This review by Levine and Berger discusses the cross-talk between the p53 protein and epigenetic programs. The p53 protein not only enforces the stability of the genome by the prevention of genetic alterations in cells but also plays an important role in regulating the epigenetic changes that occur in cells.
Keywords: epigenetics, p53, stem cells
Abstract
Epigenetic programs regulate the development and maintenance of organisms over a lifetime. These programs are carried out through chemical modifications of DNA and proteins such as histones and transcription factors. These epigenetic modifications are less stable than genetic alterations and even reversible under a variety of circumstances, such as developmental changes, regeneration of tissues, cell divisions, aging, and pathological conditions observed in many cancers. The p53 protein not only enforces the stability of the genome by the prevention of genetic alterations in cells but also plays a role in regulating the epigenetic changes that can occur in cells. The full-length p53 protein is largely inactive in stem cells but, when activated, helps to commit these cells to developmental lineages through a series of epigenetic changes. Just as p53 impacts epigenetic change, the enzyme activities that carry out epigenetic protein modifications act on the p53 protein and its splice variants in stem and progenitor cells to silence or activate its transcriptional activities. Thus, there is a great deal of cross-talk between the p53 protein and epigenetic programs. This review collects the diverse experimental evidence that leads to these conclusions. This in turn permits new ideas and directions for the treatment of cancers, reactivating developmental pathways for tissue regeneration and responses to the impact of aging.
The available evidence suggests that many cancers arise with both epigenetic and genetic alterations that contribute to the phenotype of the tumor (Jones and Baylin 2002). Unlike genetic changes, epigenetic changes are reversible. They are composed of methylation of some cytosine residues in DNA, histone methylation, acetylation, and other post-translational modifications as well as chromatin packaging into euchromatic or heterochromatic regions of chromosomes. Some tumor suppressor genes, such as p16 and VHL, are silenced by DNA methylation of their transcriptional promoter regions (Jones and Baylin 2002), demonstrating that these epigenetic changes contribute directly to tumor formation. This has led to the development of a number of drugs that block epigenetic changes or inhibit the copying of epigenetic modifications in DNA or proteins. As these drugs have been used to kill tumor cells, it has become clearer that they preferentially kill p53-deficient cancer cells and have a more attenuated impact on tumor cells with wild-type p53 genes and proteins (Yi et al. 2014). The treatment of patients with acute myelogenous leukemia with 5′aza-2′deoxy-cytodine, which blocks cytosine methylation in DNA, resulted in 21 out of 21 patients with p53 mutations undergoing bone marrow blast tumor cell elimination or reductions, while 78 patients with wild-type p53 genes and proteins responded partially or not at all to this drug (Welch et al. 2016). The fact that drugs that block DNA methylation act preferentially on cells that have a mutant p53 gene, and not in cells that have a wild-type p53 gene, suggests that the wild-type p53 protein contributes to the stability of the epigenetic state in normal and cancer cells (Nieto et al. 2004; Yi et al. 2014; Welch et al. 2016). Consistent with this observation is an experiment in which a floxed loss of DNMT1 (DNA methyltransferase-1 gene) in fibroblasts in culture resulted in the failure to copy the methylation of CpG residues in the DNA. After two generations of cell divisions in the absence of DNMT1, the cells died by a p53-dependent apoptosis (Jackson-Grusby et al. 2001). These observations are consistent with a role for p53 in regulating epigenetic stability.
Nuclear transplantation through the cytoplasm of oocytes can reset epigenetic marks in the nuclei of normal differentiated cells and produce pluripotent embryonic stem (ES) cells that can direct the development of a normal organism (Gurdon 1973; Wilmut et al. 1997), demonstrating the reversibility of epigenetic states. Yamanaka and colleagues (Takahashi et al. 2007) showed that four transcription factors—Myc, KLF-4, Oct-4, and Sox-2 (so-called Yamanaka reprogramming factors)—added to normal mouse or human fibroblasts could result in the production of induced pluripotent stem (iPS) cells that were capable of contributing to most or all tissues of a mouse when they were placed among the inner cell mass cells of a mouse blastocyst. The efficiency of this epigenetic reprograming was increased and the time to carry out this reprograming was decreased in the absence of a functional p53 protein (Yi et al. 2012), once again indicating a role for the p53 protein functions in promoting epigenetic stability.
