Zanca et al. show that heterogeneous expression of the wild-type EGFR receptor and its constitutively active mutant form, EGFRvIII, limits sensitivity to EGFR-directed therapies through an interclonal communication mechanism mediated by IL-6 cytokine secreted from EGFRvIII-positive tumor cells.
Keywords: tumor heterogeneity, glioblastoma, survivin, NF-κB, EGFR, IL-6
Abstract
In glioblastoma (GBM), heterogeneous expression of amplified and mutated epidermal growth factor receptor (EGFR) presents a substantial challenge for the effective use of EGFR-directed therapeutics. Here we demonstrate that heterogeneous expression of the wild-type receptor and its constitutively active mutant form, EGFRvIII, limits sensitivity to these therapies through an interclonal communication mechanism mediated by interleukin-6 (IL-6) cytokine secreted from EGFRvIII-positive tumor cells. IL-6 activates a NF-κB signaling axis in a paracrine and autocrine manner, leading to bromodomain protein 4 (BRD4)-dependent expression of the prosurvival protein survivin (BIRC5) and attenuation of sensitivity to EGFR tyrosine kinase inhibitors (TKIs). NF-κB and survivin are coordinately up-regulated in GBM patient tumors, and functional inhibition of either protein or BRD4 in in vitro and in vivo models restores sensitivity to EGFR TKIs. These results provide a rationale for improving anti-EGFR therapeutic efficacy through pharmacological uncoupling of a convergence point of NF-κB-mediated survival that is leveraged by an interclonal circuitry mechanism established by intratumoral mutational heterogeneity.
Glioblastoma (GBM) is the most deadly form of cancer of the human central nervous system, with a median survival of 12–15 mo and a 5-yr survival rate of <5% (Cloughesy et al. 2014). Standard of care treatment includes maximal surgical resection followed by radiotherapy concomitant with adjuvant temozolomide (Stupp et al. 2005). The aggressive nature of GBM is reflected by its invasion and disruption of normal brain parenchyma, intratumoral heterogeneity, and resistance to treatment. This heterogeneity presents phenotypically as mixed cytological subtypes within the tumor, genotypically as mixed mutations and gene amplifications, and transcriptionally as regional differences in gene expression. As a result, these multiple and spatially distinct heterotypic populations within a GBM make any lesion- or pathway-specific therapy less effective (Verhaak et al. 2010; Brennan et al. 2013; Patel et al. 2014; Louis et al. 2016). While considerable effort has been placed on understanding the interactions between heterotypic tumor cells and the surrounding normal microenvironment, much less is known about the interactions between and among heterogeneous tumor cells within these neoplasms and how these interactions might influence response to therapy.
Amplification and/or mutation of the epidermal growth factor receptor (EGFR) occurs in nearly 60% of GBM (Brennan et al. 2013), thus representing a potentially valuable therapeutic target. While inhibition of EGFR has proven efficacious in other cancer types such as lung cancer, the response of GBM patients to anti-EGFR therapy has been disappointing (Reardon et al. 2014). The heterogeneity of EGFR expression has a profound impact on GBM biology. Wild-type EGFR (wtEGFR) and the constitutively active mutant form of the receptor (EGFRvIII) establish a paracrine-mediated interclonal cross-talk circuitry in which minor populations of EGFRvIII-expressing cells within the tumor potentiate the fitness of the wtEGFR-expressing cells through the secretion of interleukin-6 (IL-6) (Inda et al. 2010). Indeed, high expression levels of IL-6 have been linked to poor prognosis, chemoresistance, and cancer progression in GBM and other cancer types (Tchirkov et al. 2007; Wang et al. 2009; Charles et al. 2011; Bent et al. 2016). Additionally, EGFRvIII, regardless of its constitutive activity, is a substrate for wtEGFR, leading to STAT signaling and tumorigenicity in a cell-intrinsic fashion (Fan et al. 2013).
While these heterotypic EGFR interactions promote enhanced tumor growth, the consequences of this genetic heterogeneity on anti-EGFR therapy in GBM have not been adequately investigated. Here we used in vitro and in vivo analysis of human and murine GBM cell lines, patient-derived xenograft (PDX) models, and clinical specimens from GBM patients to describe an active attenuation in sensitivity to EGFR tyrosine kinase inhibitors (TKIs) that is mediated by EGFRvIII-driven production of IL-6. In turn, activation of the IL-6 common receptor gp130, expressed on wtEGFR cells, promoted NF-κB activation and subsequent expression of a prosurvival gene target, BIRC5 (referred to here with the protein name survivin). This signaling axis could be efficiently uncoupled by the epigenetic regulator bromodomain and extraterminal (BET) protein family inhibitor JQ1, leading to inhibition of survivin expression and restoration of sensitivity to anti-EGFR therapy. These results suggest that, in addition to cell-intrinsic mechanisms (Lynch et al. 2004; Mellinghoff et al. 2005; Diaz et al. 2012; Nathanson et al. 2014; Wykosky et al. 2015), attenuation of response to these therapies can also occur through extrinsic cues fostered by interclonal tumor cell communication. Understanding heterogeneous tumor cell interactions, as exemplified by the EGFR mutational landscape found in GBM (Brennan et al. 2013), will better inform strategies to overcome therapy resistance commonly encountered for this tumor type.
Results
EGFRvIII-specific secreted factors promote resistance to EGFR kinase inhibitors in vitro and in vivo
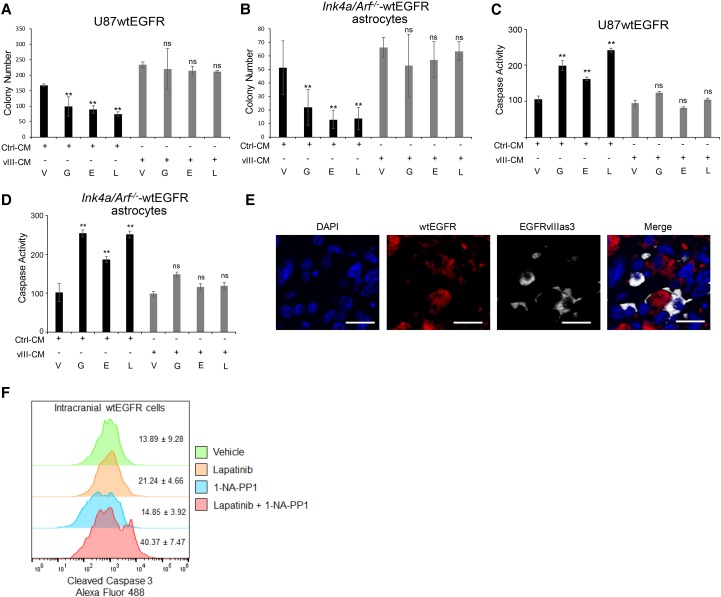
The EGFRvIII-linked secretome promotes wtEGFR phosphorylation (Supplemental Fig. S1A), the proliferation of wtEGFR+ cells, and the maintenance of wtEGFR/EGFRvIII heterogeneity in GBM (Inda et al. 2010). We sought to determine whether this secretome might also possess the ability to confer resistance to EGFR TKIs. Type I EGFR TKIs (gefitinib and erlotinib), which bind to the active conformation of EGFR, and type II EGFR TKIs (e.g., lapatinib), which bind to the inactive state of the receptor (Park and Lemmon 2012), were tested for their ability to inhibit soft agar colony growth of U87MG glioma cells expressing wtEGFR (Inda et al. 2010) incubated with conditioned medium (CM) derived from U87MG cells expressing EGFRvIII CM (vIII-CM) or parental cell CM (Ctrl-CM) as a control. As shown in Figure 1, Ctrl-CM showed significantly reduced wtEGFR colony formation in the presence of EGFR TKIs; however, vIII-CM neutralized this effect (Fig. 1A) despite the inhibition of EGFR activation (Supplemental Fig. S1A). Similar results were also observed in two-dimensional (2D) cell growth assays (Supplemental Fig. S1B) and were further substantiated by a complementary series of experiments using primary astrocytes derived from Ink4a/Arf−/− mice that overexpress wtEGFR (mAstr–Ink4a/Arf−/−-wtEGFR) (Fig. 1B; Supplemental Fig. S1A; Inda et al. 2010). We next evaluated whether the observed TKI resistance phenotype was due to modulation of apoptosis induction. By assessing both caspase 3/7 activation and annexin V cell surface expression, we determined that vIII-CM potently inhibited cell death induced by EGFR TKIs (Fig. 1C,D; Supplemental Fig. S1C). These results show that EGFRvIII paracrine factors promote resistance to anti-EGFR therapies in vitro irrespective of the source of the CM, as both human and murine cells show similar effects.
Figure 1.
EGFRvIII-secreted molecules exert anti-apoptotic action. (A,B) Soft agar colony formation assay quantification of U87wtEGFR cells treated with control medium obtained from parental U87MG cells (Ctrl-CM) or EGFRvIII cells (vIII-CM) in the presence of EGFR TKIs (A) and mAstr–Ink4/Arf−/−-wtEGFR cells treated with control medium obtained from parental mAstr–Ink4/Arf−/− cells or mAstr–Ink4/Arf−/−-vIII in the presence of EGFR TKIs (B). Colony number was determined from nine fields for each condition. (V) Vehicle; (G) gefitinib; (E) erlotinib; (L) lapatinib. (C,D) Caspase 3/7 activation assay of U87wtEGFR cells treated with control medium obtained from parental U87MG cells (Ctrl-CM) or EGFRvIII cells (vIII-CM) in the presence of EGFR TKIs (C) and mAstr-Ink4/Arf−/− wtEGFR cells treated with control medium obtained from parental mAstr–Ink4/Arf−/− cells (Ctrl-CM) or EGFRvIII cells (vIII-CM) in the presence of EGFR TKIs (D). Luminescence as relative light unit (RLU) intensity with blank subtracted and average values with standard deviations is shown. Percentage over control is reported. One-way ANOVA and two-tailed Student's t-test were used to compare samples. (**) P < 0.001; (ns) nonsignificant. (E) Intratumoral localization of EGFRvIIIas3 cells labeled with TurboFP635 (Alexa fluor 647) and U87wtEGFR cells labeled with iRFP720 (Alexa fluor 555). Bars, 20 µm. (F) Apoptotic wtEGFR cells in intracranial mixed tumors were identified by gating on EGFR antibody-stained cells (not shown) followed by detection of cleaved caspase 3. n = 4. Data are represented as mean ± SD.
