this idea, we deleted the first 19 residues of RNase II that are required for anchoring it to the membrane (Lu and Taghbalout 2013) and determined its effect on rRNA degradation. As can be seen in Figure 5, the RNase II (Δ19) strain behaves like wild type with regard to accumulation of rRNA fragments. Thus, the role of RNase II in rRNA metabolism is not compromised by removal of its putative anchor to the membrane.
FIGURE 5.
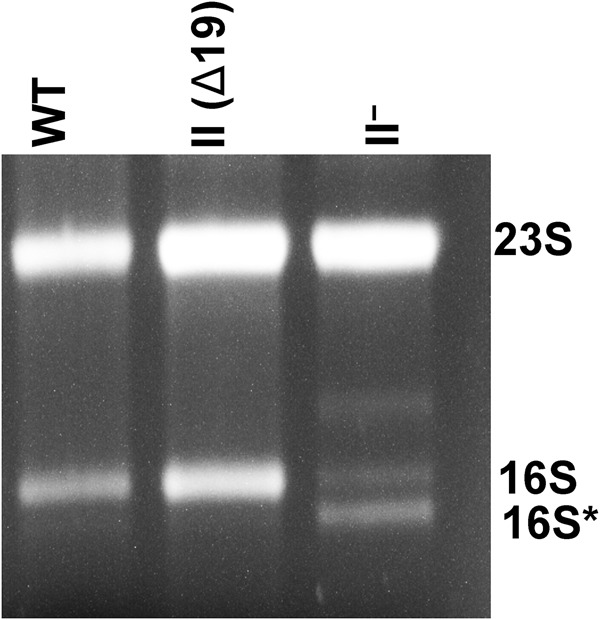
Effect of membrane-anchoring domain of RNase II on rRNA degradation during starvation. WT, RNase II−, and RNase II (Δ19) cells were first grown in M9/glucose at 31°C to early exponential phase and then starved of glucose for 4 h at 42°C. Cells were collected by centrifugation, and total RNA was extracted and resolved by Synergel/agarose gel electrophoresis, as described in Materials and Methods. The gel was stained with ethidium bromide and visualized by UV.
RNase II regulates the level of RNase PH
The data presented to this point suggest a functional relationship between RNase II and RNase PH. In wild-type cells, RNase II appears to down-regulate rRNA degradation during nutrient deprivation since when it is removed, degradation increases substantially (Basturea et al. 2011) and rRNA fragments accumulate (Fig. 3). However, these effects disappear when RNase PH is also absent (Fig. 3B; Basturea et al. 2011). Inasmuch as the action of RNase PH is known to be important for initiation of rRNA degradation (Basturea et al. 2011; Sulthana et al. 2016), these data support a mechanism in which RNase II negatively affects the action of RNase PH on rRNA. One possibility consistent with such a mechanism is that the level of RNase PH is tightly regulated during starvation to avoid too much degradation of rRNA, and that RNase II plays a role in this regulation.
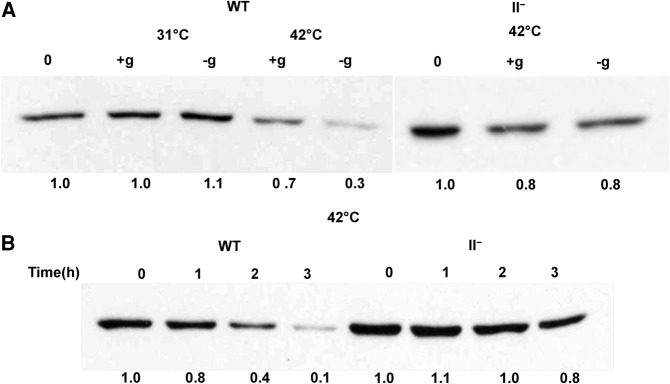
To test this hypothesis, a 2× Flag tag was fused to the N terminus of RNase PH in the chromosomal rph gene, and the amount of RNase PH present in cells grown under a variety of conditions was determined by immunoblotting using antibody directed against the Flag tag. As shown in Figure 6A, the level of RNase PH in wild-type cells was unaffected at 31°C during growth or during starvation. In contrast, the amount of RNase PH decreased substantially during starvation at 42°C compared to the levels prior to the starvation period, while the change in growing cells was relatively minor. On the other hand, the level of RNase PH was relatively unchanged in the RNase II− strain in either growth condition.