Jaenisch and colleagues (Hochedlinger et al. 2004) explored the reversal of epigenetic marks in cancer cells by passaging the nuclei of a murine melanoma through murine oocytes, converting the cancerous phenotype into a normal developmental pluripotent state. Here, the clones of nucleated oocytes developed normally into blastocysts in culture, permitting isolation of ES cells in culture with an efficiency of 1%–12% of the nuclei transplanted. These ES cells could form differentiated teratomas (benign tumors composed of disorganized tissues) when transplanted subcutaneously into mice and produced chimeric mouse embryos when injected into the inner mass cell compartment of an embryo. The gene driver in these melanomas was an inducible mutated Ras oncogene that, when turned off, permitted the reversible epigenetic changes from a tumor phenotype to a normal developmental phenotype. When the Ras oncogene was turned on, melanomas developed in these mice with an average latency of 19 d, demonstrating the reversible epigenetic state when the expression of the Ras oncogene, the genetic lesion, was turned off.
There are numerous recent observations that underscore the role of epigenetic regulation in the interrelationship between cancer and cellular reprogramming. Transient expression of the Yamanaka reprogramming factors can result in tumor-like cell development in vivo in a mouse model, and these tumor-like cells exhibit epigenetic changes in DNA methylation (Ohnishi et al. 2014); these observations indicate that epigenetic changes can drive oncogenesis. In another experiment in mice, tissue damage and senescence are promoted by expression of the Yamanaka reprogramming factors, with subsequent senescence-associated signaling factors improving cellular reprogramming (Mosteiro et al. 2016), suggesting a reversible (i.e., epigenetic) barrier to reprogramming that is lowered by senescence. Striking results in mice show that aging can be slowed or even reversed by expression of the Yamanaka reprogramming factors, and this rejuvenation correlates with epigenetic remodeling (Ocampo et al. 2016). Taken together, these experiments clearly show that cancer and aging are intimately regulated by Yamanaka reprogramming factors, strongly pointing to epigenetic regulation as a driver in disease and aging and that, importantly, epigenetic alterations are reversible under the appropriate conditions.
Teratocarcinomas
These experiments lead to the question of whether there are any tumor types that are controlled exclusively by changes in the epigenetic state, where no genetic changes are required to form a cancer? One type of tumor that apparently possesses no genetic changes in p53 (http://www.cbioportal.org) is exemplified in testicular and ovarian teratocarinomas, studied in mice and humans, which are tumors of cells forming the germline. These may represent cancers that are entirely or mostly driven by epigenetic changes and indeed express wild-type p53 (Fig. 1). In mice, the 129Sv line has a low spontaneous rate of testicular tumors (Pierce 1967; Stevens 1967), and the LT strain has a low frequency of ovarian teratocarcinomas (Stevens and Varnum 1974). These germ cell tumors are composed of two cell types: the initial tumorigenic embryonal carcinoma (EC) cell, which is then capable of differentiating into one or many benign tissue types. A single EC cell is able to form tumors composed of either many differentiated cell types that are no longer capable of tumor formation (Kleinsmith and Pierce 1964) or mixtures of EC cells and differentiated cells.
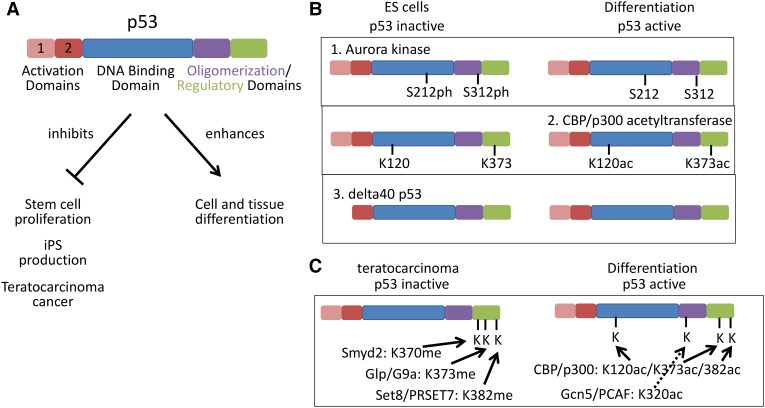
Figure 1.