We then attempted to model wtEGFR/EGFRvIII receptor cross-talk and anti-EGFR therapy resistance in in vivo experiments using a system in which we could specifically target EGFRvIII in engrafted mixed tumors. Here, U87MG cells were stably transduced to express an analog-sensitive allele of EGFRvIII (EGFRvIIIas3), which has been shown previously to be inhibited for kinase activation by compound 1-NA-PP1 (Fan et al. 2013). After confirming that 1-NA-PP1 was able to cross the blood–brain barrier, down-regulate EGFRvIII-associated signaling, and impair IL-6 secretion specifically in EGFRvIIIas3-expressing cells (Supplemental Fig. S2A–D), an equal mixture of wtEGFR and EGFRvIIIas3 tumor cells was orthotopically engrafted into the brains of immunocompromised mice (Fig. 1E). Seven days after injection, the mice were treated with 10 mg/kg 1-NA-PP1 or a control solution for the next 7 d with the addition of 200 mg/kg lapatinib throughout this regimen. Upon sacrifice, the mouse brains were dissected, and isolated tumor cells were stained for cleaved caspase 3 and TUNEL staining (Fig. 1F; Supplemental Fig. S2E), which showed that cotreatment with 1-NA-PP1 and lapatinib resulted in an increase in apoptosis detection in wtEGFR cells compared with lapatinib or 1-NA-PP1 treatment alone. Taken together, these results show that EGFRvIII exerts protective cell-extrinsic effects on neighboring wtEGFR+ cells in vivo, which attenuate sensitivity to EGFR TKI treatment.
IL-6 mediates resistance to EGFR TKIs in GBM
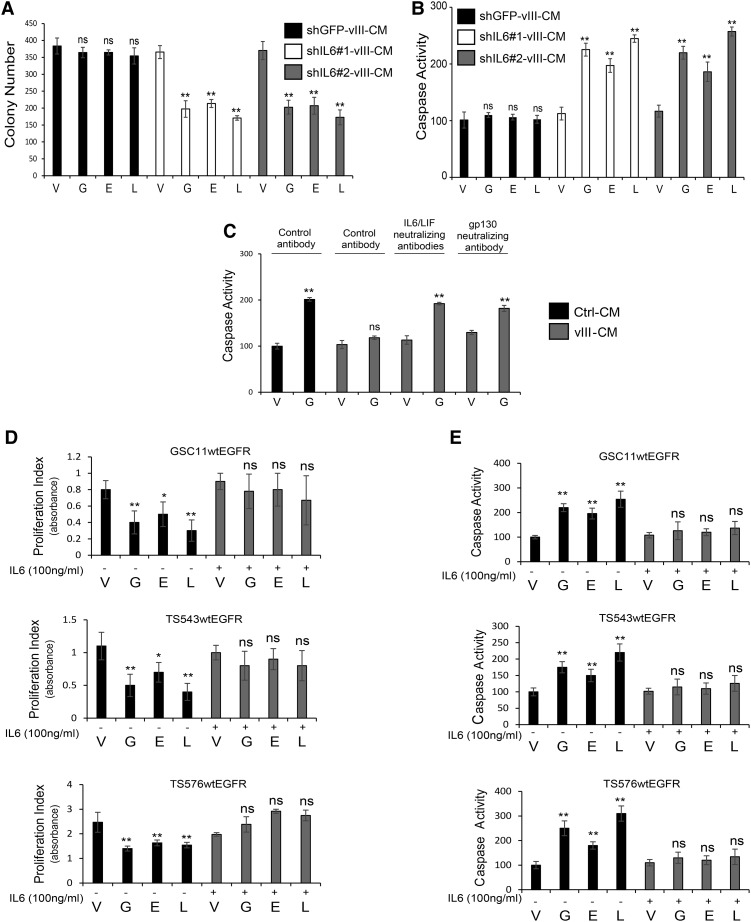
IL-6 has been reported to have a prominent role in promoting chemoresistance in GBM and other types of cancers (Tchirkov et al. 2007; Wang et al. 2009; Lee et al. 2014; Bent et al. 2016). Since EGFRvIII+ cells and tumors have been shown to secrete elevated levels of IL-6, which supports wtEGFR ligand-independent phosphorylation through gp130 activation (Inda et al. 2010), we evaluated a potential role of this EGFRvIII-prompted paracrine factor as an anti-apoptotic effector against EGFR TKIs. shRNAs targeting IL-6 were used to down-regulate its expression in U87EGFRvIII cells (Supplemental Fig. S3A), and CM from cells with reduced cytokine levels was applied to wtEGFR+ cells in the presence of EGFR TKIs. Subsequently, cell viability and cell death were measured by soft agar colony formation and caspase activation, respectively. As shown in Figure 2, CM from IL-6 knockdown cells was deficient in protecting from TKI-induced cell death when compared with CM from control shRNA cells (Fig. 2A,B). In contrast, overexpression of IL-6 in U87 parental cells resulted in CM with an increased ability to protect from EGFR TKI-mediated apoptosis (Supplemental Fig. S3B–D). To further confirm the role of IL-6 in promoting attenuated sensitivity to these inhibitors, U87wtEGFR cells were treated with vIII-CM in the presence of gefitinib with the addition of neutralizing antibodies for IL-6 and LIF, which are enriched in vIII-CM (Inda et al. 2010). Cytokine signaling neutralization resulted in an increase in apoptosis induction (as measured by caspase 3/7 activation and TUNEL assay) and a decrease in proliferation. Additionally, blocking the IL-6 and LIF coreceptor gp130, expressed on wtEGFR cells, produced similar results (Fig. 2C; Supplemental Fig. S3E). To validate the role of IL-6 in mediating resistance to EGFR TKIs, we tested PDX models GSC11, GSC23, TS543, and TS576, engineered to overexpress wtEGFR (Supplemental Fig. S3F). These PDX models were exposed to EGFR TKIs in the presence or absence of recombinant IL-6, and proliferation and apoptosis were measured as above (Fig. 2D,E). Neutralization of gp130 also resulted in an increased response to EGFR inhibition in the PDX model GSC23, which expresses high levels of endogenous IL-6 (Supplemental Fig. S3G–I). Since LIF also acts through the gp130 coreceptor and is secreted by EGFRvIII cells, we tested LIF for its ability to attenuate EGFR TKI sensitivity. GSC11wtEGFR, TS543wtEGFR, and TS576wtEGFR were stimulated with recombinant LIF and treated with EGFR TKIs followed by assessment of cell proliferation and caspase activation. Addition of LIF to the culture medium induced resistance to EGFR TKIs in PDX models (Supplemental Fig. S4A,B). Altogether, these experiments identify IL-6/gp130 as potent mediators of anti-apoptotic signaling leading to attenuation of EGFR TKI-induced cell death in GBM.
Figure 2.
Role of IL-6 in resistance to anti-EGFR therapy. (A) Soft agar colony formation assay quantification of U87wtEGFR cells treated with CM obtained from EGFRvIII cells transduced with control shRNA (shGFP-vIII-CM) or shRNAs against IL-6 (shIL6#1-vIII-CM or shIL6#2-vIII-CM) in the presence or absence of EGFR TKIs. (B) Caspase activation assay of U87wtEGFR treated with the same conditions as in A. (C) Caspase activation assay in U87wtEGFR cells in the presence of human gp130-neutralizing or IL-6/LIF-neutralizing antibodies. (D,E) Cell proliferation analysis by WST-1 assay (indicated as proliferation index) and caspase activation assay in PDX models. Data are represented as mean ± SD.
Survivin plays a major role in resistance to EGFR TKIs
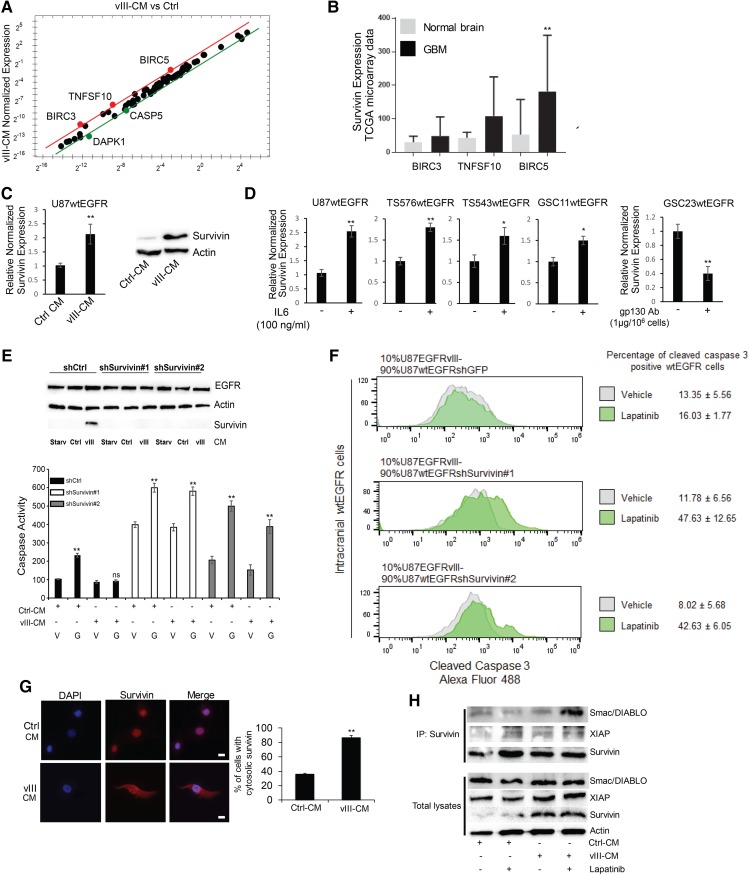
To identify the genes responsible for EGFR TKI resistance in wtEGFR+ cells induced by EGFRvIII-mediated soluble factors, a real-time quantitative PCR (qPCR) screening of apoptosis-related genes was performed on U87wtEGFR+ cells treated with Ctrl-CM or vIII-CM. BIRC3, BIRC5, and TNFSF10 were among the genes specifically altered for expression by vIII-CM treatment (Fig. 3A). The Cancer Genome Atlas (TCGA) microarray analysis showed that the anti-apoptotic protein BIRC5, referred to here as survivin, is up-regulated in GBM versus normal brains (Fig. 3B). These results were also confirmed by real-time qPCR using specific primers for survivin cDNA and Western blotting analysis in wtEGFR+ cells treated with vIII-CM (Fig. 3C). Similar results were obtained in U178 glioma cells transduced with wtEGFR and treated with vIII-CM (Supplemental Fig. S5A). To identify apoptosis-related genes directly regulated by IL-6, a real-time qPCR array consisting of these genes was performed also on TS576wtEGFR PDX cells treated with this cytokine, which confirmed that survivin was similarly up-regulated in this model (Supplemental Fig. S5B). Exposure of U87wtEGFR cells or PDX models (GSC11, TS543, or TS576 overexpressing wtEGFR) to recombinant IL-6 also induced survivin, while inhibition of IL-6 signaling by gp130-neutralizing antibody in GSC23wtEGFR cells resulted in a reduction in its expression (Fig. 3D).