FIGURE 6.
RNase II regulates the level of RNase PH during starvation. (A) WT and RNase II− cells containing the 2× Flag rph gene were first grown in M9/glucose at 31°C to early exponential phase (zero time) and then either starved of glucose for 2 h (−g) or grown in the presence of glucose for 2 h (+g) at 31°C or 42°C. Cells were collected by centrifugation, lysed, and then analyzed by immunoblotting using anti-Flag monoclonal antibody. Shown is a representative gel from an experiment carried out three times with essentially identical results. The amount of total RNase PH at zero time for each cell was set at 1.0. (B) Cells were grown at 31°C to early exponential phase (zero time) and then subjected to glucose starvation at 42°C for the indicated times. Cells were treated and RNase PH analyzed as in A. Shown is a representative gel from an experiment carried out twice with essentially identical results. The amount of total RNase PH in each cell at zero time was set at 1.0.
A more detailed examination of the time course of RNase PH disappearance in wild-type cells is shown in Figure 6B. RNase PH levels were down-regulated as much as 90% during 3 h of starvation, whereas under the same conditions, there was only a minimal change in the amount of RNase PH when RNase II was absent. These data show directly that RNase II is required for the down regulation of RNase PH. Inasmuch as RNase PH is important for the initiation of rRNA degradation during starvation, these data also suggest that elevation of RNase PH provides a reasonable explanation for the enhanced rRNA degradation and decreased cell survival observed in the RNase II− background under starvation conditions.
Regulation of RNase PH occurs at the level of protein stability
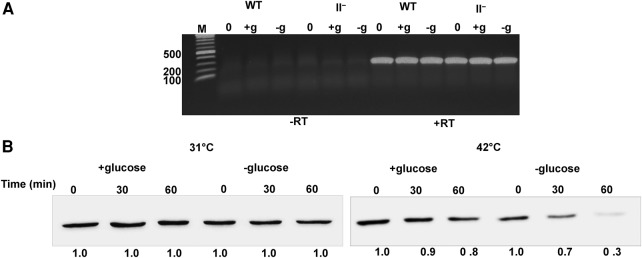
To gain additional insight into how RNase II affects the amount of RNase PH present during starvation, we first measured the levels of rph transcripts in wild-type and RNase II− cells under +glucose and −glucose conditions using RT-PCR. Total RNA was isolated from wild-type and RNase II− cells and cDNA was synthesized using rph gene-specific primers. The cDNA was then amplified by PCR, and the amounts of products were compared during the exponential phase of the PCR before reaction components became limiting. Based on this initial analysis (Fig. 7A), no difference was observed between wild-type and RNase II− cells in the amount of rph mRNA, suggesting that the amount of RNase PH is not regulated at the mRNA level.
FIGURE 7.
RNase II regulation of RNase PH occurs post-transcriptionally. (A) RNase PH transcript analysis by RT-PCR. WT and RNase II− cells were first grown in M9/glucose at 31°C to early exponential phase (zero time), and then either starved of glucose for 2 h (−g) or grown in the presence of glucose for 2 h (+g) at 42°C. Total RNA was isolated and first-strand cDNA synthesis followed by PCR amplification was carried out using rph gene-specific primers, as described in Materials and Methods. Triplicate RT (+RT) reactions were performed along with reactions in which the reverse transcriptase was omitted (−RT) to enable assessment of genomic DNA contamination in each sample. (B) Stability of RNase PH during starvation. WT cells were first grown in M9/glucose at 31°C to early exponential phase (zero time), treated with chloramphenicol, and either starved of glucose (−g) or incubated with glucose (+g) at 42°C. Cells were collected at 30 and 60 min, lysed, and then analyzed by immunoblotting using anti-Flag monoclonal antibody. Shown is a representative gel from an experiment carried out twice with essentially identical results. The amount of RNase PH at zero time of chloramphenicol addition was set at 1.0 under each condition.