The p53 protein is epigenetically modified to activate or repress. (A) p53 protein domain structure. Wild-type p53 activity controls cell growth and promotes cell differentiation. (B) p53 is inactivated in ES cells to allow proliferation by Aurora kinase phosphorylation and by protein truncation in Δ40 p53, which eliminates the first activation domain. (C) p53 is inactivated in teratocarcinoma cells by Smyd2 methylation and Set8/PR-SET7 methylation and is activated in differentiated cells by CBP/p300 and Gcn5/PCAF acetylation.
Certain attributes of these tumors suggest involvement of epigenetic pathways. In both humans and mice, spontaneous remissions from a malignant phenotype to a nontumorigenic benign phenotype (termed a teratoma) have been observed. In mice, the inherited nature of these sex-limited genetic predispositions to form testicular and ovarian teratocarcinomas (observed only in certain inbred strains of mice), along with the ability of EC cells to differentiate into a benign tumor, suggests the possibility that the predisposing genes in the 129Sv and LT mice (and possibly in human families) regulate epigenetic functions in germ cell development. Based on these observations, one possibility is that these tumors are regulated purely by epigenetic alterations in development that are reversible, producing benign tumors. Mintz and Illmensee (1975) attempted to test this idea. The 6050 testicular teratocarcinoma tumor was grown for 8 yr as a transplanted tumor in vivo in mice, generating an ascites tumor producing embryoid bodies. Embryoid bodies have an inner core of EC cells surrounded by an outer core of yolk sac epithelial cells. The EC cells from this tumor had a normal karyotype, and these cells were then removed and injected into and adjacent to the inner cell mass cell compartment of a genetically distinct blastocyst. When this blastocyst was reimplanted into a pseudopregnant female, she gave birth to a few male mosaic mice composed of cells contributed by both the EC cell nuclei and host mother nuclei. When these mosaic offspring were mated to mice of a different genotype, one gave birth to an offspring that transmitted traits from the EC cell nucleus (Mintz and Illmensee 1975). This study provides evidence that testicular teratocarcinomas could well be an example of a tumor whose cancerous phenotype is largely or solely regulated by reversible epigenetic changes. Unfortunately, these experiments have not been reproduced, presumably because the karyotypes of these tumors are not always stable, resulting in poor efficiencies of producing offspring with EC cell contributions.
What is clear and reproducible is that tumorigenic and malignant EC cells can be differentiated into benign and nontumorigenic cell and tissue types under a variety of conditions. It takes the special environment of inner cell mass cells in a blastocyst to provide a milieu for the organization of these tissues to form into a viable embryo. Information to form an embryo derives from genetic, epigenetic, and spatial signaling, and, in testicular teratocarcinomas, teratomas, or transplantable tumors, the epigenetic information is reversible and the correct spatial information is missing unless cells are placed within the inner cell mass of an embryo.
The role of the p53 family of genes in germline totipotent stem cells
In humans, the p53 gene is related to two other genes that also encode transcription factors called p63 and p73. The evolutionary ancestral family of these three genes has been elucidated (Belyi et al. 2010) by using amino acid sequence comparisons of the DNA-binding domains of these three proteins. Invertebrates have a single gene from the p53 family, studied in the choanoflagellates, sea anemones, round worms, and flies. In all cases studied, the p53 protein is expressed in the germline (sperm and eggs), and, in response to a stress signal such as DNA damage or starvation, the p53 family member initiates apoptotic cell death, eliminating clones of cells that might produce defective offspring. Indeed, for all of the organisms studied to date, the DNA sequence to which the p53 family member protein binds so as to initiate a transcriptional program for cell death is identical in sea anemones and humans, conserved over 1 billion years. Thus, the amino acid sequences, the structure of the DNA-binding domain, and the function of the p53 family members are conserved. In clams and vertebrates, the p53 family of genes expands into p53, p63, and p73. The p53 protein is now expressed in somatic as well as some germline cells, retaining its function of responding to a wide variety of stress signals by killing or repairing those cells. This presumably is the redirecting of the p53 functions from the germline to tissue-specific somatic stem and progenitor cells so that it may now function as a tumor suppressor in the entire organism. Interestingly, the p63 protein appears to take over the germline fidelity functions in female mice (Suh et al. 2006). The p53 transcription factor is activated in the uterus at the time of implantation of the fertilized egg and is responsible for the transcription of LIF, which is essential for implantation of the embryo (Hu et al. 2007). For these functions, p63 and p53 act in a sexually dimorphic, female-specific fashion. p63 is also a skin tissue-specific stem cell transcription factor. The role of the p53 family in male fertility of vertebrates is still not clear.
