This Outlook discusses the study in this issue by Zanca et al., in which mutant EGFR expression in a small population of cancer cells in high-grade brain tumors (glioblastoma) can orchestrate the complex behavior of the entire tumor.
Keywords: tumor heterogeneity, glioblastoma, survivin, NF-κB, EGFR, IL-6
Abstract
Converging evidence from numerous laboratories has revealed that malignant brain cancers are complex ecological systems composed of distinct cellular and acellular elements that collectively dictate glioblastoma biology. Our understanding of the individual contributions of each of these components is vital to the design of effective therapies against these cancers. In this issue of Genes & Development, Zanca and colleagues (pp. 1212–1227) demonstrate that one subpopulation of glioblastoma cells expressing a mutant epidermal growth factor receptor (EGFRvIII) is responsible for the survival of non-EGFRvIII-expressing tumor cells as well as for evading molecularly targeted therapy.
“Never in the field of human conflict was so much owed by so many to so few,” remarked Winston Churchill in his praise of the contributions of the Allied airmen of the Royal Air Force during the Second World War. In keeping with the power of the few, Furnari and colleagues (Zanca et al. 2017) now report that a small population of cancer cells in high-grade brain tumors (glioblastoma) can orchestrate the complex behavior of the entire tumor. Glioblastomas are deadly malignancies characterized by significant cellular and molecular heterogeneity. As such, malignant gliomas are composed of numerous distinct cell types, including various neoplastic cells (glioma stem cells and neoplastic astrocytes) and nonneoplastic cells (endothelial cells, reactive astrocytes, and tumor-associated monocytes). Moreover, even among the neoplastic cells, there exists considerable heterogeneity, with subpopulations of cancer cells exhibiting specific transcriptomal profiles and expressing different mutations (Patel et al. 2014). One of these signature mutations involves the epidermal growth factor receptor (EGFR), in which several exons encoding sequences in the extracellular domain of this receptor tyrosine kinase (RTK) are deleted (EGFRvIII mutation), resulting in an aberrantly functioning hyperactive EGFR molecule. While EGFRvIII mutant glioblastoma cells comprise only 15% of the total cancer cells in the tumor, they can serve as critical sculptors of the tumor microcosm.
In this issue of Genes & Development, Zanca et al. (2017) demonstrate that EGFRvIII mutant glioblastoma cells control the survival of wild-type (nonmutant) EGFR-expressing glioblastoma cells as well as provide a unique escape mechanism for evading therapeutic RTK inhibition. These studies identified interleukin-6 (IL-6), secreted by EGFRvIII mutant glioblastoma cells, as an anti-apoptotic effector responsible for attenuating glioblastoma cell sensitivity to EGFR tyrosine kinase inhibitor (TKI) therapy. IL-6-mediated reduced EGFR TKI sensitivity has also been reported in non-small cell lung cancer, where EGFR TKI treatment triggers IL-6-mediated feedback through activation of Stat3 and downstream prosurvival genes (Lee et al. 2014). However, in glioblastoma, IL-6 also acts in a paracrine manner and binds to a common IL-6 receptor (gp130) located on wild-type EGFR-expressing glioblastoma cells. gp130 engagement by IL-6 results in NF-kB activation, leading to expression of survivin, an anti-apoptotic protein. In this fashion, survivin induction protects glioblastoma cells from EGFR TKI-mediated apoptosis by providing an alternative way to activate the mechanistic target of rapamycin (mTOR) complex, thus bypassing TKI suppression of mTOR-mediated cell growth. Importantly, mutant EGFRvIII also increases NFkB activity, resulting in elevated IL-6 transcription. Since NF-κB is responsible for both IL-6 production in EGFRvIII mutant glioblastoma cells and IL-6-mediated survivin induction in wild-type EGFR-expressing tumor cells, inhibitors that blocked NF-κB activation were sufficient to overcome TKI resistance in these brain cancers.
Collectively, the new findings described in this report add to an ever-deepening appreciation of how a small number of cells in any given tumor can dictate the biological behavior of the entire cancer as well as how the glioblastoma ecosystem is maintained through intersecting activities mediated by one particular subpopulation of driver cells. The notion that an “Achilles’ heel” cell type exists in glioblastoma is exciting given the relative resistance of these cancers to conventional and investigational therapies. However, the dark side of this notion is that numerous different cancer cell types could evolve within the tumor biome to influence overall glioblastoma growth, thus creating opportunities to escape future designer chemotherapies.