Consequently, we focused attention on the stability of RNase PH itself. RNase PH stability was measured in wild-type cells in the presence or absence of glucose. Cells were treated with chloramphenicol to inhibit new protein synthesis, and the amount of RNase PH present was determined by immunoblotting. The data in Figure 7B reveal that at 31°C the amount of RNase PH remained constant in either the presence or absence of glucose. In contrast, at 42°C, while RNase PH was relatively stable in growing cells, it was unstable during starvation, decreasing 70% in 1 h (Fig. 7B). Inasmuch as RNase PH levels remain relatively constant in RNase II− cells (Fig. 6B), its stability was not examined in that genetic background. These data indicate that RNase PH undergoes rapid turnover when cells are starved at 42°C, and taken together with the information presented in Figure 6, strongly suggest that RNase II plays an important role in the instability of RNase PH under these conditions.
DISCUSSION
The information presented here provides the first evidence that the processive exoribonuclease, RNase II, is essential for E. coli survival during starvation and prolonged stationary phase. In its absence, starved cells die rapidly, while cells in stationary phase lose viability at a more gradual pace as they are depleted of nutrients. Although RNase II is the most active hydrolytic exoribonuclease in E. coli (Deutscher and Reuven 1991), until now it was not thought to have an essential function. However, based on this new data, it is clear that no other RNase can substitute for RNase II to maintain cell survival during nutrient deprivation. Whether RNase II has a similar function under other stress conditions is not yet known.
In earlier work (Basturea et al. 2011), we had made the puzzling observation that the absence of RNase II in starving cells led to a dramatic increase in the amount of acid-soluble material derived from RNA, implying greatly increased rRNA degradation. While the explanation for this counterintuitive finding was not understood, it nevertheless focused our attention on the role of RNase II in rRNA degradation. As we have now found, not only does the absence of RNase II lead to enhanced overall degradation, it also results in the accumulation of rRNA fragments that are known to promote the loss of cell viability (Cheng and Deutscher 2003). Taken together, these findings provide a reasonable explanation for how the absence of RNase II leads to cell death.
However, this is only part of the story since the deleterious consequences of the lack of RNase II can be reversed by also removing RNase PH. Thus, when both RNase II and RNase PH are absent, cells remain viable during starvation and prolonged stationary phase, and rRNA fragments do not accumulate. These findings support the connection between rRNA fragment accumulation and decreased cell survival. They also implicate RNase PH in the process and suggest that the effects of RNase II removal act through RNase PH. We found previously that rRNA degradation during starvation is initiated by RNase PH action at the 3′ end of 16S rRNA, and that little degradation occurs in the absence of RNase PH (Basturea et al. 2011; Sulthana et al. 2016). As a consequence, RNase II cannot exert its effect on rRNA degradation when RNase PH is absent and there is little degradation to begin with. This, of course, raised the question of how RNase II affected RNase PH action on rRNA.
Most importantly, the work presented here revealed that RNase II regulates the amount of RNase PH present during starvation, and that in turn modulates rRNA degradation. This finding adds to the growing list of RNases that are regulated under a variety of stress conditions (Deutscher 2015a) and emphasizes that RNase regulation is an important factor to consider in studies of RNA metabolism under stress conditions. Microbes live under conditions of constant environmental fluctuation and they are able to sense and respond to environmental stress, which is critical to their survival. The long-term adaptation of E. coli to various stresses has been studied in the laboratory and has provided information regarding mutations that favor bacterial growth under such conditions (Herring et al. 2006; Conrad et al. 2009; Dragosits et al. 2013). Interestingly, one of the most frequent mutations found in these adaptation experiments is in rph, encoding RNase PH. This mutation proved advantageous to stressed cells and enabled increased growth; however, the mechanism by which such a mutation facilitated growth was not understood. This work provides a possible explanation since ribosomes would be protected from their usual degradation in the absence of RNase PH. Surprisingly, RNase II activity also decreases under certain stress conditions (Cairrão et al. 2001; Song et al. 2016). We presume that in these instances either sufficient RNase II activity remains to regulate RNase PH or that RNase II protein rather than RNase II activity may be the important effector.