There are additional observations for the sexually dimorphic functions of the p53 transcription factor. In some inbred stains of mice, a deletion or knockout of the p53 gene results in up to a third of the female offspring born runted or exocephalic. These female mice have altered imprinting of the insulin-like growth factor (IGF-2) gene in their livers (Yi et al. 2012). These observations are consistent with a role for p53 in the fidelity of sexually dimorphic imprinting during development of the embryo.
The role of p53 in self-renewal and regeneration
The flat worms or planarians have a single gene that most closely resembles p53 (Pearson and Sánchez Alvarado 2010). In the laboratory, these worms do not have a sexual cycle but divide by binary fission and regeneration of body parts after being physically severed into two or more parts. When cut in half, the head section regenerates a tail, and the tail section regenerates a head. Shortly after being cut into two halves, a new stem cell called a neoblast appears. In situ hybridization of RNA in neoblasts with a p53-specific probe demonstrates no p53 RNA in neoblasts until they commit to differentiation and regeneration, when p53 appears in the neoblast and cell progeny of neoblasts. The flat worm can be fed with bacteria that contain a siRNA for the p53 gene sequence, which gets into the neoblasts, and then self-renewal and regeneration of normal tissue are disrupted, and the worms eventually die. The regeneration of both a head and a tail is blocked by p53 siRNA, and many different tissue-specific pathways are inhibited. Interestingly the elimination of p53 from the neoblast results in the continued replication of stem cells, which are eventually depleted, and the worm dies. The absence of p53 enhances stem cell production, and the presence of p53 activity promotes differentiation.
Several vertebrates, such as amphibians and fish, retain the ability to regenerate tissues after amputation. In these animals, the process of regeneration begins by the epigenetic reprograming of differentiated cells at the site of amputation, forming either a stem cell termed the blastema or lineage-restricted progenitors. In salamanders, the amputation of a limb results in dedifferentiation of muscle, cartilage, and connective tissue to form stem cells, much like the Yamanaka experiment producing iPS cells (Takahashi et al. 2007) described previously. Interestingly, as the blastema forms, p53 levels decline and are absent in these stem cells. After the replication of blastema cells, p53 levels increase, differentiation starts, and regeneration of the limb begins (Yun et al. 2013).
These studies with tissue regeneration in invertebrates and vertebrates are consistent in that p53 is absent from stem cells and that its absence results in larger stem cell populations by division. Subsequently, the expression of p53 in stem cells promotes stem cell commitment to differentiation and tissue regeneration. The dedifferentiation of cells to form stem cells occurs more rapidly and in shorter time intervals in the absence of a functional p53 (Yi et al. 2012).
p53 and mammalian stem cells
In mammalian cells, there are certain cellular states that require an altered function of p53, an altered isomer of p53, or reduced levels of p53. These states include stem cells produced during dedifferentiation of mammalian cells in culture, resulting in iPS cells that have pluripotent potentials (like inner mass cells) (Takahashi et al. 2007). These cells have enhanced efficiency of formation in the absence of a functional p53 protein (Yi et al. 2012). Of note, there is evidence that some isoforms of the p53 protein have an active role in maintaining the stem cell state via selected gene transcriptional activation of certain WNT genes that inhibit differentiation (Lee et al. 2010) and by transcriptionally repressing differentiation-specific genes via binding to and interfering with distal enhancers (Li et al. 2012). Interestingly, differentiation of mesenchymal stem cells from ES cells requires redundant transcriptional activation by p53 with family members p63 and p73 to activate Wnt3 to turn on key differentiation pathway genes.
In contrast to p53 transcriptional activity involved in maintaining pluripotency, it appears that a general mechanism of p53 regulation used in iPS and stem cells is repression of p53 function via alterations of the p53 protein. Here we discuss mechanisms that inactivate normal p53 functions in differentiated cells undergoing epigenetic reprograming and in other physiological states that maintain wild-type p53 but require reduced or altered function (Fig. 1).