Figure 3.
Identification of survivin as an EGFRvIII-induced apoptosis inhibitor that attenuates EGFR TKI sensitivity in wtEGFR cells. (A) Real-time PCR screening of apoptosis-related genes in U87wtEGFR cells treated with vIII-CM versus control. (B) Microarray analysis from TCGA GBM samples for BIRC3, BIRC5, and TNFSF10. (C) vIII-CM up-regulates survivin expression in wtEGFR cells as evaluated by Western blotting analysis (right) and real-time PCR (left). (D) Real-time PCR analysis of survivin expression upon IL-6 treatment or gp130-neutralizing antibody in GBM PDX cell lines. (E) shRNA-mediated down-regulation of survivin expression (Western; top) and caspase activation assay (bottom) in cells treated with 5 µM gefitinib. (F) EGFRvIII cells and wtEGFR cells transduced with shGFP, shsurvivin #1, or shsurvivin #2 were engrafted intracranially at 10%:90% ratio, respectively. Following tumor establishment, the mice were treated with 200 mg/kg lapatinib, and apoptosis was measured in wtEGFR cells using FACS analysis for cleaved caspase 3 staining. n = 4 mice for each condition. (G) Immunofluorescence staining for survivin (Alexa fluor 594) in U87wtEGFR cells in the presence of Ctrl-CM or vIII-CM. (H) Immunoprecipitation analysis of survivin interactors Smac/DIABLO and XIAP (top) and total lysates (bottom). Bars, 10 µm. Data are represented as mean ± SD.
To validate the role of survivin in protecting the cells from EGFR TKI-induced apoptosis, its expression was knocked down in U87wtEGFR cells by shRNAs, which resulted in sensitization to gefitinib treatment (Fig. 3E). The specificity of survivin knockdown was confirmed by reconstituting survivin using knockdown-resistant expression constructs (Supplemental Fig. S5C). Moreover, survivin knockdown was performed in PDX models expressing exogenous levels of wtEGFR or endogenous levels of EGFRvIII, which were similarly sensitized to EGFR TKI treatment (Supplemental Figs. S5D,E, S6A–C). To confirm these results in vivo, admixing experiments were performed using U87wtEGFR cells expressing control or survivin shRNAs orthotopically engrafted in the brains of immunocompromised mice at a ratio of 90% U87wtEGFR to 10% EGFRvIII (Inda et al. 2010). Seven days after the injection, mice were treated with 200 mg/kg lapatinib for an additional 7 d. Upon sacrifice, their brains were dissected, and tumors were dissociated. wtEGFR cells were identified by specific EGFR antibody staining and stained for cleaved caspase 3 and TUNEL. Mixed tumors with U87wtEGFR survivin knockdown cells treated with lapatinib had an increase in apoptotic index when compared with lapatinib treatment of shRNA control mice (Fig. 3F; Supplemental Fig. S6D). In contrast, exogenous survivin expression in wtEGFR+ cells protected the cells from EGFR TKI-induced apoptosis in vitro (Supplemental Fig. S6E). Mechanistically, vIII-CM increased survivin cytosolic localization (Fig. 3G), which is associated with a prosurvival phenotype (Chan et al. 2010). Additionally, we detected interaction with XIAP and Smac/DIABLO (Fig. 3H), particularly when survivin was localized in the cytosol (Supplemental Fig. S7A), but no association was observed with caspase 3, caspase 6, caspase 7, caspase 8, or caspase 9 (data not shown). Direct interaction between XIAP and survivin enhances XIAP caspase inhibition. Furthermore, interaction between Smac/DIABLO and survivin neutralizes Smac/DIABLO proapoptotic functions. Altogether, these findings support a role for survivin as a prosurvival mediator induced by vIII-CM in cancer cell types within the tumor microenvironment (Altieri 2008; Chan et al. 2010).
Because this phenotype was mediated by secreted factors, we investigated the possibility that these factors might also act in an autocrine fashion. U87 parental, EGFRvIII, and wtEGFR cells were serum-starved for 24 h, and survivin expression was evaluated by real-time qPCR and Western blotting, which showed that survivin was highly expressed in EGFRvIII cells compared with their parental or wtEGFR+ counterparts in a gp130-dependent fashion (Supplemental Fig. S7B). Similar results were obtained in U178MG and U373MG cells (Supplemental Fig. S7B) as well as U373MG cells expressing a tetracycline-regulated EGFRvIII construct (Wykosky et al. 2015) (Supplemental Fig. S7C). To determine whether survivin has a role in acquired resistance to EGFR TKIs in GBM, we used an established model of induced resistance to EGFR TKIs (Wykosky et al. 2015). Here, gefitinib-sensitive and gefitinib-resistant primary Ink4a/Arf−/−-EGFRvIII mouse astrocytes were screened using a real-time qPCR array of apoptosis-related genes, which revealed that survivin was among the most up-regulated genes in the resistant state (Supplemental Fig. S7D). Survivin up-regulation in resistant astrocytes was confirmed by real-time qPCR in additional Ink4a/Arf−/−-EGFRvIII gefitinib-resistant primary astrocyte cell lines (Supplemental Fig. S7E). Overall, these results suggest that survivin expression is regulated by EGFRvIII in an autocrine and paracrine fashion and is a mediator of EGFR TKI resistance in GBM.
NF-κB regulates survivin expression in GBM
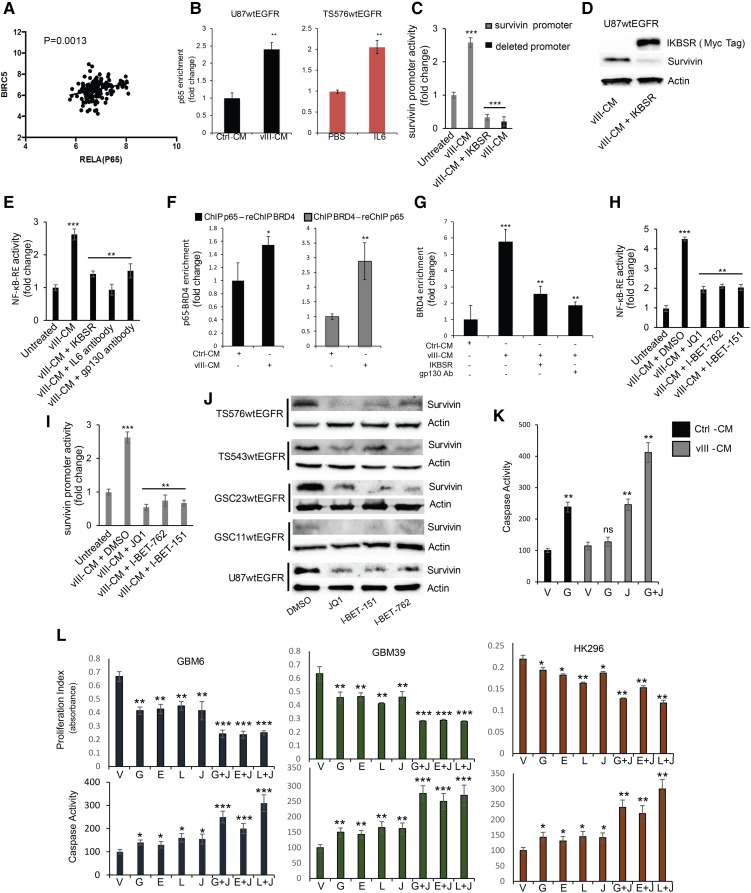
To determine the mechanism by which IL-6 is mediating survivin expression, we performed a bioinformatic analysis of transcription factor response elements in the survivin promoter, which revealed putative binding sites for STAT3 and NF-κB (Supplemental Fig. S8A). As IL-6 is known to mediate STAT3 activation (Wang et al. 2009), we first assessed whether this transcription factor was linked to survivin expression; however, an analysis of TCGA GBM transcriptomes failed to show an association between a STAT3 signature and survivin (BIRC5) expression (Supplemental Fig. S8B). Moreover, modulation of STAT3 activity using a dominant-negative mutant or a constitutively active version of STAT3 did not alter survivin expression in U87EGFRvIII cells (Supplemental Fig. S8C). In contrast, a similar analysis of GBMs with an EGFR-associated classical subgroup signature (Brennan et al. 2013) showed significant expression correlation between BIRC5 and the NF-κB subunit RelA (p65) transcription factor (Fig. 4A). Confirming a direct role for NF-κB in survivin expression, the three potential NF-κB-binding sites in its promoter region were tested by chromatin immunoprecipitation (ChIP) combined with PCR (ChIP-PCR), which showed enrichment for p65 binding upon vIII-CM treatment of U87wtEGFR cells and upon IL-6 treatment of the TS576wtEGFR PDX model when compared with the untreated control (Fig. 4B; Supplemental Fig. S8D,E). In conjunction with this result, a survivin promoter reporter construct (−1000 base pairs [bp] to +1) containing these NF-κB response elements showed that promoter activity was stimulated upon vIII-CM treatment and that its activity was efficiently inhibited by IκB superrepressor (IκB-SR) (Friedmann-Morvinski et al. 2016) or deletions of the three NF-κB-binding sites (Fig. 4C). We validated the role of NF-κB in regulating survivin expression by transiently expressing IκB-SR in U87wtEGFR in the presence of vIII-CM, which showed that IκB-SR potently represses survivin expression (Fig. 4D). To confirm NF-κB activation upon vIII-CM treatment, wtEGFR+ cells were transduced with an NF-κB reporter vector (NF-κB-RE) in the presence or absence of IκB-SR and treated with vIII-CM. Reporter activity showed that NF-κB activation was inhibited by IκB-SR or by neutralizing IL-6/gp130 signaling (Fig. 4E). In total, these results illustrate that vIII-CM potently up-regulates NF-κB activity, which in turn promotes survivin expression.