Based on the findings presented here, we propose that in wild-type cells the presence of RNase II leads to a reduction of RNase PH upon nutrient deprivation, thereby limiting the initiation of ribosome degradation and maintaining cell survival. In contrast, in cells lacking RNase II, RNase PH levels remain high, leading to excessive ribosome turnover and ultimately to cell death. At present, we do not fully understand how RNase II controls the amount of RNase PH. Our data thus far indicate that the regulation occurs at the level of RNase PH stability and that it is triggered in nongrowing cells. Inasmuch as RNase II action would be expected to be on RNA, these findings raise the possibility that an RNA molecule could be involved in RNase PH turnover. However, further work will be necessary to fully unravel this interesting regulatory process.
MATERIALS AND METHODS
Materials
Oligonucleotide primers were synthesized and purified by Sigma Genosys. M-MuLV reverse transcriptase and Taq DNA polymerase were purchased from New England Biolabs, Inc. Goat anti-mouse IgG-HRP was from Santa Cruz Biotechnology, Inc. Monoclonal anti-Flag M2 antibody produced in mouse and all other reagent grade chemicals were obtained from Sigma-Aldrich.
Bacterial strains
E. coli MG1655*(seq) I−, which is RNase PH+, was considered to be wild type for this study; MG1655 I− is the RNase PH− counterpart (Basturea et al. 2011). These are the recipient strains into which the RNase II mutation was introduced by P1 transduction (Yancey and Kushner 1990; Basturea et al. 2011). DNA encoding the 2× Flag sequence was fused to the N terminus of the chromosomal rph gene following a previously published recombineering protocol (Datsenko and Wanner 2000) using oligos PHFP (5′GGAAGTCCGTATAATGCGCAGCCACATTTGTTTCAAGCCGGAGATTTCAATGTGTAGGCTGGAGCTGCTTC-3′) and PHRP (5′CAGGGTAACGGGACGCACCTGATTATTGCTACGGCCTGCTGGACGCTTGTCATCGTCATCCTTGTAATCGATATCATGATCTTTATAATCCATATGAATATCCTCCTTA-3′). Recombinants were selected on LB–kanamycin plates and were confirmed by PCR using primers PFP (5′-AGGATAGGAATAACCGCC-3′) and PRP (5′-AGGCGGTACACAACACTTTGG-3′). The kanamycin resistance cassette was removed by plasmid pCP20 (Cherepanov and Wackernagel 1995), and the resulting gene fusion construct was confirmed by DNA sequencing. The 19 N-terminal amino acid residues of RNase II were mutated as explained above using primers RFP (5′-AGATTCGCGTAAAACTGTCAGCCGCTCTAATGGCCACCAAAATAGACAATTG TGTAGGCTGGAAGCTGCTTC-3′) and RRP (5′GCCAAAGCCTTTTTCTGTGGCTTTTACCACCCCTTCAGCGCGTGGCATATGAATATCCTCCTTA-3′). Antibiotics, when present, were at the following concentrations: kanamycin, 50 µg/mL; ampicillin, 100 µg/mL; chloramphenicol, 34 µg/mL.
Growth conditions
For starvation experiments, cultures were initially grown in 50 mL of M9 minimal medium containing 0.4% glucose at 31°C to an A600 of 0.2. Cells were collected by centrifugation for 15 min in a Sorvall SS34 rotor at 7000 rpm. The cell pellet was washed twice with M9 salts to remove glucose and resuspended in 50 mL of M9 salts. One 20 mL portion was taken and incubated with shaking at either 31°C or 42°C in the absence of any carbon source for 4 h (starvation sample). A second 20 mL portion supplemented with 0.4% glucose was incubated for 4 h at either 31°C or 42°C. The remaining 10 mL were retained as zero time controls. For stationary phase experiments, cultures were grown in 50 mL of M9 minimal medium containing 0.4% glucose at 31°C or 42°C, and samples were harvested and analyzed every 24 h.