During reprograming and formation of iPS cells, the Aurora A kinase phosphorylates the p53 protein at amino acids Ser212 and Ser312, which results in the inactivation of p53 protein activity (Lee et al. 2012). The Aurora kinases are active during mitotic and meiotic segregation of chromosomes, including the phosphorylation of histones, and thus could be repurposed in stem cells for changes in p53 protein post-translational modification. Indeed, some cancer cells harbor amplifications of these Aurora kinase genes, which could lead to inactivation of p53. In addition, the role of Aurora kinases in meiosis suggests a way to simultaneously inactivate p53 during meiosis because breakage of DNA strands for recombination would normally be expected to trigger a p53 death response (Derry et al. 2001).
A second type of post-translational modification has been described that regulates the p53 protein in human ES cells, which are derived from the inner cell mass cells (Jain et al. 2012). In these human ES cells, the p53 protein is located in the cell nucleus but is transcriptionally inactive because the lysine residues at positions 120 and 373 are not acetylated. Reduced acetylation in human ES cells occurs via Oct4 transcriptional activation of Sirt1, the NAD+-dependent deacetylase, which deacetylates p53 (Zhang et al. 2014). When retinoic acid is added to cultures of these ES cells, cell differentiation occurs along with the acetylation of these lysine residues by the CBP/p300 histone acetyltransferase. This results in the dissociation of MDM-2 and TRIM24, which are negative regulators of p53, and the increased stability and activity of p53. The activated p53 transcribes two small nuclear RNAs, mir349 and mir145, both of which negatively regulate a set of transcription factors (Oct-4, KLF-4, Lin28A, and Sox-2) that favor stem cell maintenance. Hence, differentiation involves a direct role of p53 in transcriptionally activating certain microRNA genes whose role is to target the RNAs encoding stem cell transcription factors for destruction and thus prevent return to pluripotency (Jain et al. 2012).
A third modification of p53 occurs in mouse ES cells, where the major form of the p53 protein is Δ40 p53, a protein isoform that lacks the first 40 amino acids of the full-length p53 protein (Ungewitter and Scrable 2010). This isoform results from either a splicing variant of p53 mRNA or an internal initiation of mRNA in the p53 gene. In either case, there is a functional deletion of the first transactivation domain of p53 at residues 22/23, leaving the second transactivation domain at residues 53/54 intact. The p21 gene is largely activated by the first p53 transactivation domain (residues 22/23) so that the production of p21 is compromised. The transcriptional activation of p21 by p53 blocks the cell cycle progression at the cyclin E–cdk2 phosphorylation step of RB (retinoblastoma protein), stopping cell cycle progression. It is the cessation of stem cell replication that permits the programs of differentiation. Indeed, haploinsufficiency of the Δ40 p53 gene isoform in mice causes a loss of pluripotency in ES cells and promotes a somatic or differentiated cell cycle. In contrast, an increase in the dosage of the Δ40 p53 isoform in ES cells prolongs pluripotency and blocks differentiation (Ungewitter and Scrable 2010).
Thus, it is clear that p53 is inactivated or compromised in its activity in stem cells, permitting these cells to replicate. Commitment to differentiation results in a new epigenetic program and an activation of the p53 protein activity, which stabilizes that epigenetic program, produces a cell cycle arrest via p21, and promotes a differentiated state. The inactivation of p53 in stem cells can come about by either a lack of transcription of the p53 gene, the appearance of an isoform of p53 with compromised transcriptional activity, or post-translational modifications on the p53 protein that result in an inactive p53 protein, as described above.
p53 in EC cells
The functional activity of the p53 protein in EC or teratocarcinoma stem cells is also compromised but with an interesting twist. EC cells express very high levels of the p53 protein—several-fold higher than levels obtained in normal differentiated cells. When these EC cells are promoted to differentiate into benign normal cells, the p53 protein levels drop. However, the p53 protein in the EC cells is transcriptionally inactive (Lutzker and Levine 1996). p53 normally transcriptionally activates its own negative regulator, the MDM-2 ubiquitin ligase, but, in EC cells, MDM-2 is not transcribed because p53 is not active. The MDM-2 gene is dependent on the p53 protein activity for producing optimal mRNA levels. As cells differentiate, p53 becomes transcriptionally active, and MDM-2 is produced, lowering p53 levels in those cells. In EC cells, p21 is not detected, and this low level permits reproduction of EC cells. Treatment with DNA-damaging agents activates p53 in EC stem cells and results in p21 production, differentiation, or cell death (Lutzker and Levine 1996). As a consequence of this situation, 98%–99% of testicular teratocarcinomas in humans and mice contain the wild-type form of (inactive) p53. There is no selection for p53 mutations in this cancer because p53 is inactive in the EC stem cell. Treatment of testicular teratocarcinomas with cisplatin drugs activates a wild-type p53 protein and cures the great majority of these tumors in young men.