Figure 4.
NF-κB controls survivin expression in GBM. (A) TCGA GBM transcriptomal profile association between survivin and RelA in the classical GBM subgroup. (B) ChIP-PCR analysis of p65 enrichment on the survivin promoter in U87wtEGFR cells (left) and TS576wtEGFR cells (right). The genomic region examined is located at −226 from the transcriptional start site. (C) Survivin promoter activity upon vIII-CM treatment. IKBSR- or p65-binding site deletions inhibited the activity of the promoter upon vIII-CM treatment. (D) Western blot analysis of survivin expression in U87wtEGFR transduced with IKBSR and treated with vIII-CM. (E) NF-κB response element reporter assay of U87wtEGFR with the indicated conditions. (F) Sequential ChIP analysis for bromodomain protein 4 (BRD4) and p65 on survivin promoter. (Left) The results for p65 (first antibody) and BRD4 (second antibody) ChIP analysis is shown. (Right) The results for BRD4 (first antibody) and p65 (second antibody) ChIP analysis is shown. (G) ChIP-PCR analysis of BRD4 enrichment on the survivin promoter. (H) NF-κB response element reporter assay. (I) Survivin promoter activation. (J) Survivin expression in the presence of 0.5 µM BET bromodomain inhibitors for 48 h. (K) JQ1 (0.5 µM) sensitizes cells to gefitinib-induced apoptosis. (L) JQ1 (0.5 µM) in combination with 2 µM EGFR TKIs reduced cell proliferation and induced apoptosis in EGFRvIII-positive PDX cell lines in vitro. Data are represented as mean ± SD.
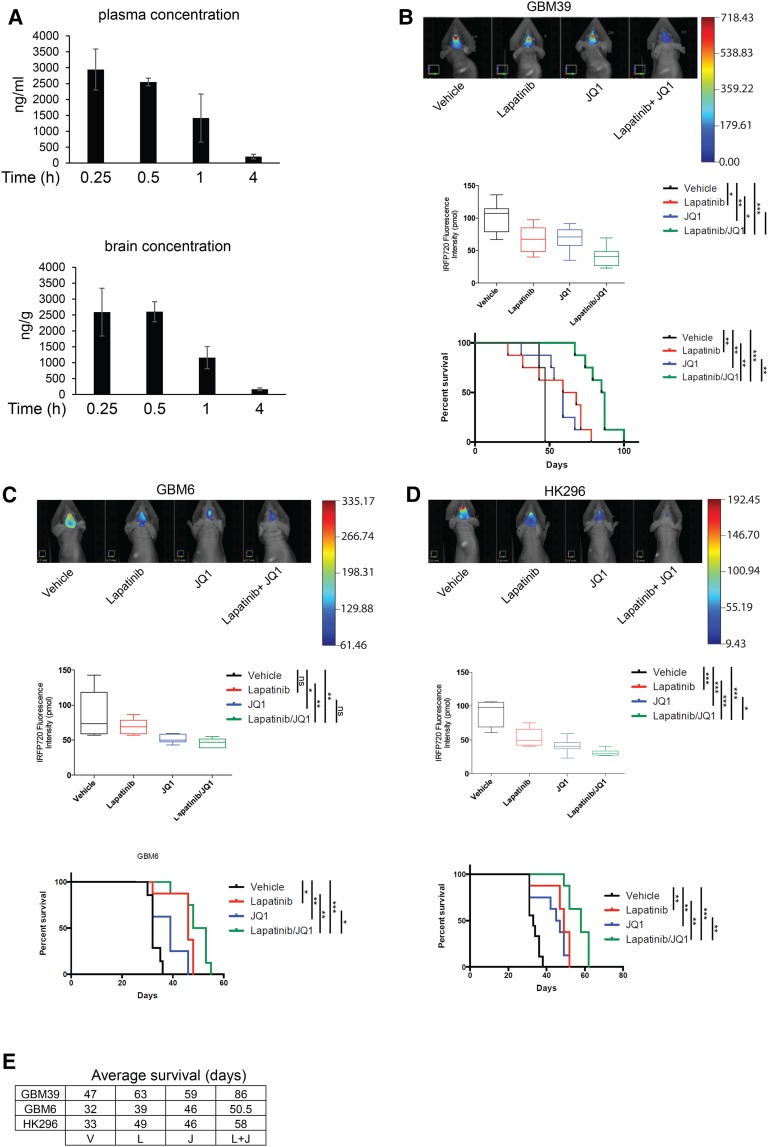
The NF-κB/survivin axis represents a potential targetable pathway for combinatorial therapy together with EGFR TKIs. It was shown recently in an atherogenic inflammatory response that bromodomain protein 4 (BRD4) can orchestrate NF-κB-dependent superenhancer formation through direct binding to NF-κB (Brown et al. 2014), which we similarly confirmed also occurs at the survivin promoter upon vIII-CM treatment (Fig. 4F). NF-κB signaling inhibition by IκB-SR or neutralizing gp130 in the presence of vIII-CM treatment reduced BRD4 recruitment on the survivin promoter (Fig. 4G). Since BET bromodomain inhibitors can inhibit BRD4 and impede other transcription factors coactivated by BRD4, such as NF-κB or MYC, we determined whether this possibility also occurred in our model by testing three such inhibitors: JQ1, I-BET-151, and I-BET-762 (Shi and Vakoc 2014). Inhibitor pretreatment of wtEGFR+ cells prior to vIII-CM and EGFR TKI addition reduced NF-κB activation (as measured by NF-κB and survivin reporter assays) (Fig. 4H,I), down-regulated survivin expression in U87wtEGFR and PDX cell lines (Fig. 4J; Supplemental Fig. S8F), and strongly induced apoptosis (Fig. 4K) without affecting p65 deposition on the survivin promoter (Supplemental Fig. S8G). Furthermore, direct down-regulation of BRD4 by shRNA did not interfere with p65 deposition on the survivin promoter (Supplemental Fig. S8H). Additionally, we confirmed that the direct stimulation of TS576wtEGFR cells with IL-6 induced BRD4 recruitment to the survivin promoter (Supplemental Fig. S9A). Next, we evaluated the effects of the combined treatment of EGFR TKIs and JQ1 in PDX models GBM6, GBM39, and HK296, which express endogenous levels of EGFRvIII. The combined treatment similarly resulted in a reduction of proliferation as well as induction of apoptosis (Fig. 4L), while survivin overexpression was sufficient to rescue GBM cells from JQ1-induced apoptosis (Supplemental Fig. S10A). GSC11wtEGFR, TS543wtEGFR, and TS576wtEGFR also showed a strong response to a lapatinib plus JQ1 combination in the presence of IL-6 (Supplemental Fig. S10B). We also tested other pharmacological inhibitors capable of blocking the NF-κB/survivin axis. Among the intracellular pathways that regulate NF-κB activity, we tested the PI3K/mTORC2 pathway for its possible role in vIII-CM-mediated NF-κB activation (Tanaka et al. 2011). Here, PI-103 and PP242 inhibitors reduced NF-κB activation and survivin expression and, in combination with gefitinib, increased apoptosis in wtEGFR cells (Supplemental Fig. S11A–D). We also tested NF-κB pathway inhibitor MLN120B (IKKβ) (Cahill et al. 2016) in combination with gefitinib, and similar results were obtained (Supplemental Fig. S11A–D). It has been described previously that BET bromodomain inhibitors also inhibit MYC activation (Shi and Vakoc 2014). To determine whether this oncogene was playing a role in our model, we transduced U87wtEGFR cells with a constitutively active MYC (T58A) (Hemann et al. 2005), which failed to increase survivin expression (Supplemental Fig. S11E). Survivin expression depended on BRD4 because shRNA down-regulation of BRD4 in U87EGFRvIII cells (Liu et al. 2015) resulted in reduced survivin levels (Supplemental Fig. S11E). Moreover, we evaluated caspase activation in U87wtEGFR cells transduced with IκB-SR and then treated with JQ1 in the presence of vIII-CM, which did not further increase induction of apoptosis when compared with IκB-SR alone, indicating that inhibition of NF-κB is primarily responsible for regulating apoptosis in response to JQ1 (Supplemental Fig. S11F). Since BET bromodomain inhibitors demonstrated the highest potential in inducing apoptosis when combined with EGFR inhibitors, we decided to test the combination of these drugs in vivo using the PDX models GBM39, GBM6, and HK296 (Nathanson et al. 2014). PDX cells expressing the near-infrared fluorescent protein iRFP720 (Supplemental Fig. S11G; Shcherbakova and Verkhusha 2013) were orthotopically engrafted into the brains of nude mice. Seven days after the injection, the mice were treated five times per week with lapatinib and/or JQ1, and tumor burden was monitored by fluorescence molecular tomography (FMT) until the appearance of neurological symptoms, at which time the mice were euthanized. As shown in Figure 5, JQ1 exhibited brain penetrance (Fig. 5A), and combined treatment was able to reduce the tumor size and prolong the lives of the animals significantly when compared with untreated or single-treatment groups (Fig. 5B–E). We also tested the possibility of BET proteins as a potential target to sensitize GBMs to inhibition of other receptor tyrosine kinases. To this aim, we used GBM8, a PDX with extrachromosomal DNA (ecDNA) PDGFRα overexpression (Supplemental Fig. S12A,B). Cells were treated with crenolanib (a PDGFRα/β inhibitor) (Supplemental Fig. S12C) and JQ1, and cell proliferation and apoptosis were measured (Supplemental Fig. S12D,E). Similar to our results above, JQ1 also sensitized GBM8 cells to crenolanib-induced apoptosis.