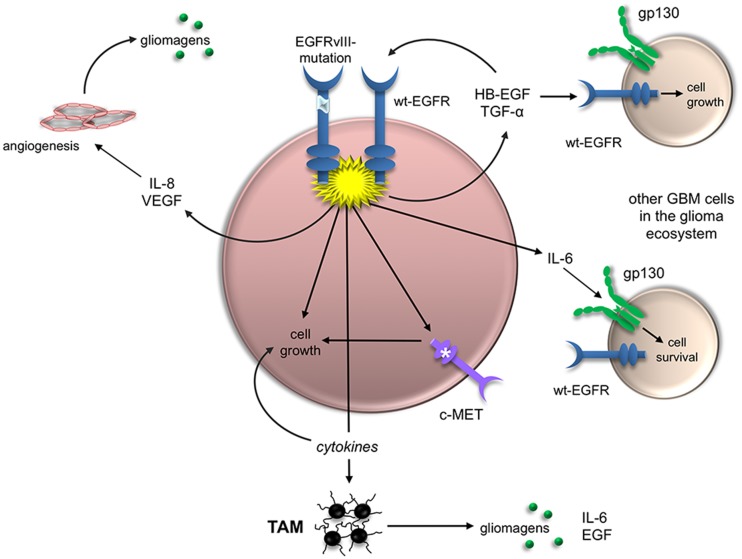
As a case in point, mutant EGFR expression in a small population of cancer cells can lead to profound changes in the glioblastoma ecosystem through a variety of distinct, nonmutually exclusive mechanisms (Fig. 1). First, EGFRvIII has the capacity to transactivate other RTKs and, in doing so, drive tumor growth or limit therapeutic responses (Huang et al. 2007; Pillay et al. 2009). For example, EGFRvIII transactivation of the hepatocyte growth factor (HGF) receptor (MET) leads to resistance to rilotumumab, an antibody therapy that blocks tumor growth by neutralizing HGF and inhibiting HGF-mediated MET signaling (Pillay et al. 2009). Similarly, EGFRvIII receptors can increase the activation of other RTKs through their inappropriate retention at the cell surface (Greenall et al. 2015) or by forming heterodimers that cause aberrant RTK activation and increased mitogenic signaling (Greenall et al. 2015).
Figure 1.
EGFRvIII is a master regulator of glioblastoma ecosystem biology. EGFRvIII mutation in a small population of glioblastoma cells can have profound effects on numerous distinct cell types and thus influence the biology of the entire tumor. First, EGFRvIII expression leads to EGFR activation and the engagement of downstream signaling pathways and transcriptional programs, leading to increased cell growth by either driving cell cycle progression or limiting programmed cell death. Second, EGFRvIII expression can indirectly promote cell expansion through the elaboration of cytokines (e.g., IL-8 and VEGF) that engage tumor-associated monocytes (TAMs) or endothelial cells (angiogenesis), resulting in the elaboration of glioma-promoting growth factors (gliomagens). Third, EGFRvIII expression can increase glioma cell growth via aberrant expression or transactivation of other RTKs (e.g., c-MET). Fourth, EGFRvIII expression can lead to the production of other growth factors (e.g., heparin-binding EGF [HB-EGF] and TGF-α), which can increase the growth of other glioblastoma cells in the tumor through activation of the wild-type EGFR. Last, EGFRvIII expression can trigger elevated levels of IL-6, which promotes cell survival by binding to the IL-6 receptor (gp130).
Second, EGFRvIII expression can directly control the growth of other tumor cells in the glioma as a consequence of paracrine secretion of cytokines and growth factors. In addition to IL-6 regulation of glioblastoma cell growth via gp130 receptor-mediated induction of survivin, EGFRvIII-expressing glioblastoma cells produce other EGFR ligands, such as heparin-binding EGF (HB-EGF) and TGF-α (Ramnarain et al. 2006), each of which can act on the wild-type EGFR receptor to drive tumor cell growth. Similarly, EGFR mutant glioblastoma cells increase IL-8 production, which additionally enhances glioblastoma stem cell growth. Moreover, chemokines (such as CCL5, secreted by glioblastoma cells) can create autocrine growth-promoting circuits in which tumor cells produce their own mitogens in a self-stimulating manner to promote cell survival (Pan et al. 2017).