The reason for the inactivation of the p53 protein in human EC cells has been elucidated recently and involves yet another type of post-translational modification of p53. p53 is methylated at lysine residues in its C terminus, leading to inactivation, and most of these methylated lysines overlap those that are acetylated to activate p53 (Berger 2010). Just as during meiosis (discussed above), where p53 activity is constrained by repressive phosphorylation so as to avoid response to programmed DNA double-strand breaks, methylation of p53 during S phase prevents a p53 response to occasional breaks caused by stress at replication forks (Loewer et al. 2010).
This methylation to restrain wild-type p53 activity during the normal cell cycle has been co-opted in EC cells to prevent p53 activation in these cancer cells (Zhu et al. 2016). The methylation of Lys370 by the SMYD2 histone methyltransferase (Huang et al. 2006) and the methylation of Lys382 by the Set8 (also known as PR-SET7) histone methyltransferase (Shi et al. 2007) result in the loss of activity of p53 in the EC stem cells (NTera2 cells) (Zhu et al. 2016). Lowering the levels of the SMYD2 and PR-SET7 histone methyltransferases in EC cells results in a functionally active p53, increased p21 transcription, and cell differentiation. Because elimination of the target residues via substitution (K370R or K382R) also activates p53, it is clear that repression is achieved by direct modification of p53 and not modification of other proteins such as histones, which are also methylated by these same methylases at transcriptionally repressed chromatin (Fig. 1). These results of K370R and K383R leading to p53 activity also suggest that p53 methylation has a specific repressive mechanism and is not simply blocking activating acetylation; elucidation of this mechanism may also reveal details of the normal function of methylation to restrain p53 during the mitotic cell cycle.
There is another important implication of this repressive methylation, as revealed initially in EC cancers. Although p53 is mutated at “hot spot” amino acid residues in the DNA-binding domain, leading to oncogenic driving functions in ∼50% of human cancers, and other cancers are p53-null, there remain a large number of human cancers where p53 is wild type. While the majority of these cancers may not overexpress p53, it nonetheless may be essential to prevent normal p53 tumor-suppressive responses. Hence, these and other repressive methylations on p53, such as Glp/G9a-mediated K373 methylation (Huang et al. 2010), could be commonly repurposed for inactivation of p53 in cancers maintaining wild-type p53.
Implications for improvement in cellular reprogramming and cancer therapy
These findings that certain epigenetic enzymes can restrain or repress activity of p53 reveal potential methods of altering p53 for therapeutic advances. As mentioned above, beyond teratocarcinomas, there are many cancers that retain wild-type p53, and oncogenic growth may involve restraining p53 activity via overexpression of one or more of the enzymes that post-translationally modify p53 to reduce activity, including Aurora A kinase to phosphorylate p53 (S212/S312) and methyltransferases to methylate p53, which comprise Smyd2 (K370), Glp/G9a (K373), and PR-Set7 (K382). Thus, decreased activity of these epigenetic enzymes could lead to reactivated p53. Interestingly, inhibitors targeting these enzymes are under pharmaceutical development, and thus it will be straightforward to determine whether utilization of inhibitors can reactivate p53, potentially leading to cell differentiation or death in common cancers.
In contrast, lowered p53 function promotes dedifferentiation into stem cells, and so efficient reprogramming of cells to pluripotency may be boosted via other manipulations of epigenetic enzymes. This would require increased activity of the repressive epigenetic enzymes (Aurora A, Smyd2, Glp/G9a, and PRSet7); however, it is exceedingly difficult to obtain enzyme agonists. Another solution would be to inhibit enzymes that activate p53 during differentiation. As mentioned above, the absence of CBP/p300 in ES cells prevents activation of p53 due to lack of acetylation at K120 and K373. There are commercially available CBP and p300 inhibitors, and additional inhibitors are under development.