Figure 5.
JQ1 sensitizes GBM cells to EGFR TKI-induced apoptosis, and the combinatorial treatment inhibits tumor growth and prolongs the survival of mice bearing orthotopic PDX models. (A) JQ1 levels in mouse plasma and brains at different time points after oral dosing as determined by liquid chromatography/mass spectrometry (LC/MS). FMT analysis of GBM39 (B), GBM6 (C), or HK296 (D) orthotopic xenograft tumor burden quantification and survival curves. n = 8. (E) Average survival (days) of nude mice bearing intracranial tumors and treated with vehicle (V), lapatinib (L), JQ1 (J), or a combination of both lapatinib and JQ1 (L + J). Data are represented as mean ± SD.
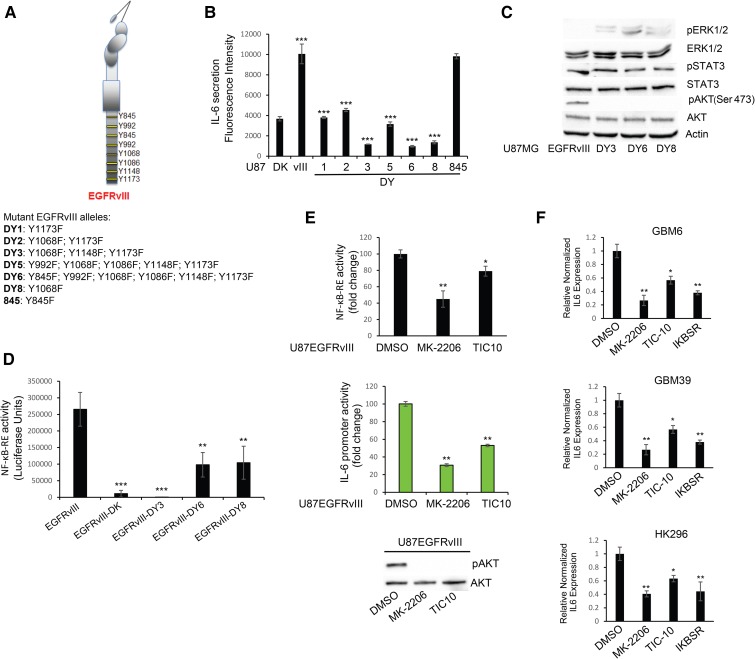
EGFRvIII mediates IL-6 secretion through AKT pathway activation
Our previous studies have shown that EGFRvIII prompted secretion of the protumorigenic and angiogenic cytokines IL-6 and IL-8 (Inda et al. 2010). To identify candidate pathways regulating cytokine production, in particular IL-6, we screened a series of EGFRvIII alleles in which tyrosine phosphorylation target residues within the C-terminal tail had been mutated singly or in combinations to phenylalanine (DY mutants series) (Fig. 6A). Such mutations ablate particular pathways of signal transduction used by the receptor (Huang et al. 2007). CM from U87MG cells expressing these various DY mutants was assessed using a Bio-Plex multiplex system for IL-6 as well as LIF, IL-8, GROα, MCP-1, and G-CSF secretion (Fig. 6B; Supplemental Fig. S13A; Inda et al. 2010). Mutant receptors DY3, DY6, and DY8 showed consistent reduction in IL-6 secretion, which was confirmed by ELISA (Supplemental Fig. S13B). To determine which intracellular pathway is responsible for cytokine production and specifically inhibited in the DY3, DY6, and DY8 mutants (Supplemental Fig. S13C), we used a Bio-Plex phosphoprotein assay platform to screen for possible candidates. Serum-starved cells harvested for phosphoprotein analysis showed that AKT activation was ablated in all DY mutants; meanwhile, STAT3 activation was not affected by such mutations (Supplemental Fig. S13D). These results, confirmed by Western blotting analysis (Fig. 6C), are consistent with the Y1068 mutation shared among DY3, DY6, and DY8, which is a phosphotyrosine site in EGFR linked to AKT activation (Huang et al. 2007). Because NF-κB has been shown to regulate IL-6 production (Iliopoulos et al. 2009), we evaluated the NF-κB activation status in the EGFRvIII-DY mutants using a NF-κB reporter construct as used in Figure 4, which showed that NF-κB activity was reduced by the DY3, DY6, and DY8 mutants (Fig. 6D). Additionally, since AKT activity can activate NF-κB (Dan et al. 2008), we evaluated the effects of inhibiting the AKT pathway on NF-κB activation. Pharmacological AKT inhibition resulted in a down-regulation of reporter activity for both NF-κB and the IL-6 promoter (Fig. 6E). Dependency of IL-6 production on NF-κB activity was further confirmed using IκB-SR (Supplemental Fig. S14A), and a similar dependency on BRD4 was measured by real-time PCR after JQ1 treatment of U87EGFRvIII cells (Supplemental Fig. S14B). We further validated NF-κB activation dependency on AKT activity using a dominant-negative (DN) AKT, which inhibited NF-κB (Supplemental Fig. S14C) and a constitutively active form of AKT, which in turn activated NF-κB (Supplemental Fig. S14D). Finally, we evaluated the dependency of IL-6 production on AKT in GBM39, GBM6, and HK296 PDX models treated with AKT pharmacological inhibitors or transduced with IκB-SR (Fig. 6F; Supplemental Fig. S14A). Of note, the effects on expression levels of IL-6 appeared in the early phase of treatment, while reduction in cell proliferation was evident after 3 d of treatment (Supplemental Fig. S14E), possibly due to the kinetics of dampening IL-6 autocrine pathway signaling. Taken together, these results suggest that EGFRvIII regulates NF-κB activity and cytokine production through the AKT pathway.
Figure 6.
PI3K/AKT signaling controls IL-6 secretion in EGFRvIII cells. (A) Schematic representation of Tyr-to-Phe substitutions in EGFRvIII DY mutants. (B) Bio-Plex analysis for IL-6 in DY mutants. (C) Western blot analysis of intracellular pathways in DY mutants. (D,E) NF-κB response element reporter assay in DY mutants (D) or EGFRvIII cells (E, top panel) treated with 0.5 µM MK-2206 or 1 µM TIC-10 for 48 h. (E, middle panel) IL-6 reporter assay in the presence of AKT inhibitors. Western blotting analysis is shown in the bottom panel of E. (F) Survivin real-time PCR analysis in PDX cell lines treated with the same inhibitors as in E. Data are represented as mean ± SD.
Survivin expression in GBM patients
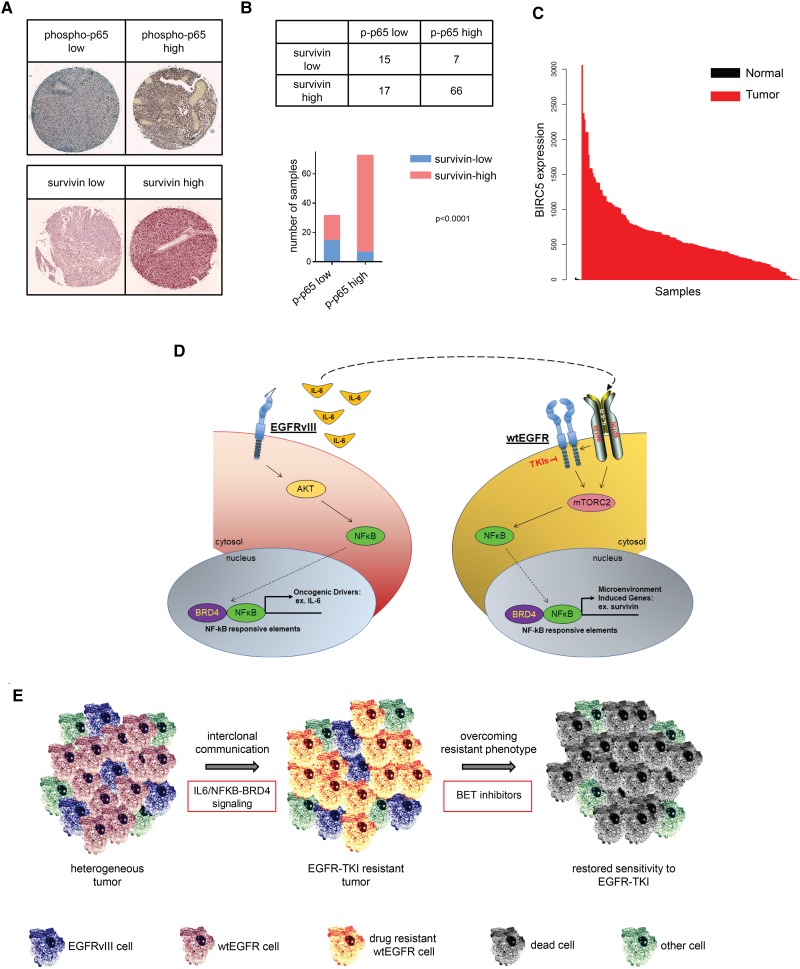
Last, to confirm that the NF-κB/survivin axis is active in human GBM clinical samples, we examined the phosphorylation levels of the p65 NF-κB subunit (as a surrogate biomarker for pathway activation) and the expression levels of survivin from 105 GBM tumors arrayed on a tissue microarray (Fig. 7A; Tanaka et al. 2011), which showed that increased phosphorylation of p65 was tightly associated with high expression of survivin (Fig. 7A,B). Moreover, analysis of TCGA data revealed that BIRC5 is elevated in glioma patients when compared with normal brains (P = 2.45 × 10−4, Wilcoxon test) (Fig. 7C). Additionally, gene set enrichment analysis (GSEA) of the transcriptomes of TCGA patients indicated significant correlations between BIRC5 expression and cell cycle regulation, apoptosis, and NF-κB signaling pathways (Supplemental Fig. S15A). Erlotinib-treated patients also show a tendency for increased BIRC5 expression (Supplemental Fig. S15B,C), similar to our results on acquired resistance in Ink4a/Arf−/−-EGFRvIII mouse astrocytes (Supplemental Fig. S7D,E). Taken together, our results show that the NF-κB/survivin axis is active in human GBM and represents a valuable target for combinatorial treatment to increase the efficacy of EGFR-directed TKIs.