RNA electrophoresis
For the RNA degradation experiments, an equal number of cells (measured by absorbance at 600 nm) were taken at regular intervals of time as described above and centrifuged at 5000 rpm for 5 min. Total RNA was extracted from these cells by hot phenol/chloroform treatment and precipitated with ethanol (Huang and Deutscher 1992). The RNA was dissolved in 20 µL of DEPC-treated water and 2 to 10 µL were separated by Synergel/agarose gel electrophoresis as previously described (Wachi et al. 1999), using 0.9% Synergel and 0.7% agarose in 0.5× TBE (45 mM Tris–borate, 1 mM EDTA, pH 8.0). Gels were stained with 1 mg/mL ethidium bromide and photographed under UV irradiation at 254 nm.
Measurement of RNase PH stability and Western blot analysis
Cells were grown in M9 minimal medium containing 0.4% glucose to an A600 of ∼0.3. A portion of the culture was collected as the zero time point and chloramphenicol was added to the remaining culture. Cells were collected at the indicated times, lysed by sonication (Chen and Deutscher 2005), and assayed by immunoblotting to determine the amount of RNase PH remaining using anti-Flag M2 mAbs (1:1000 dilution).
RT-PCR analysis
Total RNA was extracted from cells and 2 µg were used for first-strand cDNA synthesis by M-MuLV reverse transcriptase in the presence of primer PRP2 (5′CAATGGATTCGATTCCCCTCG-3′). RT-PCR analysis was performed using Taq DNA polymerase and PFP2 (5′-GTTTGCGATCTGGAATACGTTGAA-3′) and PRP2 as primers.
ACKNOWLEDGMENTS
We thank Dr. Arun Malhotra, Dr. Chaitanya Jain, and members of the laboratory for comments on the manuscript. This work was supported by grant GM16317 from the National Institutes of Health.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.060558.116.
REFERENCES
- Arraiano CM, Mauxion F, Viegas SC, Matos RG, Séraphin B. 2013. Intracellular ribonucleases involved in transcript processing and decay: precision tools for RNA. Biochim Biophys Acta 1829: 491–513. [DOI] [PubMed] [Google Scholar]
- Bandyra KJ, Luisi BF. 2013. Licensing and due process in the turnover of bacterial RNA. RNA Biol 10: 627–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basturea GN, Zundel MA, Deutscher MP. 2011. Degradation of ribosomal RNA during starvation: comparison to quality control during steady-state growth and a role for RNase PH. RNA 17: 338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairrão F, Chora A, Zilhão R, Carpousis AJ, Arraiano CM. 2001. RNase II levels change according to the growth conditions: characterization of gmr, a new Escherichia coli gene involved in the modulation of RNase II. Mol Microbiol 39: 1550–1561. [DOI] [PubMed] [Google Scholar]
- Chen C, Deutscher MP. 2005. Elevation of RNase R in response to multiple stress conditions. J Biol Chem 280: 34393–34396. [DOI] [PubMed] [Google Scholar]
- Cheng ZF, Deutscher MP. 2002. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J Biol Chem 277: 21624–21629. [DOI] [PubMed] [Google Scholar]
- Cheng ZF, Deutscher MP. 2003. Quality control of ribosomal RNA mediated by polynucleotide phosphorylase and RNase R. Proc Natl Acad Sci 100: 6388–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158: 9–14. [DOI] [PubMed] [Google Scholar]
- Cohen L, Kaplan R. 1977. Accumulation of nucleotides by starved Escherichia coli cells as a probe for the involvement of ribonucleases in ribonucleic acid degradation. J Bacteriol 129: 651–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad TM, Joyce AR, Applebee MK, Barrett CL, Xie B, Gao Y, Palsson BØ. 2009. Whole-genome resequencing of Escherichia coli K-12 MG1655 undergoing short-term laboratory evolution in lactate minimal media reveals flexible selection of adaptive mutations. Genome Biol 10: R118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci 97: 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher MP. 1990. Ribonucleases, tRNA nucleotidyltransferase, and the 3′ processing of tRNA. Prog Nucleic Acid Res Mol Biol 39: 209–240. [DOI] [PubMed] [Google Scholar]