Third, cytokines and chemokines induced by mutant EGFR expression can alter the glioblastoma microenvironment through the recruitment or activation of stromal cell types, such as endothelial cells and monocytes (macrophages and microglia). For example, mutant EGFR induces IL-8 and vascular endothelial growth factor (VEGF) secretion to increase angiogenesis in human glioblastoma xenografts (Infanger et al. 2013). In addition, monocytes are recruited by chemokines (e.g., CX3CL1, CCL5, and CCL2) produced by glioblastoma cells. These tumor-associated microglia or macrophages are induced to secrete IL-6, EGF, and likely other mitogens to establish paracrine relationships that further enhance glioblastoma growth (Zhang et al. 2012).
Taken together, the findings reported by Zanca et al. (2017) provide a conceptual framework in which the collective behavior of the tumor is defined by the adaptive mechanisms and circuits initiated and orchestrated by a small population of tumor cells. With a greater understanding of these interconnections, future therapies might emerge that target these bionetworks at multiple nodes of vulnerability.
Acknowledgments
D.H.G. is supported by a Research Program Award from the National Institute of Neurological Disorders and Stroke (1-R35-NS097211-01). Y.P. is supported by a McDonnell Foundation Institution fellowship grant.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.303453.117.
References
- Greenall SA, Donoghue JF, Van Sinderen M, Dubljevic V, Budiman S, Devlin M, Street I, Adams TE, Johns TG. 2015. EGFRvIII-mediated transactivation of receptor tyrosine kinases in glioma: mechanism and therapeutic implications. Oncogene 34: 5277–5287. [DOI] [PubMed] [Google Scholar]
- Huang PH, Mukasa A, Bonavia R, Flynn RA, Brewer ZE, Cavenee WK, Furnari FB, White FM. 2007. Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proc Natl Acad Sci 104: 12867–12872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infanger DW, Cho Y, Lopez BS, Mohanan S, Liu SC, Gursel D, Boockvar JA, Fischbach C. 2013. Glioblastoma stem cells are regulated by interleukin-8 signaling in a tumoral perivascular niche. Cancer Res 73: 7079–7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Zhuang G, Cao Y, Du P, Kim HJ, Settleman J. 2014. Drug resistance via feedback activation of Stat3 in oncogene-addicted cancer cells. Cancer Cell 26: 207–221. [DOI] [PubMed] [Google Scholar]
- Pan Y, Smithson LJ, Ma Y, Hambardzumyan D, Gutmann DH. 2017. Ccl5 establishes an autocrine high-grade glioma growth regulatory circuit critical for mesenchymal glioblastoma survival. Oncotarget 8: 32977–32989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, et al. 2014. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344: 1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillay V, Allaf L, Wilding AL, Donoghue JF, Court NW, Greenall SA, Scott AM, Johns TG. 2009. The plasticity of oncogene addiction: implications for targeted therapies directed to receptor tyrosine kinases. Neoplasia 11: 448–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramnarain DB, Park S, Lee DY, Hatanpaa KJ, Scoggin SO, Otu H, Libermann TA, Raisanen JM, Ashfaq R, Wong ET, et al. 2006. Differential gene expression analysis reveals generation of an autocrine loop by a mutant epidermal growth factor receptor in glioma cells. Cancer Res 66: 867–874. [DOI] [PubMed] [Google Scholar]
- Zanca C, Villa GR, Benitez JA, Thorne AH, Koga T, D'Antonio M, Ikegami S, Ma J, Boyer AD, Banisadr A, et al. 2017. Glioblastoma cellular cross-talk converges on NF-κB to attenuate EGFR inhibitor sensitivity. Genes Dev (this issue) 10.1101/gad.300079.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Sarkar S, Cua R, Zhou Y, Hader W, Yong VW. 2012. A dialog between glioma and microglia that promotes tumor invasiveness through the CCL2/CCR2/interleukin-6 axis. Carcinogenesis 33: 312–319. [DOI] [PubMed] [Google Scholar]