Figure 7.
The NF-κB/survivin axis is hyperactive in clinical GBM samples. (A) Immunohistochemical images of phospho-p65 and survivin. Tissue microarray samples are stained with H&E. (B) Immunohistochemical analysis of tissue microarray samples shows statistically significant correlation (Fisher's exact test) between phospho-p65 and survivin expression. (C) TCGA analysis of survivin expression in GBM versus normal brains. (D) Schematic representation of pathways activation. Intracellular signaling activation mediated by EGFRvIII or extracellular activation of the wtEGFR/gp130 complex converges upon activation of NF-κB. Upon TKI-mediated inactivation of EGFR, IL-6 activation of gp130 promotes cell survival through mTORC2/NF-κB signaling. (E) Interclonal communication of EGFR-expressing cells results in an apoptosis-resistant phenotype. Tumor cells regain sensitivity to EGFR TKIs through inhibition of NF-κB by BET bromodomain inhibitors.
Discussion
A number of factors contribute to make GBM one the most challenging cancers to treat. An ideal candidate drug is one that is capable of crossing the blood–brain barrier, selective in inducing cell death in cancer cells, and well tolerated by patients. The current standard of treatment entails a maximal surgical resection followed by DNA-damaging radiotherapy and concomitant temozolomide administration. While the complex genetics of untreated GBM are being investigated with next-generation sequencing technologies (Brennan et al. 2013), it is now understood that the genetics and epigenetics of gliomas are also impacted by therapy and tumor progression (Johnson et al. 2014; Mazor et al. 2015; Wang et al. 2016). In addition to genetics, a central aspect of the pathogenesis of GBM is its heterogeneity, which is reflected in the various histological subtypes, epigenetic and gene expression landscapes, and different cells of origin and tumor microenvironment constituents, all contributing to the therapeutically recalcitrant nature of this tumor type (Alcantara Llaguno et al. 2015; Archetti et al. 2015; Furnari et al. 2015; Caiado et al. 2016; Mazor et al. 2016). To achieve treatment success for GBM patients, it is critical to understand the contribution of each one of these tumor features. The EGFR is often amplified and/or mutated in GBM, with EGFRvIII being a common mutant that confers a particularly aggressive phenotype to the tumor (Inda et al. 2010; Furnari et al. 2015). Four subtypes (classical, mesenchymal, neural, and proneural) of GBM have been identified by an integrated genomic analysis of human glioma samples, with EGFR initially thought to be a prominent feature of the classical subtype (Verhaak et al. 2010). The complex interactive nature of wtEGFR/EGFRvIII characterizes GBM (Inda et al. 2010; Fan et al. 2013; Nathanson et al. 2014; Furnari et al. 2015; Jahani-Asl et al. 2016), which we sought to understand here to improve the efficacy of anti-EGFR therapy.
Historically, targeting EGFR has resulted in substantial therapeutic improvement for lung, breast, and colon cancer patients (Scaltriti and Baselga 2006); however, identical therapies have provided little clinical benefit for GBM patients (Reardon et al. 2014). Mechanisms of resistance to anti-EGFR therapy also arise in other cancers. Upon initial response, KRAS mutations nullify anti-EGFR therapy in colon cancer (Diaz et al. 2012), while the T790M EGFR mutation alters response to gefitinib in non-small-cell lung cancer (NSCLC) (Lynch et al. 2004). A major molecular determinant promoting lack of response to EGFR inhibition is deletion or mutation of the PTEN gene (Mellinghoff et al. 2005). Additional mechanisms are likely to take part in this cell-intrinsic resistance to anti-EGFR treatment (Nathanson et al. 2014; Reardon et al. 2014; Wykosky et al. 2015). So far, insufficient attention has been paid to the role of heterogeneity of EGFR expression in GBM in determining the responsiveness to targeted therapy. This study demonstrates the presence of a wtEGFR/EGFRvIII circuit that conveys an EGFR TKI resistance mechanism mediated by IL-6. IL-6 is a pleiotropic factor that is recognized as a molecule linking inflammation and cancer and is involved in many solid tumors, such as brain, liver, and colon cancers. It activates an immune response within the tumor burden while simultaneously fueling cancer cells with prosurvival signaling (Hodge et al. 2005). In mutationally activated EGFR-driven NSCLCs, treatment with EGFR TKIs results in IL-6-mediated feedback activation of STAT3 and downstream prosurvival genes that attenuate drug sensitivity (Lee et al. 2014). It is interesting to note that, although NSCLC and GBM, as shown here, use IL-6 as a cytokine mediator to attenuate EGFR TKI sensitivity, our study shows that STAT3 is not the critical driver of this mechanism in GBM but requires the NF-κB transcription factor, perhaps illustrating cell type or mutant receptor-specific contextual differences in pathway signaling (Huang et al. 2009).
We reported previously that EGFRvIII prompted the growth of wtEGFR+ cells through activation of the IL-6R/gp130 receptors (Inda et al. 2010). In addition to this enhanced tumorigenicity, we show here that IL-6 increased NF-κB activation and survivin expression, resulting in an anti-apoptotic phenotype. Fostered by IL-6, wtEGFR+ cells lost their potential to respond to inhibition by different EGFR TKIs (gefitinib, erlotinib, and lapatinib) (Fig. 7E; Vivanco et al. 2012). Survivin is part of the inhibitor of apoptosis protein (IAP) family, containing a single baculovirus IAP repeat (BIR) domain. It has broad anti-apoptotic action compromising both caspase-dependent and caspase-independent cell death. It is unaltered in normal human tissues and up-regulated in malignant transformation, accounting for the interest in its anti-apoptotic role in human cancers (Altieri 2008). The small molecule YM-155 is a reported survivin inhibitor, but a lack in brain penetrance makes it an unlikely drug for GBM patients (Minematsu et al. 2012). In this research, we reported that survivin expression is regulated by NF-κB. NF-κB is a family of transcription factors comprising NF-κB1 (p105/p50), NF-κB2 (p100/p52), RELA (p65), RELB, and c-Rel. In the canonical pathway of NF-κB activation, dimers of NF-κB are kept inactive by the cytosolic inhibitor IκB. When IκB is targeted for proteasomal degradation by phosphorylation, active NF-κB migrates to the nucleus and activates the transcription of genes involved in cancer progression and chemoresistance; for example, Bcl-2, Bcl-xl, Cox2, IAP, and CCND1. A variety of stimuli can activate NF-κB-mediated transcription, among which wtEGFR and EGFRvIII have been reported to activate NF-κB (Dan et al. 2008; Tanaka et al. 2011; Cahill et al. 2016). In this regard, NF-κB has been shown to be a major molecular driver of chemotherapy and radiotherapy resistance in GBM (Tanaka et al. 2011; Bhat et al. 2013; Cahill et al. 2016; Friedmann-Morvinski et al. 2016). To inhibit survivin expression, we chose the epigenetic regulator JQ1, which targets the bromodomain-containing factor BRD4, known to maintain NF-κB in an active state and regulate the enhancer-mediated function of this transcription factor (Brown et al. 2014; Zou et al. 2014). Moreover, JQ1 has been shown to abrogate an EGFRvIII-induced transcriptional regulatory network (Liu et al. 2015). In our study, JQ1 was indeed able to abrogate both IL-6-mediated EGFR TKI resistance and IL-6-induced gefitinib resistance by restoring induction of cell death mediated by receptor blockade in both wtEGFR and EGFRvIII cells. Importantly, we show that JQ1 crosses the blood–brain barrier, highlighting brain penetrance as an important consideration in GBM drug development.
In this heterogeneous scenario of cellular cross-talk between wtEGFR+ and EGFRvIII+ cells, NF-κB serves as a convergence signaling molecule shared between both cell types. Indeed, NF-κB activity is stimulated by EGFRvIII in EGFRvIII+ cells and from cytokine receptors in wtEGFR+ cells (Fig. 7D). EGFRvIII is not the only driver sustaining tumor fitness within the pathological microenvironment. Through its activity, we speculate that EGFRvIII can stimulate other cell components of the tumor, such as microglia, which can similarly secrete IL-6, thus enhancing the oncogenic effects initiated by EGFRvIII. By targeting NF-κB, we could potentially eradicate not just one cell population but all cells responding to the EGFRvIII stimuli in a heterogeneous environment as presented in GBM (Fig. 7E; Charles et al. 2011).
Deletion of the NF-κB inhibitor NFKBIA and genetic alterations of EGFR are mutually exclusive in GBM (Bredel et al. 2011)—data that support an essential role for the NF-κB signaling axis in these tumors. With wtEGFR/EGFRvIII being activators of NF-κB, the concomitant inhibition of both might be therapeutically relevant for GBM patients. In agreement with our study, application of combined lapatinib and JQ1 treatment has proven efficacious in a model of lapatinib-resistant breast cancer, where JQ1 overcomes resistance that is mediated by activation of additional receptor tyrosine kinases (Stuhlmiller et al. 2015).
It is likely that multiple inherent and induced molecular mechanisms contribute to EGFR therapy resistance. Recent data implicated EGFR amplification and mutation as initial events in the pathogenesis of GBM, suggesting that anti-EGFR treatments might be effective as an early therapeutic intervention (Wang et al. 2016). The emerging evidence of numerous EGFR variants in GBM, a mixture of point mutations and minor and major deletions involving both the extracellular and intracellular domains of the receptor, presents a variegated receptor landscape with a unique possibility for intervention once functional/operational malignant networks are identified and then uncoupled. More broadly, the consequences of tumor heterogeneity—of which EGFR mutation and amplification represent a microcosm of the full spectrum of GBM heterogeneity—are now starting to be understood. Implementation of the knowledge gained from the study of the genetic, epigenetic, and microenvironmental factors cooperating in the formation of a complex tumor landscape will ultimately dictate the direction of future therapeutic approaches for GBM patients.