- Deutscher MP. 2015a. How bacterial cells keep ribonucleases under control. FEMS Microbiol Rev 39: 350–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher MP. 2015b. Twenty years of bacterial RNases and RNA processing: how we've matured. RNA 21: 597–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher MP, Reuven NB. 1991. Enzymatic basis for hydrolytic versus phosphorolytic mRNA degradation in Escherichia coli and Bacillus subtilis. Proc Natl Acad Sci 88: 3277–3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan WP, Kushner SR. 1986. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc Natl Acad Sci 83: 120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragosits M, Mozhayskiy V, Quinones-Soto S, Park J, Tagkopoulos I. 2013. Evolutionary potential, cross-stress behavior and the genetic basis of acquired stress resistance in Escherichia coli. Mol Syst Biol 9: 643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand S, Tomasini A, Braun F, Condon C, Romby P. 2015. sRNA and mRNA turnover in Gram-positive bacteria. FEMS Microbiol Rev 39: 316–330. [DOI] [PubMed] [Google Scholar]
- Herring CD, Raghunathan A, Honisch C, Patel T, Applebee MK, Joyce AR, Albert TJ, Blattner FR, van den Boom D, Cantor CR, et al. 2006. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat Genet 38: 1406–1412. [DOI] [PubMed] [Google Scholar]
- Huang S, Deutscher MP. 1992. Sequence and transcriptional analysis of the Escherichia coli rnt gene encoding RNase T. J Biol Chem 267: 25609–25613. [PubMed] [Google Scholar]
- Hui MP, Foley PL, Belasco JG. 2014. Messenger RNA degradation in bacterial cells. Annu Rev Genet 48: 537–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain C. 2012. Novel role for RNase PH in the degradation of structured RNA. J Bacteriol 194: 3883–3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KF. 1993. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J Bacteriol 175: 3401–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnik-Habrink M, Lewis RJ, Mäder U, Stülke J. 2012. RNA degradation in Bacillus subtilis: an interplay of essential endo- and exoribonucleases. Mol Microbiol 84: 1005–1017. [DOI] [PubMed] [Google Scholar]
- Li Z, Deutscher MP. 2004. Exoribonucleases and endoribonucleases. EcoSal Plus 1 10.1128/ecosalplus.4.6.3 [DOI] [PubMed] [Google Scholar]
- Lu F, Taghbalout A. 2013. Membrane association via an amino-terminal amphipathic helix is required for the cellular organization and function of RNase II. J Biol Chem 288: 7241–7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nossal NG, Singer MF. 1968. The processive degradation of individual polyribonucleotide chains. I. Escherichia coli ribonuclease II. J Biol Chem 243: 913–922. [PubMed] [Google Scholar]
- Okamura S, Maruyama HB, Yanagita T. 1973. Ribosome degradation and degradation products in starved Escherichia coli. VI. Prolonged culture during glucose starvation. J Biochem 73: 915–922. [DOI] [PubMed] [Google Scholar]
- Song L, Wang G, Malhotra A, Deutscher MP, Liang W. 2016. Reversible acetylation on Lys501 regulates the activity of RNase II. Nucleic Acids Res 44: 1979–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulthana S, Deutscher MP. 2013. Multiple exoribonucleases catalyze maturation of the 3′ terminus of 16S ribosomal RNA (rRNA). J Biol Chem 288: 12574–12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulthana S, Basturea GN, Deutscher MP. 2016. Elucidation of pathways of ribosomal RNA degradation: an essential role for RNase E. RNA 22: 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachi M, Umitsuki G, Shimizu M, Takada A, Nagai K. 1999. Escherichia coli cafA gene encodes a novel RNase, designated as RNase G, involved in processing of the 5′ end of 16S rRNA. Biochem Biophys Res Commun 259: 483–488. [DOI] [PubMed] [Google Scholar]
- Yancey SD, Kushner SR. 1990. Isolation and characterization of a new temperature-sensitive polynucleotide phosphorylase mutation in Escherichia coli K-12. Biochimie 72: 835–843. [DOI] [PubMed] [Google Scholar]
- Zundel MA, Basturea GN, Deutscher MP. 2009. Initiation of ribosome degradation during starvation in Escherichia coli. RNA 15: 977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]