The reversibility of epigenetic changes that initiate and promote the formation of disease provides the opportunity to treat these diseases with drugs that affect epigenetic modifications in a cell. This application of epigenetic therapeutics may be efficacious as a treatment approach for cancers as well as a method to improve cellular reprogramming, tissue regeneration, remediation of aging processes, and even correction of developmental abnormalities. As we begin to understand the epigenetic changes that mediate the functions of the immune and nervous systems, the growing number of specific epigenetic drugs could be used to treat autoimmunity and cognitive diseases.
Acknowledgments
This review and experiments reported in this review were aided by grants from the National Institutes of Health to A.J.L. (5P01CA087497-15) and S.L.B. (5R01CA078831-19).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.298984.117.
References
- Belyi VA, Ak P, Markert E, Wang H, Hu W, Puzio-Kuter A, Levine AJ. 2010. The origins and evolution of the p53 family of genes. Cold Spring Harb Perspect Biol 2: a001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL. 2010. Keeping p53 in check: a high-stakes balancing act. Cell 142: 17–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derry WB, Putzke AP, Rothman JH. 2001. Caenorhabditis elegans p53: role in apoptosis, meiosis, and stress resistance. Science 294: 591–595. [DOI] [PubMed] [Google Scholar]
- Gurdon JB. 1973. Nuclear transplantation and regulation of cell processes. Br Med Bull 29: 259–263. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K, Blelloch R, Brennan C, Yamada Y, Kim M, Chin L, Jaenisch R. 2004. Reprogramming of a melanoma genome by nuclear transplantation. Genes Dev 18: 1875–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Feng Z, Teresky AK, Levine AJ. 2007. p53 regulates maternal reproduction through LIF. Nature 450: 721–724. [DOI] [PubMed] [Google Scholar]
- Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, Kubicek S, Opravil S, Jenuwein T, Berger SL. 2006. Repression of p53 activity by Smyd2-mediated methylation. Nature 444: 629–632. [DOI] [PubMed] [Google Scholar]
- Huang J, Dorsey J, Chuikov S, Pérez-Burgos L, Zhang X, Jenuwein T, Reinberg D, Berger SL. 2010. G9a and Glp methylate lysine 373 in the tumor suppressor p53. J Biol Chem 285: 9636–9641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E, et al. 2001. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet 27: 31–39. [DOI] [PubMed] [Google Scholar]
- Jain AK, Allton K, Iacovino M, Mahen E, Milczarek RJ, Zwaka TP, Kyba M, Barton MC. 2012. p53 regulates cell cycle and microRNAs to promote differentiation of human embryonic stem cells. PLoS Biol 10: e1001268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. 2002. The fundamental role of epigenetic events in cancer. Nat Rev Genet 6: 415–28. [DOI] [PubMed] [Google Scholar]
- Kleinsmith LJ, Pierce GB. 1964. Multipotentiality of single embryonal carcinoma cells. Cancer Res 24: 1544–1551. [PubMed] [Google Scholar]
- Lee KH, Li M, Michalowski AM, Zhang X, Liao H, Chen L, Xu Y, Wu X, Huang J. 2010. A genomewide study identifies the Wnt signaling pathway as a major target of p53 in murine embryonic stem cells. Proc Natl Acad Sci 107: 69–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DF, Su J, Ang YS, Carvajal-Vergara X, Mulero-Navarro S, Pereira CF, Gingold J, Wang HL, Zhao R, Sevilla A, et al. 2012. Regulation of embryonic and induced pluripotency by aurora kinase-p53 signaling. Cell Stem Cell 11: 179–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, He Y, Dubois W, Wu X, Shi J, Huang J. 2012. Distinct regulatory mechanisms and functions for p53-activated and p53-repressed DNA damage response genes in embryonic stem cells. Mol Cell 46: 30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewer A, Batchelor E, Gaglia G, Lahav G. 2010. Basal dynamics of p53 reveal transcriptionally attenuated pulses in cycling cells. Cell 142: 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzker S, Levine AJ. 1996. A functionally inactive p53 protein in embryonal teratocarcinoma cells is activated by DNA damage or cellular differentiation. Nat Med 2: 804–810. [DOI] [PubMed] [Google Scholar]