Materials and methods
Cell lines, plasmids, and reagents
Parental U87MG, U87EGFRvIII, U87DK (dead kinase), U87wtEGFR, mAstr–Ink4a/Arf−/−, mAstr–Ink4a/Arf−/−-wtEGFR, mAstr–Ink4a/Arf−/−-EGFRvIII, mAstr–Ink4a/Arf−/−-EGFRvIII-GR, U178MG, U178EGFRvIII, U178wtEGFR, U373MG, U373EGFRvIII, U373wtEGFR, and U373EGFRvIIITEToff were obtained and cultured as described previously (Inda et al. 2010; Wykosky et al. 2015). PDX sphere lines were cultured in DMEM/F12 medium supplemented with B27 (GIBCO/Life Technologies) and 20 ng/mL human recombinant EGF, 20 ng/mL bFGF, and 2 mg/mL heparin (Stem Cell Technologies). GBM6, GBM8, and GBM39 were provided by David James (Northwestern University); GSC11 and GSC23 were provided by Frederick Lang (M.D. Anderson Cancer Center); TS543 and TS576 were provided by Cameron Brennan (Memorial Sloan Kettering Cancer Center); and HK296 was provided by Harley Kornblum (University of California at Los Angeles). All cells were incubated at 37°C, 5% CO2, and 100% relative humidity in low-attachment flasks. PDX cell lines were dissociated with Accutase (Stemcell Technologies). EGFR TKIs (gefitinib, erlotinib, and lapatinib) were purchased from LC Laboratories, 1-NA-PP1 was from D-L Chiral Chemicals, and JQ1 was from MedChem Express. All other compounds were from Selleck Chemicals.
The near-infrared fluorescent protein iRFP720 cDNA construct was from Shcherbakova and Verkhusha (2013), EGFRvIIIAS3 was from Fan et al. (2013), and pLV-IκB-SR vector was a gift from Inder Verma (Salk Institute). shRNA constructs targeting survivin, IL-6, IL-8, and GFP were purchased from Sigma (Mission shRNA). pLV-EF1a-GFP-IRES-Hyg plasmid (Biosettia) was used to subclone wtEGFR for sphere transduction. Survivin overexpression in U87wtEGFR cells was achieved by subcloning survivin cDNA into the pBABE-puro backbone. The pGL4.32[luc2P/NF-κB-RE/Hygro] vector for NF-κB luciferase reporter assays was purchased from Promega. The LightSwitch luciferase assay system (Active Motif) was used to analyze IL-6 and the survivin promoter.
Intracranial injection and tumor size measurement
A total of 2.5 × 105 cells in a 5-µL volume was injected intracranially into 4- to 5-wk-old athymic nude mice using a stereotactic system. Tumors were allowed to establish for 1 wk before any treatment, and engraftment of tumors was quantitatively confirmed via FMT signal intensity at the onset of neurological symptoms in the control groups. Tumor growth was monitored using the FMT 2500 fluorescence tomography system (PerkinElmer). For drug treatment studies, vehicle (20% captisol; A Ligand Technology) or 100 mg/kg JQ1 or 200 mg/kg EGFR TKI gefitinib or lapatinib (LC Laboratories) resuspended in vehicle was administered once daily to mice via oral gavage starting at day 7 after injection. 1-NA-PP1 (10 mg/kg) was resuspended in vehicle (20%:20%:60% [v/v/v] solutol/PEG400/water) and administered twice daily by intraperitoneal injection. Mice were euthanized in accordance with our institutional guidelines for animal welfare and experimental conduct at University of California at San Diego. Survival until the onset of neurologic symptoms was used for intracranial injections.
Pharmacokinetic studies
Pharmacokinetic studies were conducted by WuXi AppTec with animals obtained from an approved vendor (Shanghai Laboratory Animal Co., Ltd. [SLAC]). Compounds in the formulations specified above were administered by either single intraperitoneal injection (1-NA-PP1) or oral gavage (JQ1) to animals following the standard operating procedures of the WuXi Apptec animal facility. 1-NA-PP1 and JQ1 dose levels were 10 mg/kg and 50 mg/kg, respectively. The dose volume of each animal was determined by the body weights of animals collected on the dosing day. Samples were collected from the blood and brains. For blood, bleeding (∼30 µL of blood per time point) was collected via the submandibular or saphenous vein. All blood samples were transferred into microcentrifuge tubes containing 4 µL of 0.5 M K2EDTA as an anti-coagulant and placed on wet ice. Samples were then processed for plasma by centrifugation at 3000g for 15 min at 4°C within 30 min of collection. Plasma samples were stored in polypropylene tubes, quick-frozen over dry ice, and kept at −70°C until liquid chromatography/mass spectrometry (LC/MS) analysis. For brains, whole brains were dissected at the designated time points after blood samples were collected. The whole-brain samples were blotted on filter paper, weighed, and then homogenized in 4 vol of water with a Polytron on ice. The homogenates were placed on dry ice until LC/MS analysis.
Tissue microarray
The tissue microarray was purchased from US Biomax and immunohistochemically stained as described previously (Tanaka et al. 2011). Briefly, a tissue array consisting of cores from formalin-fixed paraffin-embedded tumors and normal brain blocks was deparaffinized by immersion in 0.01 M citrate buffer (pH 6) for 30 min in a pressure cooker to promote antigen retrieval. Peroxidase activity was quenched with 3% hydrogen peroxide in water, and primary antibodies were applied for 16 h at 4°C followed by biotinylated secondary antibodies and the avidin–biotin complex. Primary antibodies against survivin and phospho-p65 (Cell Signaling Technologies) were used. Negative control slides received normal mouse serum (Dako) for the primary antibody step. Slides were counterstained with Harris hematoxylin. Staining intensity was scored independently by two neuropathologists based on a scale of three categories indicated as low for negatively stained specimens, medium for weakly positive samples, and high for strongly positive spots. Statistical significance between normal and tumor staining was calculated using the independent t-test. Statistical analyses were performed with the two-tailed Student's t-test to determine the statistical significance between independent samples, with P-values of <0.05 considered statistically significant.
Gene expression analysis
Normalized gene expression levels (RNA sequencing) for 20,502 genes and clinical information for 160 GBMs and five normal brains were downloaded from TCGA data portal (https://tcga-data.nci.nih.gov). Differences in expression levels between tumors treated (n = 4) and not treated (n = 156) with erlotinib were assessed using Wilcoxon test. Bootstrapping was performed by selecting four random BIRC5 expression values between the 156 tumors not treated with erlotinib 100,000 times. Mean expression value was determined for each step, and the P-value was calculated as the number of times that the observed mean BIRC5 expression value was greater than the random mean expression, divided by 100,000. Bootstrapping was performed using the same method also on 400 GBMs (18 treated with erlotinib) from TCGA whose expression levels were measured by microarrays.
Transcriptomal profiling analysis
TCGA GBM transcriptomal profiles were downloaded from TCGA data portal (http://tcga-data.nci.nih.gov/tcga). Level 3 normalized Agilent 244K gene expression mRNA array data were downloaded, and genes were median-centered. Correlative analyses between gene expression were performed using Prism (GraphPad).
GSEA
The correlation between BIRC5 and each of the other 20,501 human genes was calculated considering all 160 GBM samples. Genes ranked from the highest correlation to the lowest correlation were used as input for GSEA (Subramanian et al. 2005). Two-thousand-one-hundred-fifteen gene sets from the Molecular Signature Database (MSigDB) (Subramanian et al. 2005), including 50 hallmark (Liberzon et al. 2015), 149 Biocarta (http://www.biocarta.com), 178 Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa et al. 2016), and 512 reactome (Fabregat et al. 2016) gene sets, were analyzed and used as input to run GSEA with default parameters (1000 permutations and gene set size between 15 and 500 genes).
Statistical analysis
Experimental data were from three biological replicates. Data sets were analyzed by unpaired Student's t-test or multiple comparisons one-way ANOVA or two-way ANOVA according to the experiment using GraphPad Prism software. In all figures, a single asterisk indicates P < 0.05, double asterisks indicate P < 0.001, and triple asterisks indicate P < 0.0001. Significance was concluded at P < 0.05. Kaplan-Meier curves and comparisons of survival were analyzed using log-rank (Mantel-Cox) and Gehan-Breslow-Wilcoxon tests.
Supplementary Material
Acknowledgments
We thank Dr. David James, Dr. Frederick Lang, Dr. Cameron Brennan, and Dr. Harley Kornblum for GBM-PDX neurospheres. We thank Dr. Karen Arden for continuous support and critical evaluation of the results. We thank Dr. Robert Davis, Dr. German Gomez, Dr. Tiffany Taylor, Dr. Rachel Reed, Dr. Melissa Mcalonis, and Dr. Sora Lee for technical support. In memory of Rosa Lupo. This work was supported by the Defeat GBM Research Collaborative, a subsidiary of the National Brain Tumor Society (F.B.F. and P.S.M.), R01-NS080939 (F.B.F.), the James S. McDonnell Foundation (F.B.F.), the National Cancer Institute (2T32CA009523-29A1) (A.H.T), and 1RO1NS097649-01 (C.C.C.). C.Z. was partially supported by an American-Italian Cancer Foundation post-doctoral research fellowship. F.L. received a Gao Feng Gao Yuan Scholarship Award. T.C.G., A.K.S., P.S.M., W.K.C., and F.B.F. receive salary and additional support from the Ludwig Institute for Cancer Research.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.300079.117.
References
- Alcantara Llaguno Sheila R, Wang Z, Sun D, Chen J, Xu J, Kim E, Hatanpaa Kimmo J, Raisanen Jack M, Burns Dennis K, Johnson Jane E, et al. 2015. Adult lineage-restricted CNS progenitors specify distinct glioblastoma subtypes. Cancer Cell 28: 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altieri DC. 2008. Survivin, cancer networks and pathway-directed drug discovery. Nat Rev Cancer 8: 61–70. [DOI] [PubMed] [Google Scholar]
- Archetti M, Ferraro DA, Christofori G. 2015. Heterogeneity for IGF-II production maintained by public goods dynamics in neuroendocrine pancreatic cancer. Proc Natl Acad Sci 112: 1833–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent EH, Gilbert LA, Hemann MT. 2016. A senescence secretory switch mediated by PI3K/AKT/mTOR activation controls chemoprotective endothelial secretory responses. Genes Dev 30: 1811–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat Krishna PL, Balasubramaniyan V, Vaillant B, Ezhilarasan R, Hummelink K, Hollingsworth F, Wani K, Heathcock L, James Johanna D, Goodman Lindsey D, et al. 2013. Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell 24: 331–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredel M, Scholtens DM, Yadav AK, Alvarez AA, Renfrow JJ, Chandler JP, Yu ILY, Carro MS, Dai F, Tagge MJ, et al. 2011. NFKBIA deletion in glioblastomas. N Engl J Med 364: 627–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan Cameron W, Verhaak Roel GW, McKenna A, Campos B, Noushmehr H, Salama Sofie R, Zheng S, Chakravarty D, Sanborn JZ, Berman Samuel H, et al. 2013. The somatic genomic landscape of glioblastoma. Cell 155: 462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown Jonathan D, Lin Charles Y, Duan Q, Griffin G, Federation AJ, Paranal Ronald M, Bair S, Newton G, Lichtman AH, Kung AL, et al. 2014. NF-κB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol Cell 56: 219–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill KE, Morshed RA, Yamini B. 2016. Nuclear factor-κB in glioblastoma: insights into regulators and targeted therapy. Neuro Oncol 18: 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiado F, Silva-Santos B, Norell H. 2016. Intra-tumour heterogeneity—going beyond genetics. FEBS J 283: 2245–2258. [DOI] [PubMed] [Google Scholar]
- Chan KS, Wong CH, Huang YF, Li HY. 2010. Survivin withdrawal by nuclear export failure as a physiological switch to commit cells to apoptosis. Cell Death Dis 1: e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles NA, Holland EC, Gilbertson R, Glass R, Kettenmann H. 2011. The brain tumor microenvironment. Glia 59: 1169–1180. [DOI] [PubMed] [Google Scholar]
- Cloughesy TF, Cavenee WK, Mischel PS. 2014. Glioblastoma: from molecular pathology to targeted treatment. Annu Rev Pathol 9: 1–25. [DOI] [PubMed] [Google Scholar]
- Dan HC, Cooper MJ, Cogswell PC, Duncan JA, Ting JPY, Baldwin AS. 2008. Akt-dependent regulation of NF-κB is controlled by mTOR and Raptor in association with IKK. Genes Dev 22: 1490–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz LA Jr, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, Allen B, Bozic I, Reiter JG, Nowak MA, et al. 2012. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 486: 537–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat A, Sidiropoulos K, Garapati P, Gillespie M, Hausmann K, Haw R, Jassal B, Jupe S, Korninger F, McKay S, et al. 2016. The reactome pathway knowledgebase. Nucleic Acids Res 44: D481–D487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Q-W, Cheng Christine K, Gustafson WC, Charron E, Zipper P, Wong Robyn A, Chen J, Lau J, Knobbe-Thomsen C, Weller M, et al. 2013. EGFR phosphorylates tumor-derived EGFRvIII driving STAT3/5 and progression in glioblastoma. Cancer Cell 24: 438–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann-Morvinski D, Narasimamurthy R, Xia Y, Myskiw C, Soda Y, Verma IM. 2016. Targeting NF-κB in glioblastoma: a therapeutic approach. Sci Adv 2: e1501292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnari FB, Cloughesy TF, Cavenee WK, Mischel PS. 2015. Heterogeneity of epidermal growth factor receptor signalling networks in glioblastoma. Nat Rev Cancer 15: 302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann MT, Bric A, Teruya-Feldstein J, Herbst A, Nilsson JA, Cordon-Cardo C, Cleveland JL, Tansey WP, Lowe SW. 2005. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature 436: 807–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge DR, Hurt EM, Farrar WL. 2005. The role of IL-6 and STAT3 in inflammation and cancer. Eur J Cancer 41: 2502–2512. [DOI] [PubMed] [Google Scholar]
- Huang PH, Mukasa A, Bonavia R, Flynn RA, Brewer ZE, Cavenee WK, Furnari FB, White FM. 2007. Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proc Natl Acad Sci 104: 12867–12872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang PH, Xu AM, White FM. 2009. Oncogenic EGFR signaling networks in glioma. Sci Signal 2: pre6. [DOI] [PubMed] [Google Scholar]
- Iliopoulos D, Hirsch HA, Struhl K. 2009. An epigenetic switch involving NF-κB, Lin28, Let-7 microRNA, and IL-6 links inflammation to cell transformation. Cell 139: 693–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inda M-d-M, Bonavia R, Mukasa A, Narita Y, Sah DWY, Vandenberg S, Brennan C, Johns TG, Bachoo R, Hadwiger P, et al. 2010. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev 24: 1731–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahani-Asl A, Yin H, Soleimani VD, Haque T, Luchman HA, Chang NC, Sincennes M-C, Puram SV, Scott AM, Lorimer IAJ, et al. 2016. Control of glioblastoma tumorigenesis by feed-forward cytokine signaling. Nat Neurosci 19: 798–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, Fouse SD, Yamamoto S, Ueda H, Tatsuno K, et al. 2014. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science 343: 189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44: D457–D462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-J, Zhuang G, Cao Y, Du P, Kim H-J, Settleman J. 2014. Drug resistance via feedback activation of Stat3 in oncogene-addicted cancer cells. Cancer Cell 26: 207–221. [DOI] [PubMed] [Google Scholar]
- Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov Jill P, Tamayo P. 2015. The molecular signatures database hallmark gene set collection. Cell Syst 1: 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Hon Gary C, Villa Genaro R, Turner Kristen M, Ikegami S, Yang H, Ye Z, Li B, Kuan S, Lee Ah Y, et al. 2015. EGFR mutation promotes glioblastoma through epigenome and transcription factor network remodeling. Mol Cell 60: 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. 2016. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131: 803–820. [DOI] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. 2004. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non–small-cell lung cancer to gefitinib. N Engl J Med 350: 2129–2139. [DOI] [PubMed] [Google Scholar]
- Mazor T, Pankov A, Johnson Brett E, Hong C, Hamilton Emily G, Bell Robert JA, Smirnov Ivan V, Reis Gerald F, Phillips Joanna J, Barnes Michael J, et al. 2015. DNA methylation and somatic mutations converge on the cell cycle and define similar evolutionary histories in brain tumors. Cancer Cell 28: 307–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazor T, Pankov A, Song Jun S, Costello Joseph F. 2016. Intratumoral heterogeneity of the epigenome. Cancer Cell 29: 440–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellinghoff IK, Wang MY, Vivanco I, Haas-Kogan DA, Zhu S, Dia EQ, Lu KV, Yoshimoto K, Huang JHY, Chute DJ, et al. 2005. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med 353: 2012–2024. [DOI] [PubMed] [Google Scholar]
- Minematsu T, Sonoda T, Hashimoto T, Iwai M, Oppeneer T, Felder L, Shirai N, Miyashita A, Usui T. 2012. Pharmacokinetics, distribution and excretion of YM155 monobromide, a novel small-molecule survivin suppressant, in male and pregnant or lactating female rats. Biopharm Drug Dispos 33: 160–169. [DOI] [PubMed] [Google Scholar]
- Nathanson DA, Gini B, Mottahedeh J, Visnyei K, Koga T, Gomez G, Eskin A, Hwang K, Wang J, Masui K, et al. 2014. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science 343: 72–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Lemmon MA. 2012. Occupy EGFR. Cancer Discov 2: 398–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, et al. 2014. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344: 1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon DA, Wen PY, Mellinghoff IK. 2014. Targeted molecular therapies against epidermal growth factor receptor: past experiences and challenges. Neuro Oncol 16: p viii7–viii13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaltriti M, Baselga J. 2006. The epidermal growth factor receptor pathway: a model for targeted therapy. Clin Cancer Res 12: 5268–5272. [DOI] [PubMed] [Google Scholar]
- Shcherbakova DM, Verkhusha VV. 2013. Near-infrared fluorescent proteins for multicolor in vivo imaging. Nat Methods 10: 751–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Vakoc Christopher R. 2014. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol Cell 54: 728–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuhlmiller Timothy J, Miller Samantha M, Zawistowski Jon S, Nakamura K, Beltran Adriana S, Duncan James S, Angus Steven P, Collins Kyla AL, Granger Deborah A, Reuther Rachel A, et al. 2015. Inhibition of lapatinib-induced kinome reprogramming in ERBB2-positive breast cancer by targeting BET family bromodomains. Cell Rep 11: 390–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJB, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al. 2005. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352: 987–996. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Babic I, Nathanson D, Akhavan D, Guo D, Gini B, Dang J, Zhu S, Yang H, De Jesus J, et al. 2011. Oncogenic EGFR signaling activates an mTORC2–NF-κB pathway that promotes chemotherapy resistance. Cancer Discov 1: 524–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchirkov A, Khalil T, Chautard E, Mokhtari K, Veronese L, Irthum B, Vago P, Kemeny JL, Verrelle P. 2007. Interleukin-6 gene amplification and shortened survival in glioblastoma patients. Br J Cancer 96: 474–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RGW, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. 2010. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17: 98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivanco I, Robins HI, Rohle D, Campos C, Grommes C, Nghiemphu PL, Kubek S, Oldrini B, Chheda MG, Yannuzzi N, et al. 2012. Differential sensitivity of glioma- versus lung cancer–specific EGFR mutations to EGFR kinase inhibitors. Cancer Discov 2: 458–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Lathia JD, Wu Q, Wang J, Li Z, Heddleston JM, Eyler CE, Elderbroom J, Gallagher J, Schuschu J, et al. 2009. Targeting interleukin 6 signaling suppresses glioma stem cell survival and tumor growth. Stem Cells 27: 2393–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Cazzato E, Ladewig E, Frattini V, Rosenbloom DIS, Zairis S, Abate F, Liu Z, Elliott O, Shin Y-J, et al. 2016. Clonal evolution of glioblastoma under therapy. Nat Genet 48: 768–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykosky J, Hu J, Gomez GG, Taylor T, Villa GR, Pizzo D, VandenBerg SR, Thorne AH, Chen CC, Mischel PS, et al. 2015. A urokinase receptor–Bim signaling axis emerges during EGFR inhibitor resistance in mutant EGFR glioblastoma. Cancer Res 75: 394–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z, Huang B, Wu X, Zhang H, Qi J, Bradner J, Nair S, Chen LF. 2014. Brd4 maintains constitutively active NF-κB in cancer cells by binding to acetylated RelA. Oncogene 33: 2395–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.