- Mintz B, Illmensee K. 1975. Normal genetically mosaic mice produced from malignant teratocarcinoma cells. Proc Nat Acad Sci 72: 3585–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosteiro L, Pantoja C, Alcazar N, Marion RM, Chondronasiou D, Rovira M, Fernandez-Marcos PJ, Munoz-Martin M, Blanco-Aparicio C, Pastor J, et al. 2016. Tissue damage and senescence provide critical signals for cellular reprogramming in vivo. Science 354: aaf4445. [DOI] [PubMed] [Google Scholar]
- Nieto M, Samper E, Fraga MF, Gonzalez de Buitrago G, Esteller M, Serrano M. 2004. The absence of p53 is critical for the induction of apoptosis by 5-aza-20–deoxycytidine. Oncogene 23: 735–743. [DOI] [PubMed] [Google Scholar]
- Ocampo A, Reddy P, Martinez-Redondo P, Platero-Luengo A, Hatanaka F, Hishida T, Li M, Lam D, Kurita M, Beyret E, et al. 2016. In vivo amelioration of age-associated hallmarks by partial reprogramming. Cell 167: 1719–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi K, Semi K, Yamamoto T, Shimizu M, Tanaka A, Mitsunaga K, Okita K, Osafune K, Arioka Y, Maeda T, et al. 2014. Premature termination of reprogramming in vivo leads to cancer development through altered epigenetic regulation. Cell 156: 663–677. [DOI] [PubMed] [Google Scholar]
- Pearson BJ, Sánchez Alvarado A. 2010. A planarian p53 homolog regulates proliferation and self-renewal in adult stem cell lineages. Development 137: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce GB. 1967. Teratocarcinoma: model for a developmental concept of cancer. Curr Top Dev Biol 2: 223–246. [DOI] [PubMed] [Google Scholar]
- Shi X, Kachirskaia I, Yamaguchi H, West LE, Wen H, Wang EW, Dutta S, Appella E, Gozani O. 2007. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell 27: 636–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens LC. 1967. The biology of teratomas. Adv Morphog 6: 1–31. [DOI] [PubMed] [Google Scholar]
- Stevens LC, Varnum DS. 1974. The development of teratomas from parthenogenetically activated ovarian mouse eggs. Dev Biol 37: 369–80. [DOI] [PubMed] [Google Scholar]
- Suh EK, Yang A, Kettenbach A, Bamberger C, Michaelis AH, Zhu Z, Elvin JA, Bronson RT, Crum CP, McKeon F. 2006. p63 protects the female germ line during meiotic arrest. Nature 444: 624–628. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872. [DOI] [PubMed] [Google Scholar]
- Ungewitter E, Scrable H. 2010. Δ40p53 controls the switch from pluripotency to differentiation by regulating IGF signaling in ESCs. Genes Dev 24: 2408–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JS, Petti AA, Miller CA, Fronick CC, O'Laughlin M, Fulton RS, Wilson RK, Baty JD, Duncavage EJ, Tandon B, et al. 2016. TP53 and decitabine in acute myeloid leukemia and myelodysplastic syndromes. N Engl J Med 375: 2023–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. 1997. Viable offspring derived from fetal and adult mammalian cells. Nature 385: 810–813. [DOI] [PubMed] [Google Scholar]
- Yi L, Lu C, Hu W, Sun Y, Levine AJ. 2012. Multiple roles of p53 related pathways in somatic cell reprogramming and stem cell differentiation. Cancer Res 72: 5635–5645. [DOI] [PubMed] [Google Scholar]
- Yi L, Sun Y, Levine A. 2014. Selected drugs that inhibit DNA methylation can preferentially kill p53 deficient cells. Oncotarget 10: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun MH , Gates PB, Brockes JP. 2013. Regulation of p53 is critical for vertebrate limb regeneration. Proc Natl Acad Sci 110: 17392–17397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZN, Chung SK, Xu Z, Xu Y. 2014. Oct4 maintains the pluripotency of human embryonic stem cells by inactivating p53 through Sirt1-mediated deacetylation. Stem Cells 32: 157–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Dou Z, Sammons MA, Levine AJ, Berger SL. 2016. Lysine methylation represses p53 activity in teratocarcinoma cancer cells. Proc Natl Acad Sci 113: 9822–9827. [DOI] [PMC free article] [PubMed] [Google Scholar]